Abstract
Leprosy provides a model to investigate mechanisms of immune regulation in humans, given that the disease forms a clinical-immunological spectrum. Here, we identified 13 miRNAs that were differentially expressed in the lesions of subjects with progressive lepromatous (L-lep) vs. the self-limited tuberculoid (T-lep) disease. Bioinformatic analysis revealed a significant enrichment of L-lep-specific miRNAs that preferentially target key immune genes downregulated in L-lep vs. T-lep lesions. The most differentially expressed miRNA in L-lep lesions, hsa-mir-21, was upregulated in M. leprae-infected monocytes. Hsa-mir-21, by downregulating toll-like receptor 2/1 (TLR2/1)-induced CYP27B1 and IL1B as well as upregulating IL-10, inhibited gene expression of the vitamin D-dependent antimicrobial peptides, CAMP and DEFB4A. Conversely, knockdown of hsa-mir-21 in M. leprae-infected monocytes enhanced expression of CAMP and DEFB4A and restored TLR2/1-mediated antimicrobial activity against M. leprae. Therefore, the ability of M. leprae to upregulate hsa-mir-21 targets multiple genes associated with the immunologically localized disease form, providing an effective mechanism to escape from the vitamin D-dependent antimicrobial pathway.
Interactions between the host immune response and the invading pathogen at the site of disease are critical to the outcome of the infection. Leprosy, caused by the intracellular bacterium Mycobacterium leprae, provides an extraordinary model for studying host-pathogen interactions in humans. The disease presents as a spectrum where the clinical manifestations correlates with the level of immune response to the pathogen 1, which contributes to host defense vs. persistence and pathogenesis. At one end of the spectrum, the tuberculoid form (T-lep), the infection is self-limited, where skin lesions are typified by an adaptive immune response characterized by Th1 cytokines 2, 3 and an innate immune response characterized by macrophages programmed to express the vitamin D-mediated antimicrobial pathway 4. At the other end of the spectrum, the lepromatous form (L-lep), the infection is disseminated with lesions typified by an adaptive immune response characterized by Th2 cytokines 2, 3 and an innate immune response characterized by macrophages programmed to express a phagocytic activity 4. In order to gain insight into the mechanism(s) that regulate host defense vs. persistence in human infectious disease, we investigated miRNA expression in leprosy skin lesions.
Results
Gene and miRNA prolife in leprosy
The mRNA and miRNA expression profiles in skin lesions were determined in biopsy specimens from six T-lep and five L-lep patients collected at the time of diagnosis and classified according to the clinical and histopathological criteria of Ridley (Supplemental Fig. 1) 1. Unsupervised hierarchal clustering analysis of the mRNA profiles revealed two major groups in which the L-lep and T-lep samples were segregated (Supplemental Fig. 2). In contrast, hierarchal clustering analysis of the miRNA profiles performed on the same samples indicated two major miRNA patterns, with each group containing a mixture of both L-lep and T-lep samples (Supplemental Fig. 2). These results indicate that the principal component of the measured miRNA expression patterns in leprosy did not differentiate the lesions types.
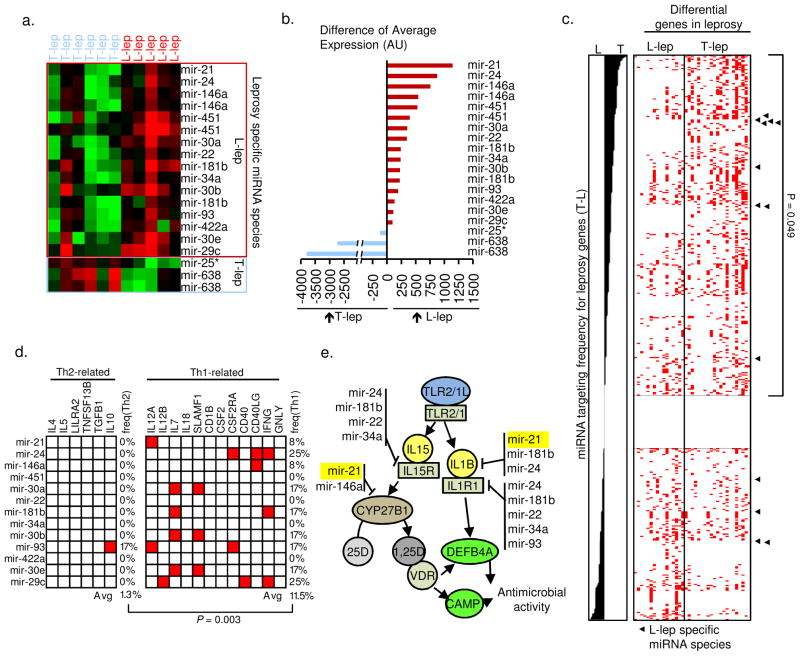
In order to identify lesion-specific differences we used a supervised approach. Differentially expressed miRNAs between the two clinical groups were identified by ranking miRNAs probes according to statistically significance (t-test) and limited to sequences present in the miRBase database (version 14). There was a five-fold greater number of differentially expressed miRNAs in the L-lep samples (16 probes representing 13 annotated miRNA species) vs. the T-lep samples (three probes representing two unique miRNA species) (Fig. 1a). In order to compare the magnitude of differential expression between these miRNA species, the un-normalized intensity values of the probes were compared. The difference in intensity of the hsa-mir-21 probe was the greatest of the miRNA species differentially upregulated in L-lep vs. T-lep lesions (Fig. 1b).
Figure 1.
MiRNA expression and targeting profile in leprosy. MiRNA probes that are differentially expressed between T-lep and L-lep lesions displayed as (a) normalized data and (b) raw expression values. (c) All miRNA species represented on the microarray platform ranged by targeting preference score. ◂ = L-lep specific miRNA species. (d) Targeting preference of the L-lep or T-lep specific miRNA species for Th2- or Th1-related genes. Red box represents a predicted target site within the 3′UTR of the indicated gene. (e) L-lep specific miRNA species and predicted targeting of genes in the TLR-induced vitamin D-dependent antimicrobial pathway.
