Abstract
Both CYP158A1 and CYP158A2 are able to catalyze an oxidative C-C coupling reaction producing biflaviolin or triflaviolin in Streptomyces coelicolor A3(2). The substrate-bound crystal structures of CYP158A2 and CYP158A1 reveal that the side chain of Ile87 in CYP158A2 points to the active site contacting the distal flaviolin molecule, however, the bulkier side chain of Lys90 in CYP158A1 (corresponding to Ile87 in CYP158A2) is toward the distal surface of the protein. These results suggest that these residues could be important in determining product regiospecificity. In order to explore the role of the two residues in catalysis, the reciprocal mutants, Ile87Lys and Lys90Ile, of CYP158A2 and CYP158A1, respectively, were generated and characterized. The mutant Ile87Lys enzyme forms two isomers of biflaviolin instead of three isomers of biflaviolin in wild-type CYP158A2. CYP158A1 containing the substitution of lysine with isoleucine has the same catalytic activity compared with the wild-type CYP158A1. The crystal structure of Ile87Lys showed that the BC loop in the mutant is in a very different orientation compared with the BC loop in both CYP158A1/A2 structures. These results shed light on the mechanism of the oxidative coupling reaction catalyzed by cytochrome P450.
Keywords: Cytochrome P450, Oxidative coupling reaction, CYP158A1, CYP158A2, Regiospecificity
The cytochrome P450 (P450 or CYP) enzymes belong to a superfamily of heme-containing monooxygenases responsible for biosynthesis of physiologically important compounds and metabolism of drugs and other toxic compounds [1-3]. P450s are distributed in all biological kingdoms from viruses, bacteria, fungi, and plants to mammals, including humans. More than 15,000 P450 genes have been identified in this superfamily (http://drnelson.uthsc.edu/cytochromeP450.html). However, for the majority of these P450s the biological function and endogenous substrates are unknown. There are 18 CYP genes in the model actinomycete Streptomyces coelicolor A3(2) [4, 5]. CYP158A2 is a member of a highly conserved operon and catalyze phenolic oxidative C-C coupling reactions of flaviolin to biflaviolin and triflaviolin [6, 7]. The flaviolin polymers are thought to provide physical protection for this soil bacterium against the deleterious effects of UV irradiation on genetic integrity [8, 9]. CYP158A1 can only produce 3,3′-biflaviolin and 3,8′-biflaviolin with quite different molar ratios compared with the products from CYP158A2, which produces three isomers of biflaviolin and one triflaviolin [6]. The substrate flaviolin-bound crystal structures of CYP158A1 and CYP158A2 reveal that two molecules of flaviolin can bind to the enzymes at the same time. Both the proximal flaviolin molecules in CYP158A1/A2 are near the heme iron, however, the distal flaviolin molecules are bound in very different locations in the CYP158A1/A2 structures. In CYP158A1, the distal flaviolin is at the entrance of the substrate access channel and 9 Å away from the proximal flaviolin. In CYP158A2, the two flaviolin molecules are about 4 Å apart from each other, which could make positional sense for a coupling reaction [6]. It is unknown how these two enzymes catalyze the oxidation of the same substrate (flaviolin) while controlling stereo- and regiospecificity. Furthermore, crystal structures of CYP158A1/A2 suggest that several key residues in the active sites associated with flaviolins are different. The most striking difference is that the side chain of Ile87 points toward the active site and contacts the distal flaviolin molecule in CYP158A2, while the bulkier side chain of Lys90 in CYP158A1 (corresponding to Ile87 in CYP158A2) is orientated away from the BC loop, toward the distal surface of the protein. The sequence alignment among the CYP158 family confirmed the above differences. Ile87 in CYP158A2 is highly conserved in all other CYP158 family members (Fig. 1). In order to explore the structural basis of stereo- and regiospecificity of phenolic coupling reactions of CYP158A1/A2, we characterized mutants Ile87Lys and Lys90Ile of CYP158A2 and CYP158A1 respectively and determined the crystal structure of the Ile87Lys mutant of CYP158A2.
FIG. 1.
B-helix and BC loop alignment of the four members of the CYP158 family with CYP158A2 from Streptomyces coelicolor A3(2). The alignment shows the variant amino acids in the BC loop secondary structural elements. Ile87 in CYP158A2 is conserved in CYP158A3 from Streptomyces avermitilis and CYP158B1 from Saccharopolyspora erythraea, but not in CYP158A1.
