Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an immature hematopoietic malignancy driven mainly by oncogenic activation of NOTCH1 signaling1. In this study we report the presence of loss-of-function mutations and deletions of EZH2 and SUZ12 genes, encoding critical components of the Polycomb Repressive Complex 2 (PRC2) complex2,3, in 25% of T-ALLs. To further study the role of the PRC2 complex in T-ALL, we used NOTCH1-induced animal models of the disease, as well as human T-ALL samples, and combined locus-specific and global analysis of NOTCH1-driven epigenetic changes. These studies demonstrated that activation of NOTCH1 specifically induces loss of the repressive mark lysine-27 tri-methylation of histone 3 (H3K27me3)4 by antagonizing the activity of the Polycomb Repressive Complex 2 (PRC2) complex. These studies demonstrate a tumor suppressor role for the PRC2 complex in human leukemia and suggest a hitherto unrecognized dynamic interplay between oncogenic NOTCH1 and PRC2 function for the regulation of gene expression and cell transformation.
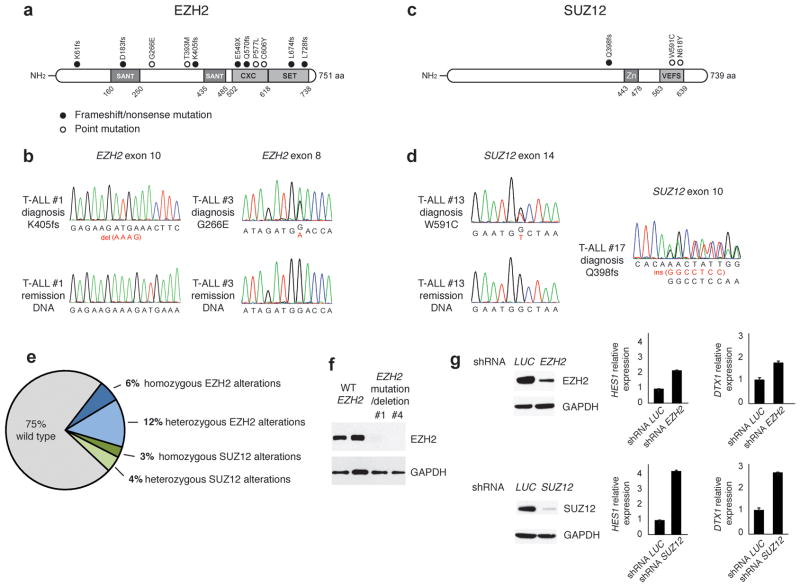
T-ALL is a hematologic malignancy5,6,7.characterized by activating mutations in the NOTCH18 gene and alterations in the FBXW79 ligase resulting in activation of Notch signaling. Although the importance of NOTCH activation in T-ALL is well established, the detailed molecular mechanisms mediating NOTCH1-induced transformation remain unknown. We hypothesized that NOTCH1 interacts with epigenetic modulators to control gene expression. In addition, we proposed that genetic alterations in key components of the epigenetic machinery could amplify oncogenic signals. To test this notion, we analyzed an extensive series of array comparative genomic hybridization (aCGH) data on adult T-ALL primary samples for the presence of recurrent deletions encompassing genes involved in epigenetic regulation. This analysis revealed the presence of recurrent deletions involving genes encoding core components of the Polycomb Repressive Complex 2 (PRC2). This complex is the “writer” of a major repressive chromatin modification, Lysine 27 trimethylation on Histone 3 (H3K27me3). We found recurrent deletions encompassing the EZH210–12 and SUZ1213,14 loci (Supplementary Fig. 1 and Supplementary Table 1). Following these results we screened primary tumor DNA samples for the presence of somatic mutations affecting the core components of the PRC2 complex15. This analysis revealed the presence of truncating or missense mutations in both EZH2 (11/68) and SUZ12 (3/68). EZH2 mutations included four non-synonymous single-nucleotide substitutions, one nonsense mutation and six frameshift-creating insertions and deletions (Fig. 1a,b, Supplementary Fig. 1 and Supplementary Table 1). SUZ12 mutations identified in T-ALL included 2 missense and 1 frameshift mutation (Fig. 1c,d). Loss of function mutations and deletions in EZH2 have been previously associated with myeloid leukemias10–12. In contrast, gain of function EZH2 mutations involved in B-cell lymphomas are typically single amino acid