Abstract
Although DNA methylation is one of the critical ways for silencing tumor suppressor and DNA repair genes during tumor initiation and progression, the mechanisms underlying DNA methylation in cancer remain unclear. Here we show that prostaglandin E2 (PGE2) silences certain tumor suppressor and DNA repair genes via DNA methylation to promote tumor growth. These findings uncover a previously unrecognized role for PGE2 in the promotion of tumor progression.
Evidence for the link between inflammation and cancer comes from epidemiologic and clinical studies showing that use of nonsteroidal anti-inflammatory drugs (NSAIDs) reduces the relative risk for developing colorectal cancer (CRC) by 40–50%. NSAIDs exert one of their anti-inflammatory and anti-tumor effects by targeting a prostaglandin-endoperoxide synthase 2 (PTGS2). The PTGS2-PGE2 signaling plays a key role in CRC progression1,2. The observations showing a positive association between PTGER2 and CpG island methylator phenotype (CIMP) in CRC and an inverse correlation between NSAIDs use and CIMP in CRC3,4 prompted us to postulate that PGE2 may promote tumor growth by affecting DNA methylation machinery in CRC.
We first examined the correlation between the levels of PTGS2, PGE2, and DNA methyltransferases (DNMTs) in human CRC and found that the PGE2 levels and PTGS2 expression are positively correlated with DNMT1 and DNMT3B expression in CRC specimens (Supplementary Fig. 1). We found that PGE2 treatment reversed the effect of a PTGS2 inhibitor celecoxib on downregulation of DNMT1 and DNMT3B in HT-29 cells (Supplementary Fig. 2a), indicating that PGE2 regulates DNMT expression. Indeed, PGE2 directly upregulated DNMT1 and DNMT3B protein expression (Fig. 1a) but not other DNMTs (data not shown) in three human CRC cell lines.
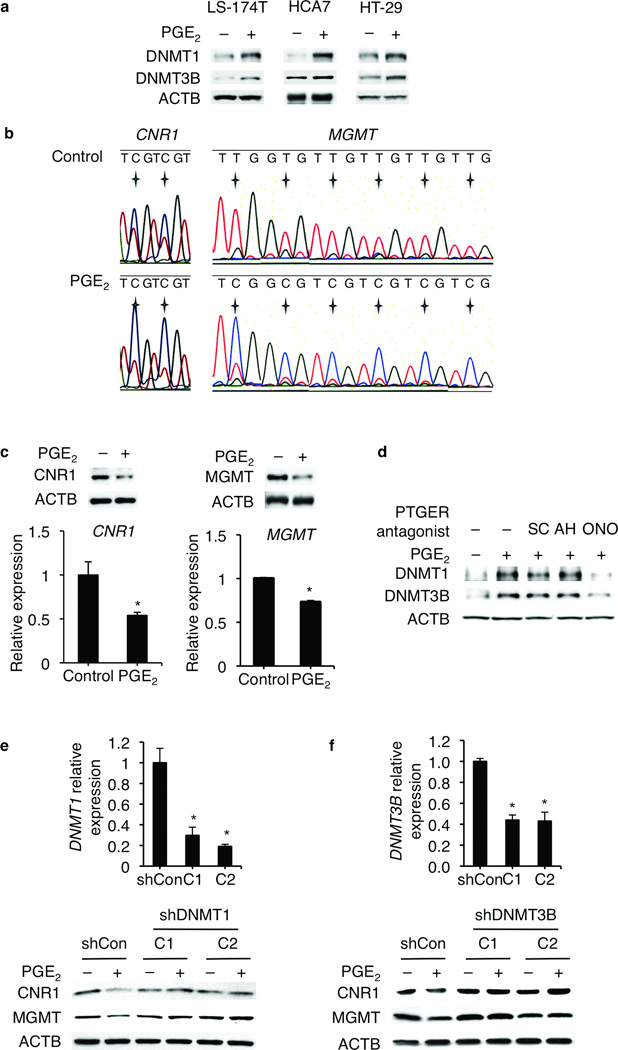
Figure 1.