Herein, we show that the Ile87Lys mutant in CYP158A2 significantly changes the ratios of the dimerization products converting CYP158A2 into a CYP158A1-like monooxygenase. However, when the corresponding Lys90 in CYP158A1 is mutated to isoleucine as found in CYP158A2, the catalytic activity of Lys90Ile does not change at all compared with the wild type CYP158A1. The crystal structure of Ile87Lys suggests that the overall BC loop topology of the mutant is very different from the BC loop in both wild type CYP158A1/A2 structures. Our results indicate that the overall active site environments and multiple residues contribute to the regiospecificity during catalysis and together shed light on the mechanism of the unusual P450-catalyzed C-C bond formation.
Experimental Procedures
Site-directed Mutagenesis of the CYP158A1 and CYP158A2 Genes
The CYP158A1/A2 proteins were genetically engineered to contain a His4-tag at the carboxy-terminus using a polymerase chain reaction (PCR)-based strategy as described previously [6, 7]. The QuikChange mutagenesis kit (Stratagene) was used for construction of site-directed mutants. The primers were: CYP158A2 Ile87Lys forward: 5′-CGCCCCCCACTTCAAGCCTGCCCGCGGCG-3′; CYP158A2 Ile87Lys reverse: 5′-CGCCGCGGGCAGGCTTGAAGTGGGGGGCG-3′; CYP158A1 Lys90Ile forward: 5′-CGCCCCGCACTTCATCCCGCGGCCCGGCTCGC-3′; CYP158A1 Lys90Ile reverse: 5′-GCGAGCCGGGCCGCGGGATGAAGTGCGGGGCG-3′. The integrity of the mutant sequences was confirmed by DNA sequencing.
Expression and Purification of Mutants of CYP158A1/CYP158A2
The expression and purification of the mutants were performed as described previously [7]. Recombinant proteins were produced in E. coli BL21 (DE3) pLysS competent cells. Briefly, the cells were cultured in LB broth containing 50 μg/ml ampicillin overnight. The transformed E. coli inoculated (1:100) in 1 liter of Terrific Broth containing 100 μg/ml ampicillin were grown at 37 °C and 240 rpm. After induction with 1 mM isopropyl-β-D-thiogalactopyranoside and the addition of 1 mM δ-aminolevulinic acid, growth was continued for an additional 20 h at 27 °C and 190 rpm. The cells were harvested by centrifugation and resuspended in 100 ml of lysis buffer. Cells were broken by freeze-thawing and the cytosol isolated following centrifugation at 100,000 × g. The soluble mutants were purified by metal (Ni2+) affinity chromatography (Qiagene) followed by S-Sepharose/Q-Sepharose (Amersham Bioscience) chromatography [7].
Spectral Substrate Binding and Activity Assays
Spectral and catalytic activity assays were carried out as reported previously for CYP158A2 using flaviolin as a substrate [7]. Absorbance difference spectra were recorded using a double beam Shimadzu UV-2401PC spectrophotometer. The interaction of flaviolin with mutants was examined by perturbation of the heme Soret spectrum. Ile87Lys and/or Lys90Ile were divided between two tandem cuvettes. Variant concentrations of flaviolin in 5% methanol were added to the sample cuvette to give a final ligand concentration in the range of 1-150 μM. An equal volume of 5% methanol was added to the reference cuvette, and the difference spectrum recorded after each titration. The Hill coefficient was obtained from a plot of log10(ΔA(382-418)/[ΔAmax-ΔA(382-418)]) vs log10[flaviolin]. Kd values were estimated by fitting plots of ΔA 382-418 vs [flaviolin]. For activity assays, Ile87Lys and/or Lys90Ile were reconstituted in 400 μl of 20 mM Tris-HCl buffer (pH 8.2) and flaviolin (0.26 μmol). Following incubation of the reaction mixture for 5 min on ice, the reconstituted enzyme solution was placed in a shaking water bath at 37 °C. The reaction was initiated by the addition of NADPH to a final concentration of 1 mM and was carried out for 2 h in a 1.5-ml test tube, at which time the reaction was terminated by addition of 4 μl of concentrated HCl. Subsequently the mixtures were extracted with ethyl acetate. The product formation was analyzed by LC/MSMS (Liquid Chromatography-Mass spectrometry) [7]. UV detection was measured at 254 nm. The kinetic parameters were calculated by a nonlinear regression fit to the Michaelis-Menten equation using the GraphPad Prism software (GraphPad Software Inc., San Diego, CA).