substitutions involving Y64116,17. Nonsense and frameshift mutations in EZH2 and SUZ12 in T-ALL are protototypical loss of function truncating alleles consistent with a PRC2 tumor suppressor role for these genes in T-cell transformation. Notably, 7 EZH2 and 3 SUZ12 mutations were heterozygous but also 4 out of 11 EZH2 and 1 out 3 SUZ12 mutations were homozygous18. In all 8/14 cases (6 EZH2 and 2 SUZ12 variants) with available matched bone marrow remission genomic DNA we confirmed the somatic origin of the EZH2 and SUZ12 mutations (Fig. 1a,c and Supplementary Table 1). The convergent findings of our re-sequencing effort and copy number analysis thus identified EZH2 and SUZ12 as novel tumor suppressor genes mutated and deleted in T-ALL. Overall, genetic lesions targeting EZH2 or SUZ12 were identified in 17/68 (25%) of primary T-ALL samples (Fig. 1e). The complete absence of EZH2 protein in both cases with combined deletion and mutation of the EZH2 gene examined (Fig. 1f) revealed that these loss of function mutations and suggested that inactivation of the PRC2 complex may constitute an important pathogenetic event in human T-ALL. Further targeted re-sequencing revealed that PRC2 genetic alterations were frequently (in 65% of the cases) associated with oncogenic NOTCH1 mutations (Supplementary Table 1). This frequency suggested that the two events could directly or indirectly co-operate. We analyzed the effects of PRC2 inactivation in the expression of prototypical NOTCH1 target genes such as HES1 and DTX1 in T-ALL cell lines harboring NOTCH1 mutations9,19. These experiments showed that silencing of both EZH2 and SUZ12 resulted in transcriptional upregulation of both target genes (Fig. 1g, Supplementary Fig. 2 and not shown), suggesting that loss of PRC2 could potentiate the NOTCH1 transcriptional program.
Figure 1.
The PRC2 complex as a tumor suppressor in T-ALL. (a) Structure of the EZH2 protein including 2 SANT DNA binding domains, the cysteine-rich CXC domain and the catalytic SET domain. Overview of all EZH2 mutations identified in primary T-ALL samples. Filled circles: nonsense and frameshift mutations, open circles: missense mutations. (b) Representative chromatograms of paired diagnosis and remission genomic DNA samples showing somatic mutations in exon 8 and exon 10 of EZH2. (c) Structure of the SUZ12 protein including a zinc finger domain and the VEFS domain. (d) Representative DNA sequencing chromatograms showing a frame-shift mutation in exon 10 of SUZ12 and of paired diagnosis and remission genomic DNA samples showing a somatic point mutation in exon 14 of this gene. (e) Pie-chart summarizing the frequencies of homozygous and heterozygous mutations of EZH2 and SUZ12 in adult TALL patients. (f) EZH2 protein levels in samples from patients (#1 and #4) with mutations and deletions on the EZH2 gene compared to WT controls. (g) Silencing of EZH2 and SUZ12 in the Jurkat human T-ALL line. HES1 and DTX1 mRNA expression levels followed silencing of either SUZ12 or EZH2. Knockdown of the luciferase (LUC) gene was used as a control.