Targeting of immune genes by leprosy specific miRNAs
Because the differentially expressed miRNA species were predominantly enriched in L-lep lesions, we hypothesized that regulation of miRNA expression at the site of the progressive disease inhibits expression of genes involved in host defense against the pathogen. This hypothesis was tested by integrating a prediction algorithm for miRNA binding sites in the three prime untranslated regions (3′UTR) of messenger RNA species with curated sets of host immune response signature genes known to be differentially expressed in leprosy lesions, including Th1 vs. Th2 related genes as well as the genes of the vitamin D pathway (Supplemental Text). All miRNA species represented on the microarray platform were ranked by their ‘targeting preference score’, calculated as the difference in frequency for targeting of the T-lep compared to L-lep signature genes (Supplemental Fig. 3 and Text). Enrichment analysis of leprosy disease-type specific miRNA species was next evaluated by the Kolmogorov-Smirnov-based permutation test. The L-lep-specific miRNA species were found to be significantly associated with the miRNAs most strongly predicted to preferentially target T-lep signature genes (P = 0.049; Fig. 1c). Thus, L-lep-specific miRNAs provide a candidate mechanism for the leprosy-induced downregulation of T-lep host immune response signature genes in L-lep lesions.
In relation to the local immune response, the L-lep specific set of miRNA species were predicted to have binding sites in the 3′UTR of Th1-related signature genes, known to be differentially expressed in T-lep vs. L-lep lesions, with an average targeting frequency of 11.5%. In contrast, the L-lep specific set of miRNAs species demonstrated a significantly (P = 0.0003) lower frequency for Th2-related genes, known to be differentially expressed in L-lep vs. T-lep lesions, with an average targeting frequency of 1.3% (Fig. 1d). Strikingly, multiple L-lep specific miRNA species targeted two key genes in the vitamin D-dependent antimicrobial pathway, cytochrome P450, family 27, subfamily B, polypeptide 1 (CYP27B1) and interleukin 1, beta (IL1B), but not the antimicrobial peptides that are induced by this pathway, cathelicidin (LL-37 encoded by CAMP) and defensin, beta 4A (DEFB4A) 5, 6 (Fig. 1e). Taken together, these results indicate that the L-lep specific miRNAs target and potentially downregulate host defense genes in leprosy.
Regulation of hsa-mir-21 in leprosy
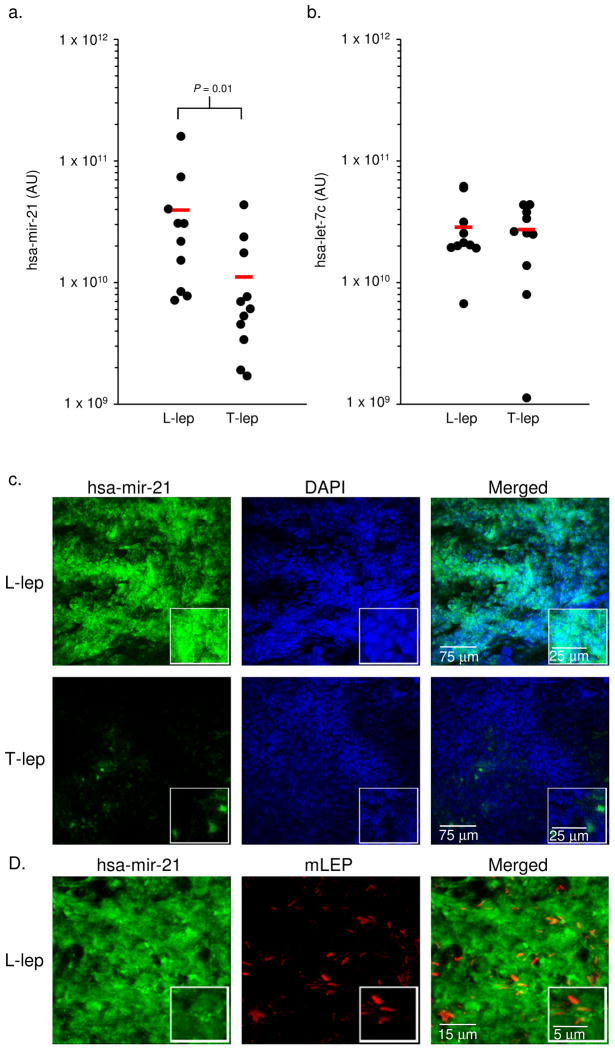
The tissue expression of the most differentially expressed miRNA, hsa-mir-21, in L-lep lesions was verified by real time PCR (qPCR) and fluorescent in situ hybridization (FISH) in additional leprosy tissue sections. By qPCR, hsa-mir-21 levels were significantly higher (3.5 fold, P = 0.01) in 10 L-lep vs. 11 T-lep lesions (Fig. 2a). An unrelated miRNA, hsa-let-7c, that was not differentially expressed in disease lesions by microarray analysis, was expressed at similar levels between the L-lep and T-lep lesions (Fig. 2b). Although the skin biopsies are comprised predominately of granulomas in the dermis, we could not rule out that the differential expression of hsa-mir-21 was due to non-immune cells. Therefore, using FISH we determined that the frequency of hsa-mir-21 positive cells in the granulomatous regions was 25-fold greater in the L-lep lesions vs. the T-lep lesions (98% vs.4% of nucleated cells, P = 0.001) (Fig. 2c). In the L-lep lesions, the hsa-mir-21 positive cells were located within the granulomas, in the same microanatomic locations as M. leprae (Fig. 2d). It was not possible to determine the frequency of cells expressing hsa-mir-21 and containing M. leprae as these are found in distinct subcellular compartments: microRNAs are located in the cytoplasm and the pathogen resides within phagosomes. A scrambled probe was used as a negative control to demonstrate the absence of non-specific binding in either lesion type (Supplemental Fig. 4a), and the positive control probe for the U6 non-coding small nuclear RNA, demonstrated equivalent RNA integrity (Supplemental Fig. 4b). Taken together, these three approaches, microarray, qPCR and FISH, provide convincing evidence for the differential expression of hsa-mir-21 in L-lep vs. T-lep lesions.
Figure 2.
Expression of hsa-mir-21 in leprosy. Expression levels of (a) hsa-mir-21 and (b) hsa-let-7c comparing L-lep vs. T-lep lesions by qPCR. The levels of hsa-mir-21 and hsa-let-7c are normalized to 36B4 levels in the same tissue and displayed as values from individual lesions (●) as well as the average (−) of 10 L-lep lesions and 11 T-lep lesions. (c) Skin biopsy sections from L-lep or T-lep subjects were probed with a hsa-mir-21 specific oligo using fluorescent in situ hybridization. Cellular nuclei were visualized using DAPI. Data shown is representative experiment for four individual L-lep samples and three individual T-lep samples. (d) Skin biopsy section derived from L-lep subjects were probed for hsa-mir-21 and M. leprae using fluorescent in situ hybridization in conjunction with a monoclonal antibody specific for M. leprae, detected by confocal microscopy. Data shown is representative experiment from three individual subjects.