Crystallization and Data Collection
Crystals of Ile87Lys of CYP158A2 were obtained using hanging-drop vapor diffusion, in which 2 μl of a 20 mg/ml protein solution was mixed with an equal volume of 0.1 M bis-Tris (pH 6.5), 1.2 M ammonium dihydrogen phosphate. At 20 °C, the ligand-free crystals appeared within a few days; to attempt to generate the substrate flaviolin-bound crystals, flaviolin (1 mM, containing 2% (v/v) methanol) was mixed with Ile87Lys protein solution before crystallization; The crystals belong to the monoclinic space group P2(1) (Table 1). Full diffraction data were collected at 100 K at the Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL. The X-ray data were processed and scaled with the HKL package programs HKL2000 [10].
Table 1.
Data Collection and Refinement Statistics
| Substrate-free | |
|---|---|
| Data collection statistics | |
| Space group | P2(1) |
| Unit Cell (Å) | 59.609 79.313 83.658 |
| Beta value (°) | 94.08 |
| Molecules/asymmetric unit | 2 |
| Data resolution (Å) | 1.76 |
| Redundancya | 3.4 (2.5) |
| Completeness %a | 98.3 (92.4) |
| I/σ(I)a | 11.2 (1.9) |
| Rmerge %a | 13.3 (45.3) |
| Refinement statistics | |
| No. of reflections used in refinement |
75480 |
| No. of water molecules | 646 |
| Protein atoms | 6220 |
| Heme atoms | 86 |
| Ligand atoms | 0 |
| Rwork % | 21.65 |
| Rfree % | 26.42 |
| Rmsd in bond lengths (Å) | 0.005 |
| Rmsd in bond angles (°) | 1.27 |
Values for the highest resolution shell in parentheses
Structure Determination
The structure of the CYP158A2 Ile87Lys mutant was determined by molecular replacement using the program PHASER [11] and the substrate-free CYP158A2 structure (Protein Data Band code: 1SE6) as a search model. The initial model was built in COOT [12] and refinement was performed using CNS1.3 [13]. There were two molecules of Ile87Lys in the asymmetric unit. Final refinement statistics are given in Table 1. The coordinates and associated structure factors have been deposited with the Protein Data Bank (accession codes: 3TZO). Figures were generated by Pymol [14]. Attempts to generate complexes with flaviolin in the active site of Ile87Lys failed because no electron densities corresponding to flaviolin were over the heme in the structure.
Results
Substrate binding and catalytic activities of Ile87Lys in CYP158A2 and Lys90Ile in CYP158A1
The crystal structures of flaviolin-bound CYP158A1 and CYP158A2 identify several key residues that are in contact with two flaviolin molecules in the active sites [6, 7]. These residues locate the flaviolin molecules in very different positions in the two enzymes. In CYP158A2, the two flaviolin molecules are stacked closely together (4 Å) over the heme plane. In CYP158A1, there is one flaviolin in the heme pocket located essentially in the same position as the proximal substrate in CYP158A2. The other is bound at the mouth of the substrate access channel [6]. To explore the regiospecificity of the oxidative coupling reaction, we have carried out site-specific mutagenesis of CYP158A1/CYP158A2 and have prepared the Ile87Lys mutant in CYP158A2 and Lys90Ile mutant in CYP158A1 (Fig. 1). Ile87 in CYP158A2 is in contact with the distal flaviolin molecule and provides the ceiling over the naphthoquinone ring on the BC loop. The Ile87Lys and Lys90Ile mutants produce normal reduced carbon monoxide difference spectra essentially identical to those of the wild type enzymes. The spectra of flaviolin binding for the two mutant proteins are typical type I P450 binding spectra, like the wild type enzymes, converting the low-spin heme Soret peak to a high-spin peak at 383 nm. The binding affinities of flaviolin for the wild type CYP158A2 and Ile87Lys are 7.3 and 43.2 μM and for wild type CYP158A1 and Lys90Ile are 10.5 and 16.9 μM, respectively. In addition, the Hill coefficients for both mutants are about 1.1 indicating that only one flaviolin molecule binds to the heme iron, when compared to that for wild type CYP158A2 (~1.7), which indicates that two flaviolin molecules are in the active site (clearly confirmed by x-ray crystallography) [7].