To further explore the role of the PRC2 complex in Notch target expression and T-ALL induction/progression we aimed to dissect the epigenetic changes associated with transformation in T-ALL. Chromatin ImmunoPrecipitation (ChIP) studies using CUTLL1 cells15, a human T-ALL line20 characterized by a Notch1 translocation showed that NOTCH1 binding on the promoter of HES1, a canonical NOTCH1 target required for NOTCH1-induced transformation5,21 (Supplementary Fig. 3 and Supplementary Table 2), peaks at −50 to −100 bp relative to the Transcriptional Start Site (TSS) followed by enrichment of RNA Polymerase II (POL II) (Supplementary Fig. 3). No binding for NOTCH1 or POL II was observed in a NOTCH1-negative T-ALL cell line (Supplementary Fig. 3b,c). Inhibition of the Notch1 signaling using a γ-secretase inhibitor (γSI)20 (Supplementary Fig. 4a) abrogated NOTCH1 binding on the HES1 promoter and led to decreased levels of HES1 mRNA expression (Supplementary Fig. 4b,c). Subsequent γSI removal restored high levels of NOTCH1, POL II and the activating mark acetylation of Lysine 9 of Histone 3 (H3K9ac) on the HES1 promoter as well as HES1 expression (Supplementary Fig. 4b–e).
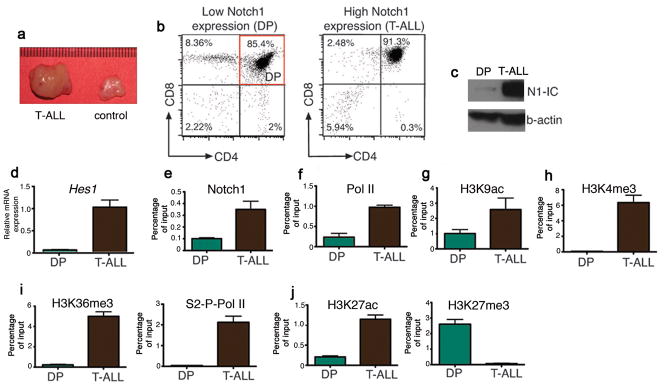
To further test the interplay between activation of NOTCH1 and epigenetic regulation we used a Notch1-IC-induced T-ALL animal model22, which recapitulates most of the features of human T-ALL (Fig. 2a and Supplementary Fig. 5a-c). Most Notch1-induced leukemias show a double positive (DP) phenotype characterized by the expression of both CD4 and CD8 co-receptors (Fig. 2b). To study the transcriptional and epigenetic changes in Hes1 during Notch1 driven leukemogenesis we compared FACS-sorted DP Notch1-transformed cells (T-ALL) to normal DP thymocytes, which show low levels of Notch1 and Hes1 activation (Fig. 2b,c). Scanning of the murine Hes1 promoter revealed significant enrichment for Notch1 binding in T-ALL compared to DP (Fig. 2e) accompanied by enrichment of PolII on the TSS (Fig. 2f). Moreover, ChIP experiments showed enrichment of the activating acetylation of lysine 9 on H3 (H3K9ac)23 and trimethylation of lysine 4 on H3 (H3K4me3)24 (Fig. 2g,h) around the Hes1 TSS, followed by enrichment for the transcriptional elongation-associated form of Pol II (Serine 2-Phosphorylated Pol II, S2-P-Pol II) and trimethylation of lysine 36 on histone 3 (H3K36me3) on the gene body (Fig. 2i). One of the most prominent differences, however, was the loss of the repressive H3K27me3 from the Hes1 promoter in the leukemic cells that was accompanied by gain of acetylation of the same residue (Fig. 2j), an activating epigenetic mark25. Similar results were obtained for Deltex1 (Dtx1), which is also a direct Notch1 target gene (Supplementary Fig. 6), but not on the Gapdh locus, used as a control. These results demonstrated that Notch1-mediated oncogenic transformation was coupled to epigenetic changes, including the loss of the H3K27me3 histone mark from Notch target gene promoters.
Figure 2.