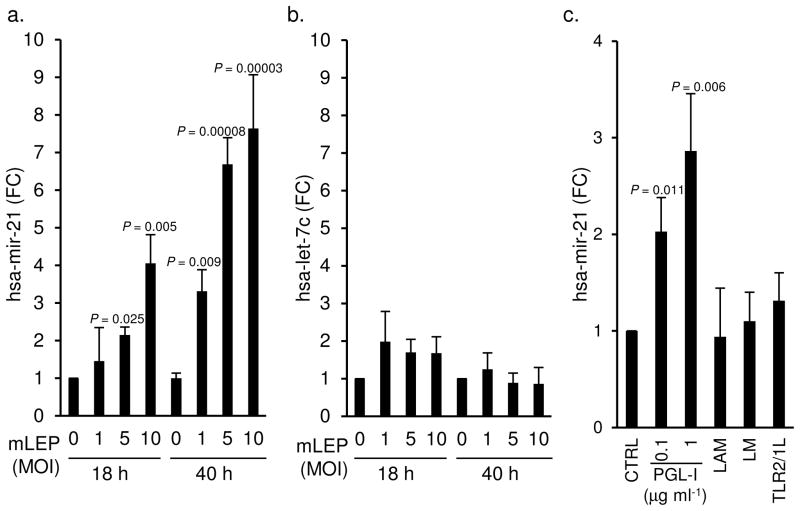
Given that M. leprae and hsa-mir-21 were both identified in the granulomas, we hypothesized that M. leprae induced hsa-mir-21 expression in monocytes/macrophages, the predominant cell type in a granuloma and the primary cell infected by the pathogen. Human peripheral blood monocytes were infected with live M. leprae at different multiplicities of infection (MOI) for 18 and 40 h, and hsa-mir-21 levels measured by qPCR. Infection of monocytes with M. leprae triggered an upregulation of hsa-mir-21 in a dose-dependent and time-responsive manner, with a 4.1-fold change (P = 0.005) at 18 h, and 7.6-fold change (P = 0.00003) at 40 h, both at a MOI of 10 (Fig. 3a). In contrast, M. leprae infection of monocytes did not result in detectable upregulation of hsa-let-7c, (Fig. 3b).
Figure 3.
Regulation of hsa-mir-21 levels in primary human monocytes by M. leprae. Levels of (a) hsa-mir-21 and (b) hsa-let-7c in primary human monocytes infected with M. leprae at MOI of 0, 1, 5, and 10 for 18h and 40h. Data shown is mean of fold change compared to no infection control ± SEM, n = 3–5. Levels of hsa-mir-21 (c) in primary human monocytes after treatment with PGL-I, LAM (10μg ml−1), LM (10μg ml−1), and TLR2/1L for 18 h. Data shown is mean fold change compared to the vehicle control treated cells ± SEM, n = 3–10.
To explore the mechanism by which M. leprae infection induces hsa-mir-21, the ability of several key cell wall biomolecules to trigger hsa-mir-21 was compared. Treatment of monocytes with phenolic glycolipid-I (PGL-I) induced a 2.9-fold change in hsa-mir-21 expression, whereas the M. leprae lipoarabinomman (LAM) and lipomannan (LM), as well as a synthetic triacylated lipopeptide (a toll-like receptor 2/1 ligand, TLR2/1L) did not significantly induce hsa-mir-21 (Fig. 3c). Together, these data demonstrate i) hsa-mir-21 is present at the site of disease in leprosy, ii) is associated with the progressive and disseminated form (L-lep) of the disease, iii) is specifically induced in monocytes by M. leprae infection; and iv) is triggered by an M. leprae specific glycolipid, PGL-I. It is therefore likely that M. leprae infection of macrophages induces the upregulation of hsa-mir-21 at the site of infection.
Regulation of the vitamin D pathway by hsa-mir-21
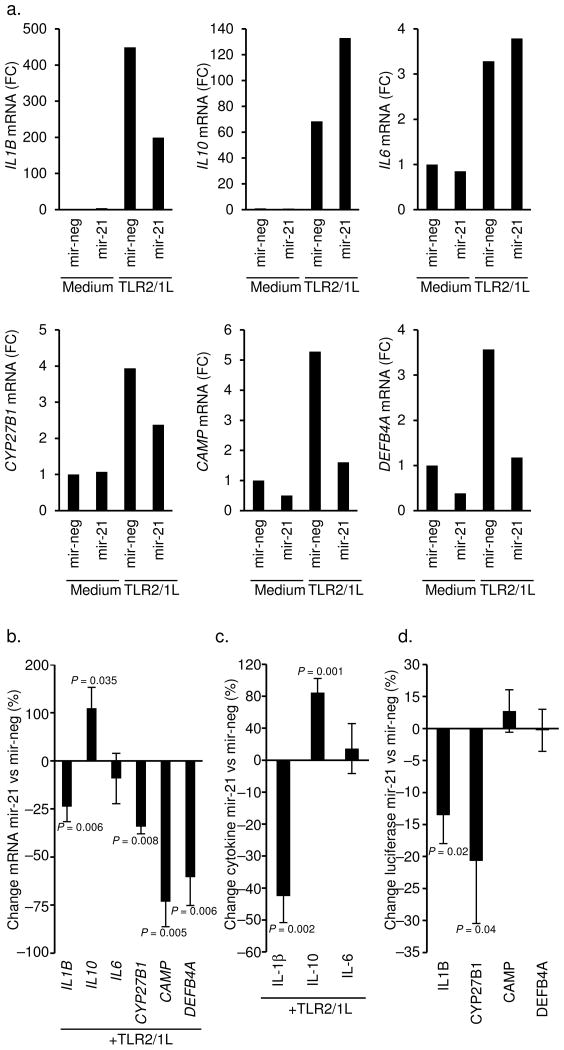
It was noteworthy that of all the L-lep specific miRNAs, only hsa-mir-21 had the potential to target both IL1B and CYP27B1 (Fig. 1e), both required for TLR-induced, vitamin D-dependent expression of CAMP and DEFB4A, which encode antimicrobial peptides 5, 6. The ability of hsa-mir-21 to regulate the expression of these antimicrobial genes was investigated by transfecting primary human monocytes with either the mature hsa-mir-21 oligo or a non-targeting control oligo, followed by activation using the TLR2/1L. As a control for targeting specificity, we determined that overexpression of hsa-mir-21 downregulated IFN-γ induced interleukin 12A (IL12A) mRNA, a direct target (Supplemental Fig. 5 and Ref 7). The presence of hsa-mir-21 during TLR2/1L activation of monocytes resulted in the downregulation of IL1B mRNA by 24% (P = 0.006, representative experiment Fig. 4a and averaged Fig. 4b). Despite the absence of predicted hsa-mir-21 target sites in the 3′UTR of interleukin 10 (IL10), transfection of hsa-mir-21 enhanced toll-like receptor 2/1 (TLR2/1)-induced IL10 mRNA levels by 110% (P = 0.035, Figs. 4a and 4c), consistent with studies in murine cells 8. In contrast, IL6, another cytokine without hsa-mir-21 target sequences, was not affected (Figs. 4a and 4c). TLR2/1-induced IL-1β secretion was reduced by 45% (P = 0.003), IL-10 release was enhanced by 85% (P = 0.001), and IL-6 levels did not change (representative experiment Supplemental Fig. 6 and averaged Fig. 4c). Therefore, the effect of hsa-mir-21 on TLR2/1-induced cytokine mRNAs and secreted proteins were consistent.