PGE2 silences certain tumor suppressor and DNA repair genes by enhancing their promoter CGI methylation in human CRC cell lines. (a) PGE2 increased DNMT1 and DNMT3B protein expression in LS-174T, HCA7, and HT-29 cells. (b) Bisulfite PCR sequencing analysis showed that PGE2 increased CGI methylation in the promoters of CNR1 and MGMT in LS-174 cells. For CNR1 promoter, a region (−370 to −160) that contains 24 CpGs was examined. Two representative CpGs were presented. For MGMT promoter, a region (+27 to +342) that contains 29 CpGs was examined. Six representative CpGs were presented. The asterix indicates the locations of CpGs. (c) PGE2 downregulated the expression of CNR1 (CB1) and MGMT at both protein (upper panels) and mRNA (lower panels) levels in LS-174T cells. Error bars indicate s.d. * P < 0.05 (two-tailed unpaired Student’s t test). (d) Blockade of PTGER4 (EP4) attenuated the upregulation of DNMT1 and DNMT3B by PGE2 in LS-174T cells. SC19220 (SC): PTGER1 (EP1) antagonist; AH6809 (AH): PTGER1-3 (EP1-3) antagonist; ONOAE-208 (ONO): PTGER4 (EP4) antagonist. (e,f) Knockdown of DNMT1 or DNMT3B by shRNAs attenuated PGE2-induced downregulation of CNR1 (CB1) and MGMT in LS-174T cells. Knockdown efficiency was examined by Q-PCR in two clones (C1 and C2) along with a non-silencing shRNA transfected control (shCon) (upper panels). Error bars indicate s.d. * P < 0.05 (two-tailed unpaired Student’s t test). CNR1 (CB1) and MGMT protein expression was examined by western blotting in these two clones (C1 and C2) and control ShCon cells (lower panels).
Based on the observations that the CGI hypermethylation is detected in the promoters of certain tumor suppressor and DNA repair genes in human CRC5,6, we examined and found that PGE2 enhanced the CGI methylation in the promoters of cannabinoid receptor 1 (CNR1) and O-6-methylguanine-DNA methyltransferase (MGMT) (Fig. 1b) as well as CDKN2B and MutL homolog 1 (MLH1) genes (Supplementary Fig. 2b,c) in LS-174T cells. CNR1 is silenced by CGI methylation in human CRC and acts as tumor suppressor in vivo7. PGE2 also increased CGI methylation in the promoters of BAX, CHEK2, NOTCH1, CAV1, NHS, MYOD1, and TMEFF2 (data not shown). As expected, PGE2 downregulated the expression of CNR1 and MGMT (Fig. 1c) as well as CDKN2B and MLH1 (Supplementary Fig. 2d) at both mRNA and protein levels in LS-174T cells. Subsequently, we found that only a PTGER4 antagonist (ONOAE-208) blocked the effect of PGE2 on DNMT1 and DNMT3B expression but not a PTGER1 antagonist (SC19220) or a PTGER1-3 antagonist (AH6809) (Fig. 1d). Moreover, knockdown of DNMT1 or DNMT3B by shRNAs attenuated the PGE2-induced downregulation of CNR1, MGMT, CDKN2B, and MLH1 in LS-174T cells (Fig. 1e,f and Supplementary Fig. 2e). Collectively, these results demonstrate that PGE2 silences certain tumor suppressor and DNA repair genes by enhancing their promoter CGI methylation via a PTGER4-DNMT pathway in vitro.
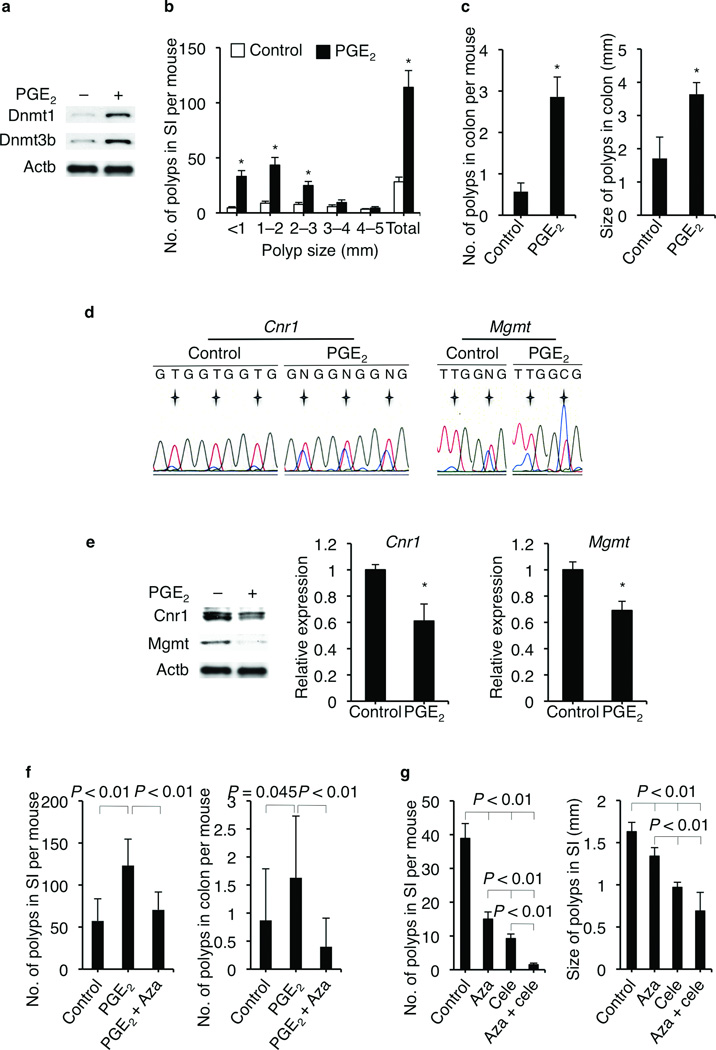