Notch1-induced epigenetic changes in an in-vivo model of T-ALL. (a) Comparison of the size of the thymus in leukemic animals and control littermates. (b) Cell populations used in this study. (c) Western blot showing the expression of N1-IC in DP and T-ALL cells. (d) Hes1 cDNA levels between DP and T-ALL. (e, f) ChIP of Notch1 and Pol II on the Hes1 promoter in the indicated cell populations. (g-j) ChIP of the indicated histone marks and S2-P-Pol II in DP and T-ALL cells.
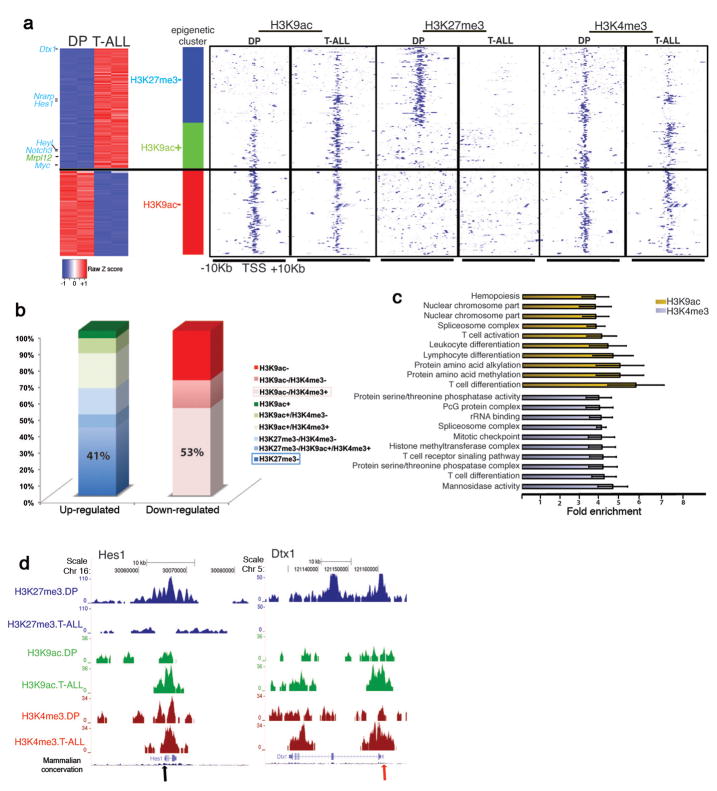
To test whether the Hes1 behavior is part of a wider Notch1 driven epigenetic reprogramming in leukemic cells, we performed whole transcriptome profiling (Supplementary Tables 3,4) and ChIP-Sequencing (ChIP-Seq, Supplementary Table 5) for H3K4me3, H3K9ac and H3K27me3 (Fig. 3a,b and Supplementary Fig. 7). Computational validation of Chip-Seq results showed high correlation of the biological replicates (Supplementary Fig. 7a) and consistency with gene expression (Supplementary Fig. 8,9). Enrichment analyses (Supplementary Tables 6–9) revealed that the regulated (and marked by modifications) genes belong in functional categories related to normal T cell differentiation and T cell transformation (Fig. 3c and Supplementary Fig. 10). Genes up-regulated in Notch1-driven leukemic cells compared to normal DP thymocytes were primarily characterized by loss of H3K27me3 (P= 6.91x10−22). Unexpectedly, gain of H3K9ac in these genes was much less significant (P=1.52x10−4) (Fig. 3a,b and Supplementary Tables 10,11), suggesting that loss of H3K27me3 is the most prominent epigenetic change coupled to gene activation. This in turn, provided evidence for a central role of PRC2 in T-ALL. On the contrary, down-regulated genes showed primarily loss of H3K9ac (P=2.79x10−19, Supplementary Tables 11,12). Most notably, the loss of H3K27me3 from the TSS region of T-ALL up-regulated transcripts was not due to lower total levels of the H3K27me3 in T-ALL (Supplementary Fig. 11 and Supplementary Table 5). Changes in H3K4me3 seemed to have a lesser role related to context-dependent and fine-tuning regulation of gene expression (Fig. 3a,b and Supplementary Table 11). Targets of Notch1, such as Hes1, Dtx1, Pre-TCR alpha (Ptcra) and c-Myc, exhibited loss of H3K27me3, whereas the changes of H3K4me3 and H3K9ac were more subtle (Fig. 3d and Supplementary Fig. 12), suggesting a functional interaction between Notch1 and loss of H3K27me3, generally associated with decreased activity of the PRC2 complex.