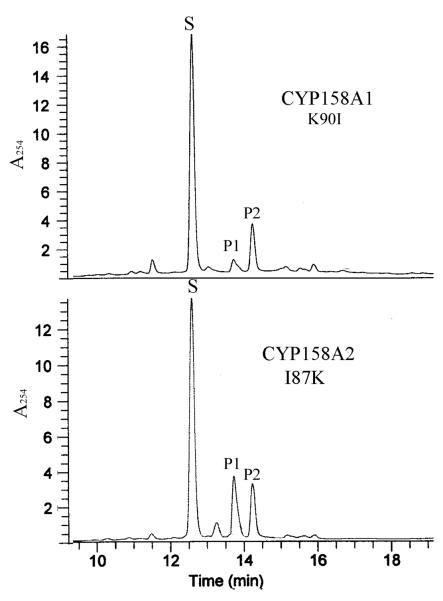
Electrospray mass spectrometry analysis of product formation from flaviolin by the Ile87Lys mutant shows that only two isomers of biflaviolin with a mass of 410 (MH+ 411, P1 tR 13.8 min; P2 tR 14.2 min) are generated and there are no detectable products for the third biflaviolin isomer (P3) and triflaviolin (P4) which are generated by wild type CYP158A2 (Fig. 2). The products were identified, as described previously, based on their retention time and mass spectrometry data [6, 7] and a few minor peaks are observed on the chromatographic trace which are not related with the oxidation by CYP enzymes as described previously [7]. These results indicate that the Ile87Lys mutant can catalyze the oxidative C-C coupling reaction as the wild type CYP158A2. However, it can only produce two isomers of biflaviolin identical to those obtained with the wild type CYP158A1. Isomer distribution from the Ile87Lys mutant (P1:P2 = 60:40) showed a significant increase in product P1 and decrease in product P2, which are opposite of the product ratios of wild type CYP158A1 (P1:P2 = 10:90). Interestingly, the catalytic reaction of the Lys90Ile mutant with flaviolin shows a very similar product profile to that of wild type CYP158A1 (Fig. 2). Thus, these results indicate that the Ile87Lys and Lys90Ile mutants can accommodate the same substrate (flaviolin) and generate the same products (P1 and P2), but with very different molar ratios. Each enzyme has its own catalytic specificity, which suggests that there are some differences in the overall topologies of the active sites.
FIG.2.
Catalytic activity of Lys90Ile of CYP158A1 and Ile87Lys of CYP158A2. Oxidation reactions were carried out as described under Experimental Procedures. Product profile obtained following flavodoxin and flavodoxin reductase supported reactions using Lys90Ile and/or Ile87Lys (1 nmol) and flaviolin (0.26 μmol) at 37°C for 2 h. The products are noted as P1, P2 and flaviolin as S. P1 and P2 have been identified as 3,8′-biflaviolin and 3,3′-biflaviolin respectively, by both HPLC and MS analysis.
Ile87Lys mutant crystal structure
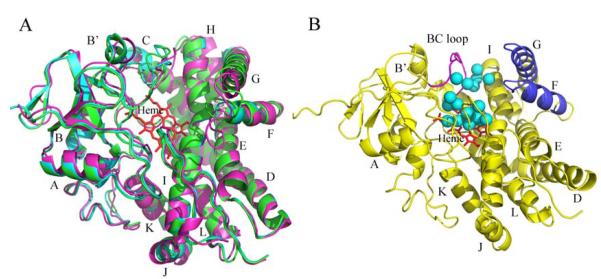
While Ile87Lys can produce two biflaviolin isomers during catalysis, when flaviolin was added for the co-crystallization experiments there is no electron density responsible for the flaviolin substrates in the active site pocket as found in the wild type CYP158A2 flaviolin-bound complex. This may be related with the relatively high binding constant for the mutant compared with the wild type enzyme. Thus, the mutant structure is a ligand free structure showing the traditional P450 fold as seen in CYP158A1/A2 [6, 7] and other P450 structures [15]. The overall structure exhibits the “open” conformation also observed for ligand-free CYP158A2 (Fig. 3A). There are two molecules in the asymmetric unit and they are almost identical except that the F/G loop in molecule A is a little longer than that in the molecule B because there is one more alpha helical turn in the G helix in molecule B. This may suggest that the G helix is dynamic and could lead to a change in the conformation of the substrate access channel. A big cleft exists between the BC loop and the FG loop creating an opening on the protein distal surface about 18 Å wide (Fig. 3B). The BC loop is flipped up towards to the protein surface to build the left wall and the F/G helices are rotated out of the active site to form the right wall of the cleft so that the heme and the I helix are exposed to the solvent. This opening on the distal surface should allow substrate access to the active site of the enzyme [16]. Additionally, owing to the absence of substrate, the substrate access channel is filled with eighteen water molecules. Two water molecules hydrogen bond with Arg288 which is a key substrate binding residue in both wild type CYP158A1/A2 enzymes [6, 7] and five water molecules are just over the heme, one of them coordinating with the heme iron at the sixth position at a distance of 2.22 Å. This is in the normal range for most other P450s [17]. When flaviolin enters into the active site, some of those water molecules would be pushed out and some other active site waters would be rearranged into positions that take part in a proton relay system helping dioxygen activation [18]. The side chains of Arg71, His101, Arg105, Arg295, Tyr318, and His351 hydrogen bond with the heme propionate moieties to stablize the heme ligation with the protein [19]. R71 also plays an important role in substrate binding to make the hydrogen bonding connection with the ketone of the proximal flaviolin molecule [6, 7].