Figure 4.
The ability of hsa-mir-21 to regulate the innate immune response in human monocytes. Primary human monocytes were transfected with the mature hsa-mir-21 (mir-21) oligo or a non-targeting control (mir-neg) then treated with TLR2/1L for 18 h and 24 h. Gene expression of (a) IL1B at 18 h, IL10 at 24 h, IL6 at 16 h, CYP27B1 at 16 h, CAMP at 24 h and DEBF4A at 24 h, were evaluated by qPCR. Data shown are representative experiments from greater than five individual donors. (b) Change in gene expression comparing mir-21 to mir-neg transfected cells following TLR2/1L simulation. Data shown is average percent change mir-21 vs mir-neg ± SEM, n = 3–11 (c) Change in cytokine protein levels in culture supernatants comparing mir-21 to mir-neg transfected cells following TLR2/1L stimulation. Data shown in average percent change mir-21 vs mir-neg ± SEM, n = 4–6. Representative experiment is shown in Supplemental Fig. 6. (d) Change in luciferase activity of cells co-transfected with a 3′UTR luciferase reporter construct (IL1B, CYP27B1, CAMP, DEFB4A) and either mir-21 or mir-neg. Data shown is mean percent change of each individual 3′UTR construct comparing mir-21 vs mir-neg ± SEM, n = 4–6. Representative experiment is shown in Supplemental Fig. 8.
Transfection of hsa-mir-21 also resulted in a 34% decrease in TLR-induced expression of CYP27B1 (P = 0.008, Figs. 4a and b). Given that hsa-mir-21 downregulated TLR2/1-induced IL1B and CYP27B1, the effect of hsa-mir-21 on TLR2/1-induced antimicrobial peptide gene expression was examined. Strikingly, TLR2/1L induction of CAMP and DEFB4A mRNAs was significantly inhibited by transfection of hsa-mir-21, by 73% (P = 0.005) and 60% (P = 0.006) respectively (Figs. 4a and 4b). Given that hsa-mir-21 upregulated IL-10, the effect of rIL-10 on TLR-induced gene expression was investigated. The addition of rIL-10 inhibited TLR-induced CAMP by 26% and DEFB4A by 35%, whereas the inhibition of interleukin 12B (IL12B) was 76% (Supplemental Figs. 7a and 7b). Therefore, hsa-mir-21 enhancement of IL-10 induction may partially contribute to the inhibition of antimicrobial gene expression.
The direct binding of hsa-mir-21 to the TLR-induced, vitamin D dependent antimicrobial pathway genes was assessed using a 3′UTR reporter assay. Hsa-mir-21 was determined to directly bind the 3′UTR of both CYP27B1 and IL1B, but did not bind to the 3′UTR of either CAMP or DEFB4A (Fig. 4d, Supplemental Fig. 8 and Supplemental Text). These data indicate that hsa-mir-21 inhibits TLR2/1-mediated CAMP and DEFB4A expression by directly regulating key epigenetic targets including CYP27B1 and IL1B, as well as indirectly through induction of immune modulatory cytokine IL-10.
Role of hsa-mir-21 in the response to infection
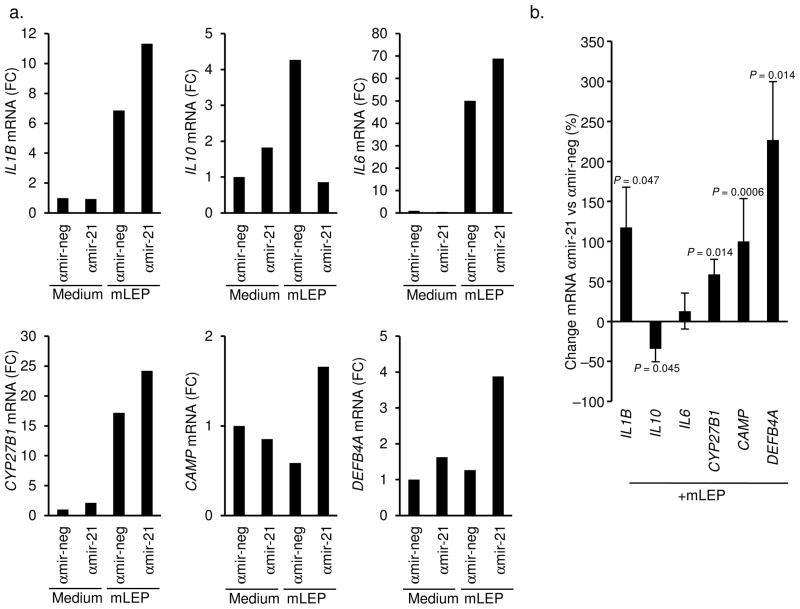
Given the ability of hsa-mir-21 to downregulate key genes in the TLR-induced antimicrobial pathway, and the observation that M. leprae induced hsa-mir-21 in monocytes, we investigated whether hsa-mir-21 contributes to inhibition of the innate immune response during M. leprae infection. This was accomplished by transfecting monocytes with a hsa-mir-21 specific antisense oligo (αmir-21). Transfected monocytes were infected with live M. leprae 9 for 18 h, and then mRNA expression was measured. Presence of αmir-21 vs. a control oligo (αmir-neg), followed by M. leprae infection resulted in a significant reduction of hsa-mir-21 levels by 70% (P = 0.00002, Supplemental Figs. 9a and 9c). Consistent with the hsa-mir-21 overexpression experiment, the αmir-21 enhanced IL12A mRNA expression in the M. leprae infected monocytes (P = 0.006, Supplemental Figs. 9b and 9c).