Figure 3.
Characterization of T-ALL epigenetic landscape using ChIP-Seq for H3K9ac, H3K4me3 and H3K27me3. (a) Cluster of the major gene expression changes between T-ALL and DP and the accompanied epigenetic changes. Left part: Expression heatmap representing up (red)- and down (blue)-regulated genes with significant epigenetic changes. Right panel: Heatmap representation of the epigenetic marks in T-ALL and DP in TSSs of selected genes. “+”:gain and “−”:loss in the levels of epigenetic mark in T-ALL versus DP. Loss of H3K27me3 (P=6.91x10−22, blue bar) and gain of H3K9ac (P=1.52x10−4, green bar) are enriched in up-regulated genes, whereas loss of H3K9ac (P=2.79x10−19, red bar) is enriched in down-regulated genes. (b) Bar graphs indicate the percentage of genes characterized by each modification in T-ALL cells. The plus and minus signs are used as above. Pink and blue frames: prevalent epigenetic clusters in down-regulated and up-regulated genes, respectively. (c) Functional annotation of epigenetic changes (T-ALL vs DP) in H3K9ac (yellow bars) and H3K4me3 (gray bars) shows enrichment in specific biological processes. (d) ChIP-Seq results for two well-characterized Notch1 targets, Hes1 and Dtx1 (arrows denote the TSS).
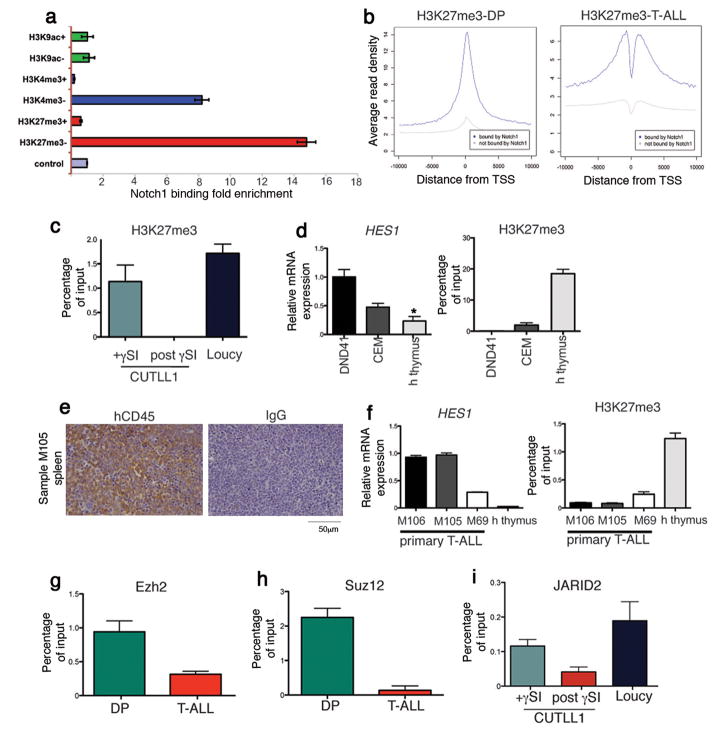
To further explore the role of Notch1 in driving the loss of H3K27me3 we performed ChIP-Seq for Notch1 (Fig. 4a and Supplementary Fig. 13). Whereas no significant peaks were detected in DP, analysis of Notch1-transformed T-ALL lymphoblasts revealed a large number of direct Notch1 binding events. Importantly, H3K27me3 loss in T-ALL was broadly overlapping with direct Notch1 binding in TSS regions (Fig. 4a,b). The lack of enrichment of H3K9ac gain or loss suggested that Notch1 binding is highly specific to H3K27me3 loss (Fig. 4a). The observed loss of H3K27me3 in Notch1 targets is mainly localized in a narrow region around TSSs (Fig. 4b). Loss of H3K27me3 was observed specifically on Notch1 targets and not in the whole T-ALL genome (Fig. 4a and Supplementary Figs 14 and 15). These combined data suggested that significant loss of H3K27me3 is a hallmark of the oncogenic function of Notch1 in T-ALL.