Our in vitro studies were confirmed in vivo. Treatment of ApcMin/+ mice with PGE2 increased Dnmt1 and Dnmt3b protein expression in colonic tumor epithelial cells (Fig. 2a) and accelerated intestinal adenoma growth (Fig. 2b,c). Moreover, PGE2 enhanced the CGI methylation of Cnr1 and Mgmt (Fig. 2d) as well as Cdkn2b and Mlh1 (Supplementary Fig. 3a) in the colonic tumor epithelial cells isolated from ApcMin/+ mice. As expected, PGE2 also downregulated the expression of Cnr1, Mgmt, Cdkn2b, and Mlh1 at both the mRNA and protein levels in the colonic tumor epithelial cells from ApcMin/+ mice (Fig. 2e and Supplementary Fig. 3b,c). Importantly, treatment of ApcMin/+ mice with 5-aza-2'-deoxycytidine (5-Aza-dC) reversed the effect of PGE2 on promoting adenoma growth (Fig. 2f) and inducing the CGI methylation of Cdkn2b (Supplementary Fig. 4a), demonstrating that PGE2 accelerates intestinal adenoma growth via regulating CGI methylation. Intriguingly, combined treatment with both celecoxib and 5-Aza-dC more effectively reduced the tumor burden in ApcMin/+ mice than either agent alone (Fig. 2g and Supplementary Fig. 4b). Furthermore, treatment of ApcMin/+ mice with PGE2 reversed the effects of celecoxib on inhibiting small intestinal adenoma growth (Supplementary Fig. 4c), demonstrating that the tumor inhibitory effect of celecoxib depends on PGE2. Collectively, these results suggest that PGE2 promotes intestinal tumor growth by silencing tumor suppressor and DNA repair genes via its effects on CGI methylation.
Figure 2.
PGE2 promotes intestinal tumor growth via upregulating CGI methylation in ApcMin/+ mice. (a) Treatment of ApcMin/+ mice with PGE2 increased Dnmt1 and Dnmt3b protein expression in the colonic tumor epithelial cells. (b,c) PGE2 increased intestinal polyp number and size in ApcMin/+ mice. Error bars indicate s.e.m (n = 7 for each group). * P <0.05 (Wilcoxon Rank Sum test). No.: number; SI: small intestine. (d) Treatment of three ApcMin/+ mice with PGE2 increased the promoter CGI methylation of Cnr1 and Mgmt in the colonic tumor epithelial cells as compared to three ApcMin/+ mice treated with vehicle. For Cnr1 promoter, a region (−369 to −34) that contains 35 CpGs was examined. Three representative CpGs were presented. For Mgmt promoter, a region (−458 to −243) that contains 6 CpGs was examined. Two representative CpGs were presented. Asterix indicates the locations of CpGs. (e) Treatment of ApcMin/+ mice with PGE2 decreased the expression of Cnr1 and Mgmt at both protein levels (left panel) and mRNA levels (middle and right panels) in the colonic tumor epithelial cells. One representative result from three mice was shown. Error bars indicate s.d. * P < 0.05 (two-tailed unpaired Student’s t test). (f) Inhibition of CGI methylation by 5-Aza-dC attenuates PGE2-induced tumor growth in male ApcMin/+ mice. Error bars indicate s.e.m. (n = 15 for each group. Wilcoxon Rank Sum test). (g) Combination treatment with celecoxib and 5-Aza-dC more efficiently inhibited tumor growth. Error bars indicate s.e.m. (n = 12, 14, 14, and 7, respectively. Wilcoxon Rank Sum test).
Our in vitro and in vivo results are of potential clinical relevance because the levels of PGE2, PTGS2, DNMT1, and DNMT3B are positively associated with CGI methylation in the CNR1, MGMT, and MLH1 promoters in human CRC specimens, respectively (Supplementary Fig. 5a). The correlation of these genes to the CGI methylation of each individual gene was also significant except for MLH1 (Supplementary Table 1). The correlation of MLH1 to PGE2 and DNMT1 didn’t reach significance although we observed positive trends. Moreover, the expression levels of CNR1, MGMT, and MLH1 are negatively correlated with their respective levels of CGI methylation (Supplementary Fig. 5b).