Relevant to the vitamin D-dependent innate immune pathway, αmir-21 increased IL1B mRNA expression in the M. leprae infected monocytes by 118% (P = 0.047, Figs. 5a and 5b). In contrast, IL10 mRNA was downregulated by 34% (P = 0.045) and there was no significant change in IL6 mRNA levels (Figs. 5a and 5b). Importantly, knockdown of hsa-mir-21 resulted in the significant increase of CYP27B1 (59%, P = 0.014), CAMP (100%, P = 0.0006), and DEFB4A (227%, P = 0.014) (Figs. 5a and 5b). These results provide evidence that the monocyte/macrophage can detect M. leprae infection and trigger the vitamin D-dependent antimicrobial pathway; however, this response is inhibited by the ability of the pathogen to upregulate hsa-mir-21.
Figure 5.
Role of hsa-mir-21 expression during M. leprae infection. Primary human monocytes were transfected with the antagomir against hsa-mir-21 (αmir-21) oligo or a non-specific control (αmir-neg) then infected with live M. leprae at an MOI of 10 for 18 h. Gene expression of IL1B, IL10, IL6, CYP27B1, CAMP and DEFB4A were evaluated by qPCR. Data shown (a) are representative experiments from greater than four individual donors. (b) Change in gene expression comparing αmir-21 to αmir-neg transfected cells following M. leprae infection for 18 h. Data shown is average percent change αmir-21 vs. αmir-neg ± SEM, n = 4–8.
Effects of hsa-mir-21 on innate antimicrobial activity
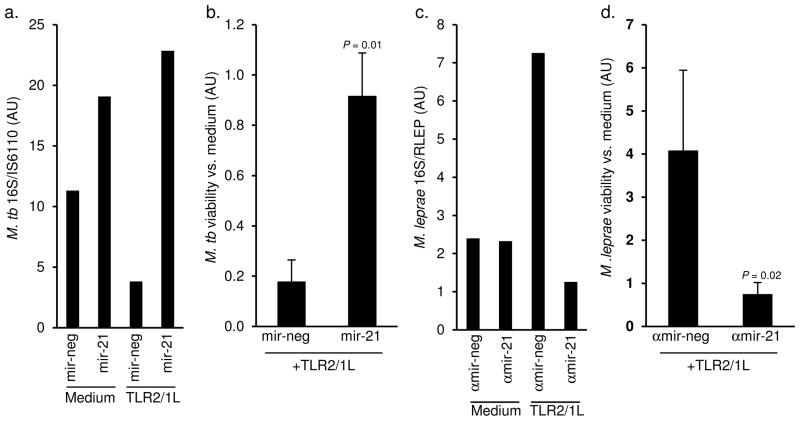
The role of hsa-mir-21 in regulating the TLR2/1-induced macrophage antimicrobial activity was investigated by overexpressing hsa-mir-21 expression during M. tuberculosis infection. For these experiments the avirulent M. tuberculosis H37Ra strain was used since it does not contain a PGL-I homologue and failed to induce expression of hsa-mir-21 in monocytes upon infection (Supplemental Fig. 10a), despite induction of IL-6 mRNA in the same cells (Supplemental Fig. 10b). Monocytes were transfected with hsa-mir-21 or a control oligo, then infected with M. tuberculosis H37Ra overnight, subsequently treated with the TLR2/1 ligand for 3 d and bacterial viability assessed by qPCR according to the ratio of 16S RNA to IS6110 DNA 10. TLR2/1L induced an antimicrobial activity against M. tuberculosis in monocytes transfected with a control oligo (Fig. 6a), at a level consistent with previous studies using the standard CFU assay 6. However, overexpression of hsa-mir-21 blocked the antimicrobial response and resulted in an increase of M. tuberculosis viability in TLR2/1L activated cells (Fig. 6a). Also, in unstimulated cells, hsa-mir-21 increased bacterial viability. Overall, M. tuberculosis viability in TLR2/1L-treated as compared to control monocytes was significantly higher in the presence of hsa-mir-21 (P = 0.01, Fig. 6b).
Figure 6.
Role of hsa-mir-21 in TLR2/1-mediated antimicrobial activity. Primary human monocytes were transfected with the mature hsa-mir-21 (mir-21) oligo or a non-targeting control (mir-neg) then infected with live M. tuberculosis H37Ra at an MOI of 0.5 for 18 h. The monocytes were then treated with the TLR2/1L (10μg ml−1) for 3 d. Levels of 16S rRNA and IS6110 DNA levels were assessed by qPCR. Data shown is the ratio of 16S rRNA to IS6110 DNA levels as (a) a representative experiment from three donors. (b) Fold change in M. tuberculosis viability comparing TLR2/1L vs. medium treated monocytes. Data shown is the mean fold change ± SEM, n = 3. Primary human monocytes were transfected with the antagomir against hsa-mir-21 (αmir-21) oligo or a non-specific control (αmir-neg) then infected with live M. leprae at an MOI of 10 for 18 h. The monocytes were then treated with the TLR2/1L (10μg ml−1) for 3 d. Levels of 16S rRNA and RLEP DNA levels were assessed by qPCR. Data shown is the ratio of 16S rRNA to RLEP DNA levels as (c) a representative experiment from five donors. (d) Fold change in M. leprae viability comparing TLR2/1L vs. medium treated monocytes. Data shown is the mean fold change ± SEM, n = 5.
To address the role of M. leprae-induced hsa-mir-21 in regulation of TLR2/1-induced antimicrobial activity, monocytes were transfected with αmir-21 or αmir-neg, then infected with live M. leprae. The transfected and infected cells were treated with the TLR2/1L for 3 d and antimicrobial activity assessed by qPCR by measuring the ratio of 16S RNA to RLEP DNA 10. In αmir-neg transfected cells, TLR2/1-activation increased bacterial viability, consistent with previous findings indicating enhanced M. tuberculosis growth in TLR2/1-stimulated cells in the absence of CAMP and DEFB4A 6. Strikingly, in αmir-21 transfected monocytes, TLR2/1-activation resulted in decreased bacterial viability (Fig. 6c). The αmir-21 oligo had no effect on M. leprae viability in unstimulated monocytes (Fig. 6c). In five donors tested, M. leprae viability in TLR2/1-stimulated cells was significantly lower in the presence of αmir-21 (P = 0.02, Fig. 6d). Together, the data from these infection experiments demonstrate the biologic relevance of hsa-mir-21 in innate host defense: the expression of hsa-mir-21 is sufficient to block TLR2/1-induced antimicrobial responses and the silencing of hsa-mir-21 induction restores TLR2/1-mediated antimicrobial activity.