Figure 4.
Notch1 binding mediates loss of H3K27me3 and eviction of PRC2 in T-ALL. (a) Enrichment of Notch1 binding sites around TSSs characterized by each indicated histone mark. (b) H3K27me3 average signal profiles around TSS areas (blue line: Notch1-bound genes, gray line: genes not-bound by Notch1). (c) ChIP for H3K27me3 in a T-ALL cell line (CUTLL1) treated with γSI. The Loucy T-ALL line is used as a negative control. (d) HES1 expression in the indicated cell lines and normal human thymocytes and ChIP for H3K27me3 in the indicated cell lines and primary cells. (e) High leukemogenic potential of the human T-ALL samples in xenograft models. Spleen sections of recipient mice stained with an hCD45 antibody or an IgG control. (f) qPCR for HES1 expression and the levels of H3K27me3 in primary human T-ALL samples and human thymocytes (P<0.0001 between M69 and the human thymus). (g) ChIP experiments for Ezh2 on the Hes1 promoter in DP (green) and T-ALL (red). (h) Suz12 binding on the Hes1 promoter. (i)γSI-mediated changes of the N1-IC levels modulate JARID2 recruitment to the HES1 promoter (P=0.059).
The rapid increase of Notch1-IC levels in human T-ALL lines upon γSI removal (Supplementary Fig. 4) resulted in a dynamic and rapid loss of the H3K27 (Fig. 4c and Supplementary Fig. 16), further proving the inverse correlation of the two events. This led us to further investigate this relationship in additional human T-ALL cell lines and primary T-ALL samples. Initially we screened additional T-ALL lines (DND41 and CEM), exhibiting high N1-IC and HES1 expression, and normal (HES1low) human thymocytes (Fig. 4d). The levels of H3K27me3 were once more inversely correlated with HES1 expression (Fig. 4d). To exclude the possibility that these results were due to cell line artifacts, we studied primary samples whose high leukemogenic potential was evaluated using transplantation (Fig. 4e and Supplementary Fig. 17). The primary T-ALL leukemic blasts exhibited higher levels of HES1 compared to normal human thymocytes and the levels of H3K27me3 were inversely correlated with HES1 expression (Fig. 4f). These studies demonstrated that the correlation between oncogenic NOTCH1 binding and loss of H3K27me3 is a universal characteristic of T-ALL.
