Abstract
SorCS1 and SorL1/SorLA/LR11 belong to the sortilin family of vacuolar protein sorting-10 (Vps10) domain-containing proteins. Both are genetically associated with Alzheimer's disease (AD), and SORL1 expression is decreased in the brains of patients suffering from AD. SORCS1 is also genetically associated with types 1 and 2 diabetes mellitus (T1DM, T2DM). We have undertaken a study of the possible role(s) for SorCS1 in metabolism of the Alzheimer's amyloid-β peptide (Aβ) and the Aβ precursor protein (APP), to test the hypothesis that Sorcs1 deficiency might be a common genetic risk factor underlying the predisposition to AD that is associated with T2DM. Overexpression of SorCS1cβ-myc in cultured cells caused a reduction (p = 0.002) in Aβ generation. Conversely, endogenous murine Aβ40 and Aβ42 levels were increased (Aβ40, p = 0.044; Aβ42, p = 0.007) in the brains of female Sorcs1 hypomorphic mice, possibly paralleling the sexual dimorphism that is characteristic of the genetic associations of SORCS1 with AD and DM. Since SorL1 directly interacts with Vps35 to modulate APP metabolism, we investigated the possibility that SorCS1cβ-myc interacts with APP, SorL1, and/or Vps35. We readily recovered SorCS1:APP, SorCS1:SorL1, and SorCS1:Vps35 complexes from nontransgenic mouse brain. Notably, total Vps35 protein levels were decreased by 49% (p = 0.009) and total SorL1 protein levels were decreased by 29% (p = 0.003) in the brains of female Sorcs1 hypomorphic mice. From these data, we propose that dysfunction of SorCS1 may contribute to both the APP/Aβ disturbance underlying AD and the insulin/glucose disturbance underlying DM.
Introduction
Rare, early-onset familial Alzheimer's disease (EOFAD) is believed to begin with the accumulation of oligomeric forms of the 42 amino acid amyloid β peptide (Aβ42) in the hippocampus and cerebral cortex (for review, see Lublin and Gandy, 2010). EOFAD is often caused by mutations in genes that directly influence Aβ metabolism, most commonly the amyloid β precursor protein (APP), presenilin 1 (PS1), or presenilin 2 (PS2) (for review, see Gandy, 2005). Genetic studies of late-onset Alzheimer's disease (LOAD) point to a number of risk factor genes, including several that belong to one of three classes of molecules: (1) the apolipoprotein family, the most notable, apolipoprotein E (APOE) (Corder et al., 1993; Saunders et al., 1993); (2) the low-density lipoprotein receptor (LDLR) family (Kang et al., 1997; Lendon et al., 1997); and (3) the vacuolar protein sorting-10 (VPS10) domain-containing receptor family. Of note, SORL1 belongs to both the LDLR family and the VPS10-domain protein family and is genetically associated with AD (Rogaeva et al., 2007; Liang et al., 2009). A deficiency in SorL1 protein has been observed in the brains of patients suffering from LOAD and is believed to underlie the mechanism of the linkage of SORL1 with AD (Scherzer et al., 2004; Dodson et al., 2006; Sager et al., 2007). APP and SorL1 are frequently colocalized to the same subcellular compartments, and SorL1 has been demonstrated to modulate Aβ generation (Andersen et al., 2005; Offe et al., 2006; Nielsen et al., 2007; Schmidt et al., 2007) via an interaction with the core component of the retromer complex, Vps35 (Andersen et al., 2010), as proposed by Small and Gandy (2006). Human studies have shown that Vps35 and other components of the retromer complex are deficient in the brains of AD patients (Small et al., 2005), and animal model studies have established that retromer deficiency recapitulates key features of the human disease (Muhammad et al., 2008).