Discussion
Interactions between the pathogen and the host determine the outcome of the immune response to microbial infection. The present data provides evidence that the human pathogen, M. leprae, regulates the microRNA profile at the site of infection in subjects with leprosy and interferes with the host antimicrobial response. We employed a novel bioinformatic strategy, combining an enrichment analysis of leprosy disease-type specific miRNA species ranked by 3′UTR mRNA targeting preference and evaluated by the Kolmogorov-Smirnov-based permutation test. Together, this led to the identification of hsa-mir-21 as being differentially expressed in the progressive L-lep form of leprosy with the potential to target genes in the vitamin D antimicrobial pathway. Infection of human monocytes with live M. leprae, or treatment with the mycobacterial virulence factor PGL-I, induced expression of hsa-mir-21. Next. hsa-mir-21 was demonstrated to functionally downregulate the TLR2/1-induced vitamin D antimicrobial pathway, by directly targeting CYP27B1 and IL-1β, as well as the indirect induction of IL-10, all leading to the inhibition of antimicrobial peptides CAMP and DEFB4A. Silencing of hsa-mir-21 during M. leprae infection led to the enhanced expression of vitamin D pathway genes. Finally, introduction of hsa-mir-21 into monocytes was sufficient to block TLR2/1-induced antimicrobial activity against M. tuberculosis, and the silencing of hsa-mir-21 induction restored TLR2/1-mediated antimicrobial activity against M. leprae. Therefore, these data identify an evasion strategy, in which a microbial pathogen regulates the host microRNA profile at the site of infection to inhibit the antimicrobial response.
Although M. leprae was the first human pathogen discovered 11, it still cannot be grown in the laboratory, providing a major obstacle to investigation of the immunology of leprosy. To our knowledge, it has not been possible to demonstrate immune-mediated antimicrobial activity against M. leprae in primary human cells 12. Comparison of antimicrobial responses in mouse and human macrophages demonstrated that the combination of lipopolysaccharide and interferon-γ reduced the viability of intracellular M. leprae in mouse but not human macrophages 12. Here, we successfully demonstrate that immune activation of M. leprae-infected human monocytes decreases bacterial viability, finding that TLR2/1 activation induces a four-fold reduction in M. leprae viability only when hsa-mir-21 was silenced. In addition, overexpression of hsa-mir-21 blocked the TLR2/1-induced antimicrobial activity against M. tuberculosis resulting in a five-fold increase in bacterial viability. Taken together, these data indicate the biological relevance of hsa-mir-21 in the host antimicrobial response.
Insight into the mechanism by which M. leprae induces a specific miRNA immune regulatory profile at the site of infection was revealed by identifying that hsa-mir-21 was induced in monocytes following M. leprae infection or by treatment with M. leprae-derived PGL-I. Previously, PGL-I has been shown to inhibit monocyte responses 13, 14, as well as associate with mycobacterial virulence 15. Further studies are needed to elucidate the mechanism of induction and functional role of those microRNAs differentially expressed in L-lep lesions. Given that the degree of genetic diversity in M. leprae clinical isolates is not as broad as compared with other human pathogens 16, it is not likely that species subtypes differentially induce single miRNAs as has been shown for Francisella tularensis 17. Complementing the study of miRNA profiles in disease lesions as shown here, additional insight can be obtained by profiling the miRNAs induced by a pathogen in an isolated cell type 18. It should be possible to learn whether the ability of a pathogen to induce a single or set of miRNA species that targets and inhibits host immune responses provides a potential virulence mechanism that contributes to the pathogenesis of infectious disease 19.
Our data demonstrate that a single miRNA species, by both directly and indirectly regulating immune modulatory genes, can affect the downstream effectors of an innate immune triggered antimicrobial pathway. Specifically, hsa-mir-21 inhibited TLR-induced CYP27B1 and IL1B gene expression, as well as enhanced IL-10 expression, thereby preventing upregulation of the CAMP and DEFB4 mRNAs which encode antimicrobial peptides. These factors are all key to the outcome of the vitamin D antimicrobial pathway: i) CYP27b1 converts inactive to active vitamin D leading to antimicrobial activity, ii) IL-1β is required for DEFB4 induction, and iii) IL-10 is known to inhibit TLR-induced responses 20. Consequently, hsa-mir-21 inhibits the innate immune response by its distinct gene regulatory activities: the indirect upregulation of an immunosuppressive cytokine and direct targeting of epigenetic components required for the TLR-induced, vitamin D dependent antimicrobial pathway 5, 6. Consistent with this model, the genes directly targeted by hsa-mir-21, CYP27B1 and IL1B are downregulated in L-lep vs. T-lep lesions 2, 4. Although the expression of microRNAs and gene targets in disease lesions is correlative, the demonstration that hsa-mir-21 is induced in human primary monocytes 18 h after M. leprae infection and its effect on the TLR-induced antimicrobial response suggests a role in disease pathogenesis. The investigation of the effect of a single miRNA in leprosy provides a framework for analyzing the set of miRNAs that are differentially expressed at the site of disease to determine their cumulative role in regulating the host immune response, including autophagy and antimicrobial pathways.
The ability of αmir-21 to enhance the vitamin D-dependent antimicrobial pathway provides a potential therapeutic strategy to intervene in human infectious disease. In leprosy, the vitamin D antimicrobial pathway may contribute to disease outcome based upon: i) the preferential expression of antimicrobial pathway genes in the T-lep vs. L-lep form 4, ii) the correlation of VDR single nucleotide polymorphism in L-lep subjects 21; and, iii) the reported successful use of vitamin D as a therapeutic adjuvant in the treatment of leprosy 22. Potentially, the combination of vitamin D supplementation with miRNA targeted therapy could provide an optimal treatment approach to leprosy and other chronic infectious diseases in which the cellular immune response is dysregulated. This type of approach may be particularly worth exploring in the clinical setting of drug resistant pathogens including MDR, XDR and TDR (totally drug resistant) tuberculosis, in which antimicrobial therapy is becoming increasingly problematic. Finally, our findings may be relevant to other diseases including infectious 23, 24, autoimmune 25 and neoplastic 26, 27, in which vitamin D sufficiency has been shown to be required for optimal host immunity.