We then focused on the relationship between oncogenic NOTCH1 with the PRC2 complex. Initially, the analysis revealed that Notch1 binding sites are enriched for PRC2 targets (4.3-fold enrichment and P=8.45x10−110, Supplementary Table 9). Moreover, we analyzed the effects of Notch1 activation on the occupancy of Notch1 target genes by the EZH2 catalytic subunit of PRC2. These studies demonstrated that Notch1 binding led to significant Ezh2 eviction from the Hes1 promoter (Fig. 4g). This could not be attributed to lower EZH2 expression in the cancer cells (Supplementary Fig. 18a). ChIP analysis for SUZ12 binding yielded identical results (Fig. 4h). EZH2 eviction and H3K27 loss was not only a feature of the Notch1-IC model utilized, as identical results were obtained using “weaker” human Notch1HD/PEST alleles in in vivo disease models (Supplementary Fig. 19). These epigenetic effects were observed even at the very early stages of the disease (Supplementary Fig. 20). Moreover, down-regulation of N1-IC levels byγSI treatment, led to a marked decrease of EZH2 binding on the HES1 or DTX1 promoters (Supplementary Fig. 18). Also, the binding of JARID226,27, one of the recruiters of the PRC2 complex to DNA, on the HES1 promoter was also inversely correlated to Notch1 binding (Fig. 4i). These responses were rapid, as significant changes in Notch1 binding and PRC2 recruitment were detected as early as 30min upon γSI removal (Supplementary Fig. 21). The reverse correlation between Notch1-PRC2 binding and H3K27me3 levels was found in all T-ALL lines studied (Supplementary Fig. 22). Identical epigenetic changes were also noted when the Notch pathway was inhibited using a dominant negative form of MAML1 (Supplementary Fig. 23).
The presented mechanistic interaction between NOTCH1 and the PRC2 complex suggested a potential role for PRC2 mutations in NOTCH-induced transformation, although Notch-independent effects are also possible. To start addressing pathway interaction, we used a Drosophila Notch-driven tumor model28 to evaluate the impact of knockdown of E(z) in cells that express weak activating alleles of Notch (Supplementary Fig. 24). We were able to show that the combination of Notch activation and E(z) loss resulted in eye tumor overgrowth28,29 in approximately 50% of the progeny (n= 64)30. In agreement with such a notion of cooperation between Notch and PRC2 loss we were able to show that: EZH2 silencing resulted in decreased apoptosis triggered by γSI inhibition of Notch signaling in human T-ALL lines (Supplementary Fig. 25). Moreover, EZH2 silencing increased the in vivo tumorigenic potential of T-ALL cells and led to enhanced mortality in transplantation experiments (Supplementary Fig. 26). These studies suggested a striking conservation of the Notch:PRC2 pathway interaction in tumorigenesis and further established the role of the PRC2 complex as a tumor suppressor in T-ALL, although the exact mechanisms of function have to be detailed further. We believe that our studies offer new therapeutic avenues for the treatment of T cell leukemia31,32 as inhibitors of H3K27 demethylases33, alone or in combination with targeted anti-Notch1 therapies, could antagonize oncogenic Notch1 function and be further exploited for the treatment of T-ALL.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
Acknowledgments
We would like to thank Drs. R. Bonasio and D. Reinberg (HHMI and NYU School of Medicine) for critical reading of the manuscript and sharing reagents; C. Siebel (Genentech) for critical reagents; P. Ballerini, C. Deswartes, T. Leblanc and A. Baruchel (Services d’Hématologie Pédiatrique, Hôpital Saint-Louis, Hôpital Robert Debré and Hôpital Trousseau, AP-HP, Paris, France) for providing primary T-ALL patient samples. M. Gialitakis, L. Parida and G. Stolovitzky for comments on the manuscript; J. Zavadil, B. Berrin and the NYU Genome Technology Center (supported in part by NIH/NCI P30 CA016087-30 grant) for expert genomic assistance. The NYU Flow Cytometry facility (supported in part by NIH/NCI 5 P30CA16087-31) for expert cell sorting. Also, the NYU Histology Core (5P30CA16087-31), and the Transgenic Mouse Core (NYU Cancer Institute Center Grant (5P30CA16087-31). J.N. was supported by the Damon Runyon Cancer Research Foundation. I.A. was supported by the National Institutes of Health (RO1CA133379, RO1CA105129, R21CA141399, RO1CA149655, and RO1GM088847), The Leukemia & Lymphoma Society, The V Foundation, the American Cancer Society (RSG0806801) and the Dana Foundation. The Laboratory is also supported by a Feinberg Lymphoma Pilot grant. Also, his study was supported by the Fund for Scientific Research (FWO) Flanders (P.V.V. and K.D.K); the National Library of Medicine (1R01LM010140-01 to R.R.); the ECOG tumor bank; a Northeast Biodefence Center ARRA award (U54-AI057158 to R.R.); the National Institutes of Health (R01CA120196 and R01CA155743 to A.F.); the Stand Up To Cancer Innovative Research Award (A.F.), the Chemotherapy Foundation (I.A. and A.F.); the Rally Across America Foundation (A.F) and the Swim Across America Foundation (A.F.). P.V.V is an ASH Scholar and I.A. and A.F. are Leukemia & Lymphoma Society Scholars. MD is supported by grants from Spanish Ministerio de Ciencia e Innovación (BFU2009-09074 and MEC-CONSOLIDER CSD2007-00023), Generalitat Valenciana (PROMETEO2006/134) and a European Union Research Grant (UE-HEALH-F2-2008-201666). P.F is supported by the Institut du Cancer (INCA), the Association Laurette Fugain, the Ligue National Contre le Cancer, and also by INSERM, CEA, and StemPole. S.P. is supported by a fellowship by INCA. I.A. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
Accession codes. Microarray data are available in the Gene Expression Omnibus under accession code GSE34554.