FIG. 3.
Overlay ribbon diagrams of Ile87Lys (green), CYP158A2 ligand-free (cyan) and CYP158A1 ligand-free (magenta) structures. A, All three crystal structures show a typical cytochrome P450 fold. Heme is the red stick model. B, The cleft between the BC loop and the FG helices is shown. There are eighteen water molecules in the empty active site pocket shown as cyan spheres. The BC loop is highlighted in pink and FG region in blue.
The Active site of Ile87Lys mutant
The overall structure of the Ile87Lys mutant is very similar to that of ligand free CYP158A2 except for the BC loop and FG loop regions. In wild type CYP158A2, the key active site residues for substrate recognition and binding of flaviolin are Arg71, Ile87, Leu179, Ile241, His287, Arg288, Leu293, Leu393 [7]. In the mutant structure, both the side chains of Arg71 and Arg288 are oriented toward the active site pocket of the enzyme, and the guanidinium group of Arg288 forms hydrogen bonds with two ordered water molecules. The imidazole ring of His287 has a weak hydrogen bonding interaction at 3.5 Å with the Ala245 carbonyl in the I helix. Interestingly, the nitrogen atom from the imidazole ring of H287 forms a hydrogen bond at 2.8 Å with the 5-hydoxyl group of the proximal flaviolin through a water molecule (WAT505) in the CYP158A2 flaviolin complex [7]. WAT505 is not present in the ligand free structure and is repositioned during the substrate binding. Thus, after the proximal flaviolin binding to the active site, WAT505 is reordered in the active site so that it can induce the rotation of the imidazole ring of His287 for the hydrogen bonding interaction. In addition, the side chain methyl group of Leu293 in both the ligand free and the flaviolin-bound structures is rotated away from the active site. In the flaviolin complex, the methyl group of Leu293 is 4.2 Å from the carbonyl C1 of the distal flaviolin. The orientation of the side chain of Leu293 would make more room for distal flaviolin binding. However, in the mutant structure, the side chain of Leu293 is flipped to the opposite direction compared with the wild type enzyme. This orientation reduces the space for binding of the distal flaviolin. If the distal flaviolin is bound to the Ile87Lys in the same position as in the wild type enzyme, the side chain of Leu293 in Ile87Lys would conflict sterically with the distal flaviolin.