Another member of the VPS10 family, SORCS1, has recently been genetically associated with LOAD (Liang et al., 2009). SORCS1 resides at a quantitative trait locus for type 2 diabetes mellitus (T2DM) in mice and rats (Clee et al., 2006; Granhall et al., 2006) and is associated via genome-wide association studies to both T1DM and T2DM in humans (Goodarzi et al., 2007; Paterson et al., 2010). Because SORCS1 is additionally associated with a risk for AD, we hypothesized that one action of SorCS1 might involve modulation of APP metabolism. Herein, we report that overexpression of SorCS1 in cultured cells lowers Aβ generation. Consistent with this finding, Aβ40 and Aβ42 levels are increased in the brains of female Sorcs1 hypomorphs. Coimmunoprecipitation experiments revealed SorCS1:APP and SorCS1:Vps35 protein:protein complexes from both transfected cells and nontransgenic mouse brain. Furthermore we have determined that brain total protein levels of Vps35 and SorL1 are decreased (49% and 29%, respectively) in the brains of female Sorcs1 hypomorphs. These data point to the sortilin/retromer axis as a point of convergence in the pathogenesis of both AD and DM.
Materials and Methods
Antibodies.
α-Myc (Cell Signaling Technology), α-GFP (Roche), α-Vps35 (Abcam), α-SorL1 (BD Biosciences), and anti-mouse, anti-rabbit, and anti-goat HRP conjugates (Vector Laboratories) were purchased. pAb369 (C-terminal APP antibody) was used to detect human and mouse holoAPP and C-terminal fragments (Buxbaum et al., 1990). Anti-SorCS1/3 (this study) recognizes endogenous SorCS1 but, under certain circumstances, also reacts with SorCS3.
Cell culture studies.
HEK293t cells were cultured at 37°C/5% CO2 in growth medium (DMEM, 10% FBS, 1% penicillin/streptomycin, 1% l-glutamine, Invitrogen). 293t cells were transfected with human APP695, GFP, and pCDNA4 (empty vector) or human APP695, GFP, and murine SORCS1cβ-myc cDNA (Nielsen et al., 2008), using LipoD293t (SignaGen) at a ratio of 1:4 cDNA:LipoD, according to the manufacturer's instructions. Forty-eight hours after transfection, cells were collected in ice-cold PBS and centrifuged at 55 × g at 4°C for 15 min, and the media were collected and snap frozen. Cells were subsequently harvested in RIPA buffer (50 mm Tris HCl pH 7.5, 100 mm NaCl, 1 mm EDTA, 1 mm DTT, 1% NP40, 0.2 mm PMSF, 0.2 mm Na3VO4, 50 mm NaF, 10 mm Na4P2O7 plus Roche complete EDTA-free protease inhibitor tablet) using 5 cycles of 20 s vortex/5 min ice incubation. Cell debris was removed by centrifugation at 4°C at 10,000 × g for 15 min. Protein concentrations from cell lysates and media were determined using the Bio-Rad Protein Determination Kit. Absorbance was read at 595 nm using a Bio-Rad Microplate Reader (680XR) and analyzed using Microplate Manager v5.2.1. Samples were subsequently prepared in 5× Laemmli buffer and boiled at 95°C for 5 min. Equal amounts of total protein were loaded onto 12% Bis Tris SDS-PAGE gels for electrophoresis and transferred to PVDF membranes. The membrane was analyzed by Western blot using pAb369 (APP C-terminal) to detect APP holoprotein (holoAPP) and presumptive α and β C-terminal fragments (CTFs) referred to as α/βCTFs. SorCS1/3, GFP, and actin were detected with the respective relevant primary antibodies, followed by HRP-conjugated, species-specific secondary antibodies. Signals were detected using enhanced chemiluminescence (Pierce). Digital images were captured using LAS3000 (FujiFilm) and analyzed using Multi Gauge v3.1 software.
Immunoprecipitations.
HEK293t cells were transiently transfected with cDNAs, and lysates were prepared as described above. For brain extracts, C57BL/6J hemibrains were homogenized in RIPA buffer using Fisher Scientific Power Gen 1000 homogenizer for 2 cycles of 10 s on ice. A 300 μg aliquot of cell lysate or a 1000 μg aliquot of brain protein was used for immunoprecipitation using A/G plus agarose beads (Santa Cruz Biotechnology) with either (1) 2 μg of the appropriate primary (cells); (2) 1:10 dilution of α-SorCS1/3 (brain); or (3) IgG control antibody according to the manufacturer's instructions.