METHODS
Statistical analysis
Percent change due to miRNA or antagomir were analyzed against no change using an unpaired Student’s t-test. Gene or miRNA induction studies were analyzed using an unpaired Student’s t-test against the medium control of each experiment. L-lep specific miRNA targeting of Th1- and Th2- related genes was analyzed using an unpaired Student’s t-test. The miRNA targeting preference was determined using the Kolmogorov-Smirnov (KS)-based permutation analysis as noted in the Supplemental Text. Error bars represent the standard error between individual donor values.
Leprosy biopsy specimens
The acquisition of all specimens was approved by the committees on investigations involving human subjects of the University of California, Los Angeles, more details can be found in the Supplemental Methods. Scalpel or punch skin biopsy specimens were obtained after informed consent from patients with tuberculoid leprosy and patients with lepromatous leprosy at the time of diagnosis; therefore, all samples are representative of untreated disease.
Microarray analysis
For gene and miRNA expression profiling, the RNA from skin biopsy specimens were processed and analyzed by the UCLA Clinical Microarray Core Facility using the Affymetrix U133 Genechip and Asuragen using the Discovarray platform, respectively. Additional details pertaining hierarchical clustering, cluster dendrograms, and heatmaps are included in the Supplemental Methods.
In situ hybridization
Leprosy skin biopsy specimens were snap frozen and sectioned to a thickness of 10 μm, and then mounted onto a glass slide. The protocol has been previously described 28 and adapted for current use. Briefly, biotinylated hsa-mir-21 specific, U6 and non-specific control probes were purchased (Exiqon) and were hybridized to the tissue at 0.1 pg μl−1 for 1–4 h followed by incubation with streptavidin-horseradish peroxidase (SA-HRP). Then, the sections were incubated with the TSA™ Plus Fluorescein System (Perkin Elmer) according to the manufacturer’s instructions. A coverslip was sealed to the slides with ProLong® Gold with DAPI (Invitrogen), left to dry at 4°C in the dark overnight, and imaged using a Leica FLIM confocal microscope (Leica).
Live M. leprae
Live and viable M. leprae bacteria were generously donated by Dr. James L. Krahenbuhl of US National Hansen’s Disease Programs, Health Resources Service Administration, Baton Rouge, LA. Additional information is included in the Supplemental Methods.
Quantitative PCR
For miRNA analysis, qPCR was performed using the TaqMan® MicroRNA Cells-to-CT kit in conjunction with the TaqMan® MicroRNA Assay for hsa-mir-21 (Applied Biosystems) or the NCODE miRNA cDNA Synthesis and qPCR Kit (Invitrogen) according to the manufacturers’ recommended conditions. For mRNA studies, total RNA was isolated from monocytes by TRIzol® (Invitrogen), and cDNA libraries were made using the iScript cDNA synthesis kit (BioRAD). qPCR reactions were carried out using the iQ SYBR Green qPCR Master Mix (BioRAD) according to the manufacturers recommended conditions. The primer sequences for h36B4, CAMP, DEFB4, and CYP27B1 were previously published 5, 6, other primer sequences and calculations are included in the Supplemental Methods.
Transfection of monocytes
Monocytes were enriched from PBMCs using a Percoll (GE Healthcare) gradient as previously described 6, and then transfected with either the mature miRNA the antagomir oligos using the Amaxa Nucleofector system with the Human Monocyte Nucleofector transfection kit (Lonza) according to the manufacturer’s recommended protocol. Additional details are included in the Supplemental Methods.
miRNA Direct Targeting Analysis
MiRNA targeting plasmids were prepared with endotoxin free conditions using the Qiagen Endofree Maxi Kit (Qiagen) according to the manufacturer’s recommended protocols. The constructs were co-transfected into HEK-293 cells with either hsa-mir-21 mature oligo or a non-targeting control oligo using the Amaxa Nucleofector Transfection Cell Line V kit (Lonza) according to the manufacturer’s optimized protocol. Following transfection the cells are rested for 2 h and then washed to replace the medium. The transfected cells are then incubated 37°C for 16 h and then luciferase activity was measured using the Dual Glo-Luciferase Assay System (Promega) according to the manufacturer’s recommended protocols. The miRNA effect is calculated as a ratio of the fire fly to renilla luciferase activities.
Antimicrobial assays
In order to assess M. leprae and M. tuberculosis H37Ra viability from infected macrophages, we adapted the previously described real time PCR based method for the assessment of bacterial viability, which compares 16S RNA levels to a genomic DNA levels as a predictor of bacterial viability (Supplemental Text) 10. Experimental details are included in the Supplemental Methods. The 16S and bacterial DNA values were calculated using the ΔΔCT analysis, with the bacterial DNA value serving as the housekeeping gene. The M. leprae 16S and M. leprae repetitive genomic element (RLEP) primers used were as previously described 10, other primer sequences are included in Supplementary Methods.
Other methods
Other methods, including reagent information, can be found in the Supplemental Methods.
Supplementary Material
Acknowledgments
We would like to thank G. Cheng, R. O’Connell, J. Krahenbuhl, R. Lahiri and B. Bloom for their helpful discussions. The live M. leprae is provided by the US National Hansen’s Disease Programs through the generous support of the American Leprosy Missions and Society of St. Lazarus of Jerusalem. This work was supported by NIH grants AI 022553, AI 047868 and AI 073539. PL is supported by NIH K22 Career Development Award AI 85025. KE is supported by a postdoctoral grant from the Wenner-Gren Foundations (Sweden). We would like to thank M. Schibler and the UCLA CNSI Advanced Light Microscopy Core Facility for their assistance with the confocal studies.
AUTHOR CONTRIBUTIONS
PTL performed the experiments, supervised the project, analyzed the data, and wrote the manuscript, MW performed the in situ hybrization experiments, and a portion of the M. leprae infection, antimicrobial and monocyte transfection experiments, RT performed the microarray experiments, EK performed the bioinformatics analysis of the microarray data, KE performed the IL-10 related experiments, BF performed a portion of the M. leprae infection, antimicrobial and monocyte transfection experiments, MDM performed a portion of the M. leprae infection and ligands experiments, AV performed a portion of the monocyte transfection experiments, THR diagnosed leprosy patients and collected skin biopsy specimens, ENS diagnosed leprosy patients and collected skin biopsy specimens, TGG designed, supervised and performed bioinformatics analysis, and RLM supervised the project and wrote the manuscript.