The major differences between Ile87Lys and the wild type CYP158A2 are located in the BC loop (Fig. 4). In wild-type CYP158A2, the residues between Leu82 and Gly94 in the BC loop form a reverse “W” shape and the Ile87 is located in the middle of the “W” pointing into the active site. However, in the mutant structure, the same residues (Leu82 to Gly94) stretch to produce a straight loop structure and form a reverse “U” shape away from the active site. Although the BC loop is the most variable region in all P450s [20], it is surprising to observe that the overall BC loop topology is so different from that in the wild type CYP158A2 due to the single amino acid change. Together, these results may confirm the hypothesis that there would be steric collision with other active site residues if the bulkier side chain of lysine served a similar role in the mutant Ile87Lys as Ile87 does in CYP158A2. This substitution is the main reason why the BC-loop conformation does not dip into the active site in the mutant structure. The differences in the BC loop structures between the mutant and wild type imply that the loop topology is only related with the intrinsic property of the specific residue and is not triggered by substrate binding since both are substrate free structures. Furthermore, these structural features in the mutant Ile87Lys open up the ceiling of the substrate binding pocket and build substantially different active site environments compared with the wild type CYP158A2. The topologies of the FG loop between the mutant and the wild type are very similar. The flipped BC loop in the mutant structure may make more room for the FG loop to swing back and forth so that the FG loop in the mutant structure shows small differences in the alpha helical turn and moves more toward the BC loop than in the wild type structure. These changes in the mutant active site may explain why the binding constant of Ile87Lys is almost seven fold lower than that of the wild type enzyme and also why it is difficult to obtain the substrate bound complex. The quite different BC loop structures between the mutant and wild type CYP158A2 result in substantial topology changes of the active site so that Ile87Lys can only generate two dimers instead of three isomers of biflaviolin.
FIG. 4.
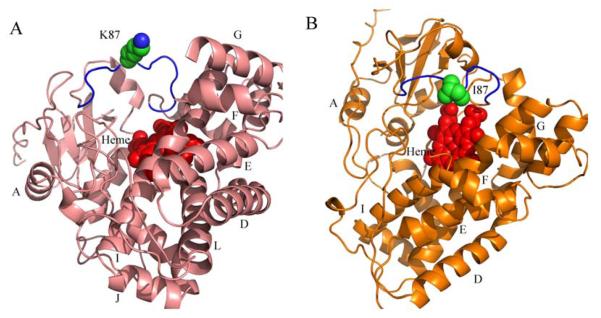
The overall BC loop topology in Ile87Lys and wild type CYP158A2. A, In wild-type CYP158A2 (salmon), the residues in the BC loop form a reverse “W” shape and the Ile87 located in the middle of the “W” points into the active site. Ile87 is colored in green and is a sphere model. B, in the mutant structure (orange), the residues in the same three dimension positions make a reverse “U” shape and the BC loop due to the substitution Ile87Lys is opened and flipped away from the active site. Lys87 is colored in green as sphere model. Both hemes are in a red sphere model.
Detailed Comparison with CYP158A1
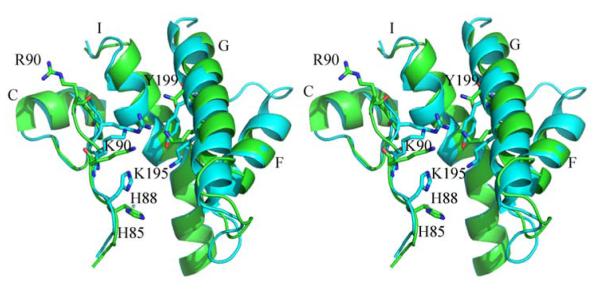
Some structural differences between wild type CYP158A1 and CYP158A2 were reported earlier [6]. However, the most interesting structural feature of the Ile87Lys mutant is whether the second flaviolin binding site would be the same as in CYP158A1. The second flaviolin binding site in CYP158A1 includes Lys90/His88 in the BC-loop and Lys195/Tyr199 from the G helix (Fig. 5). These residues form a small hydrophilic cavity between the BC loop and FG helical region to trap the ligand on the distal surface of the protein. However, in the mutant structure, as shown in Fig. 5, the side chains of residues 86-95 in the BC loop region show different orientations from those in CYP158A1. The side chains of Lys87 and Arg90 are rotated away from the active site toward the B helix in the mutant, while the side chains of Lys90 and Arg92 in CYP158A1are orientated toward the G helix. Although His85 in the mutant structure occupies the same position as His88 in CYP158A1, the imidazole ring of His85 points in the opposite direction compared with that of His88 in CYP158A1. In addition, Tyr199 in CYP158A1 corresponds to Asp196 in CYP158A2. These differences indicate that there is no second flaviolin binding cavity located in the mutant structure even though the BC loop in Ile87Lys is away from the active site.
FIG. 5.
Stereo view of the superimposition of BC loop residues and the second flaviolin binding pocket in the Ile87Lys (green) mutant and the wild type CYP158A1 (cyan); Residues are presented as stick models and are colored cyan in CYP158A1 and green in Ile87Lys.