Dysregulation of DNMT expression is associated with human cancer progression8,9. Particularly, DNMT3B expression is positively associated with CIMP in CRC10 and colorectal adenomas11, while overexpression of DNMT1 is also associated with CIMP in CRC12. One study revealed similar profiles of DNMT3B-methylated genes between mouse colon and human CRC13. In ApcMin/+ mice, modulation of Dnmt3b expression affected colon adenoma growth14,15. Consistent with these findings, our results provide the first evidence that PGE2 promotes intestinal adenoma growth by silencing certain tumor suppressor and DNA repair genes via induction of DNMT1 and DNMT3B. Although Dnmt3b was not responsible for CGI methylation at the Mgmt locus in ApcMin/+ mice15, Dnmt1 may mediate PGE2-enhanced CGI methylation in the Mgmt promoter in our studies. Our data further showed that inhibition of CGI methylation by 5-Aza-dC dramatically suppressed PGE2-induced intestinal adenoma growth in ApcMin/+ mice, suggesting demethylating agents could serve as anti-tumor agents. However, these agents have been reported to activate oncogenes and promote cancer cell proliferation, migration, and invasion in some in vitro studies16–18. Moreover, global and locus-specific hypomethylation are associated with a poor prognosis of some CRC patients19,20. Therefore, further investigation is needed before considering demethylating agents as a possible treatment for CRC.
In summary, our findings not only significantly improve our understanding of the intricate roles of PGE2 in cancer progression and of how DNA methylation machinery is regulated in cancer but also provide a rationale for considering the development of a novel combination treatment employing PTGS2 inhibitors and demethylating agents for prevention and possibly future therapy in the appropriate subsets of patients.
Supplementary Material
Acknowledgments
We want to thank D. Menter and P. Yang for their suggestions and their help in PGE2 measurement. This work is supported, in part, by the NIH MERIT award R37 DK47297, RO1 DK62112, NCI P01 CA77839, and CPRIT RP100960. We also thank the National Colorectal Cancer Research Alliance (NCCRA) for its generous support (RND) and a cancer prevention fellowship (DX) supported by the NCI grant R25T CA57730 PI: Shine Chang, Ph.D.
Footnotes
Author Contributions
R.N.D., D.W., and D.X. designed this research project; D.X. performed most of the experiments; S.H.K. contributed to establish the DNMT1 and DNMT3B knockdown stable cell lines and H.K. conducted the DNA methylation analysis for human tissues samples; D.X., and D.W. conducted the data analyses; D.W. wrote the manuscript with D.X.’s help; and R.N.D. supervised the project.
Competing financial interests
All authors do not have any competing financial interests.
References
- 1.Wang D, DuBois RN. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D, DuBois RN. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba Y, et al. Cancer Epidemiol Biomarkers Prev. 19:822–831. doi: 10.1158/1055-9965.EPI-09-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slattery ML, et al. Int J Cancer. 2007;120:656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 5.Kim MS, Lee J, Sidransky D. Cancer Metastasis Rev. 29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 6.Kim YH, et al. Genes Chromosomes Cancer. 2006;45:781–789. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, et al. Cancer Res. 2008;68:6468–6476. doi: 10.1158/0008-5472.CAN-08-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Marzo AM, et al. Cancer Res. 1999;59:3855–3860. [PubMed] [Google Scholar]
- 9.Schmidt WM, et al. Mol Carcinog. 2007;46:766–772. doi: 10.1002/mc.20307. [DOI] [PubMed] [Google Scholar]
- 10.Nosho K, et al. Clin Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim AE, et al. Gut. 2011;60:499–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 12.Kanai Y, Ushijima S, Kondo Y, Nakanishi Y, Hirohashi S. Int J Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Steine EJ, et al. J Clin Invest. 2011;121:1748–1752. doi: 10.1172/JCI43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin H, et al. Mol Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linhart HG, et al. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shteper PJ, et al. Oncogene. 2003;22:7737–7749. doi: 10.1038/sj.onc.1207056. [DOI] [PubMed] [Google Scholar]
- 17.Pakneshan P, Szyf M, Farias-Eisner R, Rabbani SA. J Biol Chem. 2004;279:31735–31744. doi: 10.1074/jbc.M401669200. [DOI] [PubMed] [Google Scholar]
- 18.Hamm CA, et al. PLoS One. 2009;4:e8340. doi: 10.1371/journal.pone.0008340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogino S et al. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn JB, et al. Cancer. 2010;117:1847–1854. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.