ACCESSION NUMBERS
| Gene | NCBI Accession |
|---|---|
| CAMP | NM_004345 |
| DEFB4A | NM_004942 |
| CYP27B1 | NM_000785 |
| IL1B | NM_000576 |
| IL6 | NM_000600 |
| IL10 | NM_000572 |
| IL12A | NM_000882 |
| IL12B | NM_002187 |
| 36B4 | NM_001002 |
| miRNA | miRBase Accession |
|---|---|
| hsa-mir-21 | MI0000077 |
| hsa-let-7c | MI0000064 |
| mRNA Array (GEO Title) | GEO Accession |
|---|---|
| T-lep1 (BT1) | GSM443590 |
| T-lep2 (BT6) | GSM443591 |
| T-lep3 (BT10) | GSM443678 |
| T-lep4 (BT3) | GSM443592 |
| T-lep5 (BT4) | GSM443622 |
| T-lep6 (BT7) | GSM443625 |
| L-lep1 (LL1) | GSM443583 |
| L-lep2 (LL4) | GSM443586 |
| L-lep4 (LL3) | GSM443585 |
| L-lep5 (LL7) | GSM443588 |
| L-lep6 (LL9) | GSM443589 |
Footnotes
The authors have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
References
- 1.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr. 1966;34:255–273. [PubMed] [Google Scholar]
- 2.Yamamura M, et al. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science. 1991;254:277–279. doi: 10.1126/science.254.5029.277. [DOI] [PubMed] [Google Scholar]
- 3.Salgame P, et al. Differing lymphokine profiles of functional subsets of human CD4 and CD8 T cell clones. Science. 1991;254:279–282. doi: 10.1126/science.254.5029.279. [DOI] [PubMed] [Google Scholar]
- 4.Montoya D, et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe. 2009;6:343–353. doi: 10.1016/j.chom.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu PT, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 6.Liu PT, et al. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS ONE. 2009;4:e5810. doi: 10.1371/journal.pone.0005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheedy FJ, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 9.Adams LB, et al. The study of Mycobacterium leprae infection in interferon-gamma gene--disrupted mice as a model to explore the immunopathologic spectrum of leprosy. J Infect Dis. 2002;185(Suppl 1):S1–8. S1–S8. doi: 10.1086/338002. [DOI] [PubMed] [Google Scholar]
- 10.Martinez AN, et al. Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J Clin Microbiol. 2009;47:2124–2130. doi: 10.1128/JCM.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen GA. Undwersogelser angaende spedalskhedens arsager. Norsk Mag Laegevid. 1874;4:1–88. [Google Scholar]
- 12.Pena MT, et al. Expression and characterization of recombinant interferon gamma (IFN-gamma) from the nine-banded armadillo (Dasypus novemcinctus) and its effect on Mycobacterium leprae-infected macrophages. Cytokine. 2008;43:124–131. doi: 10.1016/j.cyto.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vachula M, Holzer TJ, Andersen BR. Suppression of monocyte oxidative response by phenolic glycolipid I of Mycobacterium leprae. J Immunol. 1989;142:1696–1701. [PubMed] [Google Scholar]
- 14.Neill MA, Klebanoff SJ. The effect of phenolic glycolipid-1 from Mycobacterium leprae on the antimicrobial activity of human macrophages. J Exp Med. 1988;167:30–42. doi: 10.1084/jem.167.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabouret G, et al. Mycobacterium leprae phenolglycolipid-1 expressed by engineered M. bovis BCG modulates early interaction with human phagocytes. PLoS Pathog. 2010;6:e1001159. doi: 10.1371/journal.ppat.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark-Curtiss JE, Walsh GP. Conservation of genomic sequences among isolates of Mycobacterium leprae. J Bacteriol. 1989;171:4844–4851. doi: 10.1128/jb.171.9.4844-4851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cremer TJ, et al. MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS ONE. 2009;4:e8508. doi: 10.1371/journal.pone.0008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, et al. Up-regulated microRNA-146a negatively modulate Helicobacter pylori-induced inflammatory response in human gastric epithelial cells. Microbes Infect. 2010;12:854–863. doi: 10.1016/j.micinf.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Sinsimer D, et al. Mycobacterium leprae actively modulates the cytokine response in naive human monocytes. Infect Immun. 2010;78:293–300. doi: 10.1128/IAI.00816-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krutzik SR, et al. Activation and regulation of Toll-like receptors 2 and 1 in human leprosy. Nat Med. 2003;9:525–532. doi: 10.1038/nm864. [DOI] [PubMed] [Google Scholar]
- 21.Roy S, et al. Association of vitamin D receptor genotype with leprosy type. J Infect Dis. 1999;179:187–191. doi: 10.1086/314536. [DOI] [PubMed] [Google Scholar]
- 22.HERRERA G. Vitamin D in massive doses as an adjuvant to the sulfones in the treatment of tuberculoid leprosy. Int J Lepr. 1949;17:35–42. [PubMed] [Google Scholar]
- 23.Rook GAW. The role of vitamin D in tuberculosis. Am Rev Resp Dis. 1988;138:768–770. doi: 10.1164/ajrccm/138.4.768. [DOI] [PubMed] [Google Scholar]
- 24.Crowle AJ, Ross EJ, May MH. Inhibition by 1,25(OH)2-vitamin D3 of the multiplication of virulent tubercle bacilli in cultured human macrophages. Infect Immun. 1987;55:2945–2950. doi: 10.1128/iai.55.12.2945-2950.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munger KL, Levin LI, Hollis BW, Howard NS, Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296:2832–2838. doi: 10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- 26.Lappe JM, Travers-Gustafson D, Davies KM, Recker RR, Heaney RP. Vitamin D and calcium supplementation reduces cancer risk: results of a randomized trial. Am J Clin Nutr. 2007;85:1586–1591. doi: 10.1093/ajcn/85.6.1586. [DOI] [PubMed] [Google Scholar]
- 27.Ahn J, et al. Vitamin D-related genes, serum vitamin D concentrations and prostate cancer risk. Carcinogenesis. 2009;30:769–776. doi: 10.1093/carcin/bgp055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silahtaroglu AN, et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.