Another difference is that the I helix of both the mutant and wild type CYP158A2 is significantly longer than that of CYP158A1. There are two less helical turns at the C-terminal end of the I helix in CYP158A1, which results in a longer loop motif between the H and I helices. This longer loop is located between the BC loop and H helix and substantially decreases the interactions between the BC loop region and the I helix. The C-terminal end of the I helix in CYP158A1 is bent toward the BC loop compared to the straight I helix in the mutant structure. Perhaps, it is a compromise due to the lack of helical turns at the C-terminus of the I helix in CYP158A1. In addition, there is a helical kink in the I helix between Gln240 and Gly241 in the mutant structure, which is not present in CYP158A1. Thus, the side chain of Gln243 in CYP158A1 provides a hydrogen bond with the hydroxyl oxygen of Ser161 in the E helix and the side chain of Tyr218 in the H helix. In contrast, the side chain of Gln240 in the mutant does not interact with the corresponding residues Cys158 in the E helix and Thr215 in the H helix.
Discussion
Oxidative phenolic coupling is of great importance in natural product chemistry and involves enzymes such as peroxidases, laccases, and β-tyrosinases which are known to catalyze oxidative phenolic coupling of many aromatic substrates [21-23]. Secondary metabolites including alkaloids, cyclic peptides, and glycopeptide antibiotics are biosynthesized via enzyme-catalyzed C-O or C-C bond formation. More recently, molecular biology and genomics have provided additional understanding of the mechanisms underlying biosynthesis of these compounds. Some cytochrome P450 enzymes have been shown to carry out the phenolic coupling reactions: CYP158A1/A2 involved in cross linking flaviolin monomers [6, 7], CYP165B1 and CYP165C1 from the vancomycin producer Amycolatopsis orientalis which have been proposed to catalyze oxidative coupling reaction in vancomycin biosynthesis [24, 25], and CYP80A1 from Berberis stolonifera involved in berbamunine biosynthesis via intermolecular C–O phenol-coupling [26].
As already noted, Ile87 in CYP158A2 interacts with the distal flaviolin. Substitution with a longer side chain residue changes the product regiospecificity. These results confirm that the C-C oxidative coupling of flaviolins must take place in the active site or the substrate access channel within the enzyme. It is not possible that the radical or cationic intermediates could be released from the active site and react to form products in the solvent outside of the P450. Therefore, the regiospecificity of the dimerization products is probably actively driven by the active site topology reorganizing key substrate contact residues. Probably the proximal flaviolin is activated to a radical or cationic intermediate [7] by a very similar reaction in CYP158A2. This intermediate must contact the distal neutral flaviolin molecule somewhere within the active site or substrate access channel or the intermediate may bind somewhere in the active site waiting for the second flaviolin molecule to be activated. The regiospecificity will depend on the orientation or position of each flaviolin molecule or intermediate in the active site. So far the detailed mechanism catalyzed by CYP158A1/A2 enzymes through a radical mechanism or cationic intermediate is unclear. At this stage we cannot exclude any of the possible mechanisms mentioned above or that this enzyme may proceed through a combination radical and cationic mechanism.
While CYP158A1 contains about 61% amino acid sequence identity to CYP158A2, the secondary structural elements show significant differences, such as in the BC loop region, F helix and I helix. Orientations of the same residues are also quite different, for example Arg90 in CYP158A2 and Arg92 in CYP158A1 (Fig. 5). These significant differences result in differences in the active site topology so that it is not surprising to observe the altered substrate selectivity and product regiospecificity. The catalytic activity and structural features of Ile87Lys shed light on this point. Although, the biochemical results reveal that CYP158A1/A2 can catalyze oxidative C-C coupling reactions thereby polymerizing flaviolin, it is puzzling why these two enzymes show such different flaviolin binding modes and quite different product patterns if they indeed have the same biological function. We can not rule out that CYP158A1 might have a biological function different from that of CYP158A2.
Highlights.
>We generate two mutants Ile87Lys of CYP158A2 and Lys90Ile of CYP158A1. >We compare each mutant catalytic activity with corresponding wild type enzyme. >We determine the crystal structure of Ile87Lys of CYP158A2. It shows that the BC loop in the mutant structure is in a very different orientation compared with the BC loop in both CYP158A1/A2 structures.>These results shed light on the mechanism of the oxidative coupling reaction catalyzed by cytochrome P450.