Author Contributions
I.A. and P.N. conceived the studies, directed research, analyzed the results and wrote the manuscript. P.N. performed xenograft experiments, isolated and characterized mouse samples and performed and analyzed the biochemical experiments helped by T.T. and J.S. A.T. directed research, analyzed data, developed computational methods and wrote the manuscript. J.N. isolated and characterized mouse samples, helped to project design and wrote manuscript. S.B. Z.T. and T.H. helped with the analysis of the genome-wide data. P.A. helped with the design and execution of the biochemical experiments. F.U. created the resource website. P.V.V., M.H., I.R., J.B.S. and J.P. performed mutation analysis of SUZ12, EZH2 and EED. R.L. designed and supervised sequence analysis. P.V.V. performed xenograft experiments. P.V.V. and K.D.K. performed array-CGH analysis. R.R. analyzed array-CGH data. M.S.F. performed the genetic silencing studies of the PRC2 complex. E.P., J.R. and J.M.R. provided samples and correlative clinical data from ECOG. S.P. and F.P. performed and supervised experiments related to NOTCH activation into primary T-ALL samples and provided crucial materials. A.F. designed the studies, directed research and wrote the manuscript. D.F-M, V.dR., and M.D. designed and performed the Drosophila tumor experiments.
References
- 1.Aster JC, Blacklow SC, Pear WS. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J Pathol. 2011;223:262–273. doi: 10.1002/path.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 4.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 6.Paganin M, Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2010 doi: 10.1016/j.blre.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 9.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 11.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 12.Nikoloski G, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature genetics. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 13.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–2618. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 16.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morin RD, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulis ML, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wendorff AA, et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010;33:671–684. doi: 10.1016/j.immuni.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Buonamici S, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature. 2009;459:1000–1004. doi: 10.1038/nature08020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo MH, et al. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 24.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 25.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010 doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landeira D, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferres-Marco D, et al. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- 29.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 31.Lopez J, Percharde M, Coley HM, Webb A, Crook T. The context and potential of epigenetics in oncology. Br J Cancer. 2009;100:571–577. doi: 10.1038/sj.bjc.6604930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 33.Natoli G, Testa G, De Santa F. The future therapeutic potential of histone demethylases: A critical analysis. Curr Opin Drug Discov Devel. 2009;12:607–615. [PubMed] [Google Scholar]
- 34.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creyghton MP, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klinakis A, et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci U S A. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tu Y, Stolovitzky G, Klein U. Quantitative noise analysis for gene expression microarray experiments. Proc Natl Acad Sci U S A. 2002;99:14031–14036. doi: 10.1073/pnas.222164199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsirigos A, Haiminen N, Bilal E, Utro F. GenomicTools: a computational platform for developing high-throughput analytics in genomics. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr646. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.