Acknowledgment
We thank personnel at the SER-CAT 22-BM beamline at APS Argonne National Laboratory for expert technical assistance. This work was supported by National Institutes of Health Grants GM69970 (M. R. W.), and ES00267 (M. R. W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- [2].Montellano P.R.O.d. Cytochrome P450: Structure, Mechanism, and Biochemistry. Kluwer Academic/Plenum Publishers; New York: 2005. [Google Scholar]
- [3].Coon MJ. Annu Rev Pharmacol Toxicol. 2005;45:1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030. [DOI] [PubMed] [Google Scholar]
- [4].Lamb DC, Skaug T, Song HL, Jackson CJ, Podust LM, Waterman MR, Kell DB, Kelly DE, Kelly SL. J Biol Chem. 2002;277:24000–24005. doi: 10.1074/jbc.M111109200. [DOI] [PubMed] [Google Scholar]
- [5].Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- [6].Zhao B, Lamb DC, Lei L, Kelly SL, Yuan H, Hachey DL, Waterman MR. Biochemistry. 2007;46:8725–8733. doi: 10.1021/bi7006959. [DOI] [PubMed] [Google Scholar]
- [7].Zhao B, Guengerich FP, Bellamine A, Lamb DC, Izumikawa M, Lei L, Podust LM, Sundaramoorthy M, Kalaitzis JA, Reddy LM, Kelly SL, Moore BS, Stec D, Voehler M, Falck JR, Shimada T, Waterman MR. J Biol Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- [8].Funa N, Funabashi M, Ohnishi Y, Horinouchi S. J Bacteriol. 2005;187:8149–8155. doi: 10.1128/JB.187.23.8149-8155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cortes J, Velasco J, Foster G, Blackaby AP, Rudd BA, Wilkinson B. Mol Microbiol. 2002;44:1213–1224. doi: 10.1046/j.1365-2958.2002.02975.x. [DOI] [PubMed] [Google Scholar]
- [10].Otwinowski Z, Borek D, Majewski W, Minor W. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- [11].Storoni LC, McCoy AJ, Read RJ. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- [12].Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr D Biol Crystallogr. 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- [14].The PyMOL Molecular Graphics System. Version 1.2r3pre Schrödinger, LLC; [Google Scholar]
- [15].Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- [16].Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Proc Natl Acad Sci U S A. 2001;98:12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. Proc Natl Acad Sci U S A. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao B, Guengerich FP, Voehler M, Waterman MR. J Biol Chem. 2005;280:42188–42197. doi: 10.1074/jbc.M509220200. [DOI] [PubMed] [Google Scholar]
- [19].Ghosh D, Griswold J, Erman M, Pangborn W. J Steroid Biochem Mol Biol. 118:197–202. doi: 10.1016/j.jsbmb.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Johnson EF, Stout CD. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- [21].Huang KX, Fujii I, Ebizuka Y, Gomi K, Sankawa U. J Biol Chem. 1995;270:21495–21502. doi: 10.1074/jbc.270.37.21495. [DOI] [PubMed] [Google Scholar]
- [22].Schlauer J, Ruckert M, Wiesen B, Herderich M, Assi LA, Haller RD, Bar S, Frohlich KU, Bringmann G. Arch Biochem Biophys. 1998;350:87–94. doi: 10.1006/abbi.1997.0494. [DOI] [PubMed] [Google Scholar]
- [23].Niemetz R, Schilling G, Gross GG. Phytochemistry. 2003;64:109–114. doi: 10.1016/s0031-9422(03)00280-2. [DOI] [PubMed] [Google Scholar]
- [24].Zerbe K, Pylypenko O, Vitali F, Zhang W, Rouset S, Heck M, Vrijbloed JW, Bischoff D, Bister B, Sussmuth RD, Pelzer S, Wohlleben W, Robinson JA, Schlichting I. J Biol Chem. 2002;277:47476–47485. doi: 10.1074/jbc.M206342200. [DOI] [PubMed] [Google Scholar]
- [25].Puk O, Huber P, Bischoff D, Recktenwald J, Jung G, Sussmuth RD, van Pee KH, Wohlleben W, Pelzer S. Chem Biol. 2002;9:225–235. doi: 10.1016/s1074-5521(02)00101-1. [DOI] [PubMed] [Google Scholar]
- [26].Kraus PF, Kutchan TM. Proc Natl Acad Sci U S A. 1995;92:2071–2075. doi: 10.1073/pnas.92.6.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]