Indirect immunohistochemistry and microscopy.
HEK293t cells were cultured in eight-well chamber slides for 24 h before transfection with low levels of APP and SorCS1cβ-myc cDNA. Forty-eight hours after transfection, cellular localization of proteins was detected by sequential scanning confocal immunofluorescence microscopy using Alexa 488- or Texas Red-conjugated secondary antibodies and the Leica TCS DMI 63× oil objective lens. Images were processed using ImageJ. Images shown are representative fields.
Construction of Sorcs1 hypomorphic mice.
Mouse genomic DNA for the Sorcs1 vector was cloned from C57BL/6J (B6) BAC DNA. The vector was generated by insertion of fragments with 5′ homology and 3′ homology on either side of genomic Sorcs1 DNA. LoxP sites and an FRT-flanked neomycin cassette were added between these homologous arms. The final targeting construct contained LoxP sites flanking part of the Sorcs1 promoter, all of exon 1 (containing the translational start site), and a portion of intron 1.
The targeting vector was linearized and electroporated into 129/SV embryonic stem cells. Following neomycin selection, the targeted cells were microinjected into B6 blastocysts and implanted into B6 pseudopregnant female mice. Chimeric offspring were bred to B6 mice to test for germline transmission of the targeting construct. Homozygous floxed 129/SV mice were bred with B6 EIIa-Cre mice (The Jackson Laboratory) to generate germline Sorcs1 hypomorphic mice.
Verification of Sorcs1 mRNA levels in Sorcs1 hypomorphic mice.
RNA was harvested and purified using RNeasy spin columns (Qiagen). cDNA was synthesized from 1 μg of total RNA using the SuperScript III first-strand cDNA synthesis kit and oligo-dT and random hexamer primers (Invitrogen). Sorcs1 gene expression was quantified using the Realplex real-time PCR system (Eppendorf) and TaqMan Universal PCR Master Mix (Applied Biosystems). A 50-fold reduction in Sorcs1 mRNA expression compared to wild-type brain was observed (TaqMan probe Mm00491259) and is therefore referred to as a Sorcs1 hypomorphic mice rather than a knock-out mice. The housekeeping gene Actb was used as control (TaqMan probe Mm0060793).
Preparation of brains for analysis.
Mice were killed with CO2 and brains were rapidly removed, hemisected, and snap frozen. Each frozen hemibrain was then processed via differential solubilization (Kawarabayashi et al., 2001). Protein concentrations were determined and 100 μg of total protein lysate was analyzed by SDS-PAGE and Western blot, as described above. Aβ40 and Aβ42 levels were determined by the mouse Aβ (40 and 42) ELISA kit (WAKO), according to the manufacturer's instructions. Absorbance was read at 450 nm using a Bio-Rad microplate reader. Results were normalized to wet brain weight and expressed as picomoles per gram.
Statistical analysis.
Densitometric analysis of Western blot bands (integrated density) was performed using Multigauge v3.1 software (Fujifilm). Levels of holoAPP and α/βCTF were normalized to actin and expressed as percentage of control. Total Aβ levels were analyzed by Western blot, and bands were normalized to percentage of control (empty vector). Absolute Aβ40 and Aβ42 concentrations were quantitatively determined by sandwich ELISA (Wako), and Aβ42/Aβ40 ratios were calculated. Certain intergenotype and intragenotype comparisons were not biologically relevant, precluding the use of 2 × 2 matrices for statistical design (i.e., the comparison of Sorcs1−/− males vs Sorcs1+/+ females). In all instances, Shapiro–Wilk test of normality and Levene's test for homogeneity of variance were used for inclusion in parametric tests (p > 0.05 for Levene's and Shapiro–Wilk tests). Independent-samples t tests (parametric design) or Mann–Whitney U tests (nonparametric design) were used to determine significant mean differences between groups. Significance for t tests are reported with a p < 0.05 using two-tailed tests with an α level of 0.05. All statistical analysis was performed using SPSS v18.0.
Results
Overexpression of SorCS1cβ decreases Aβ generation
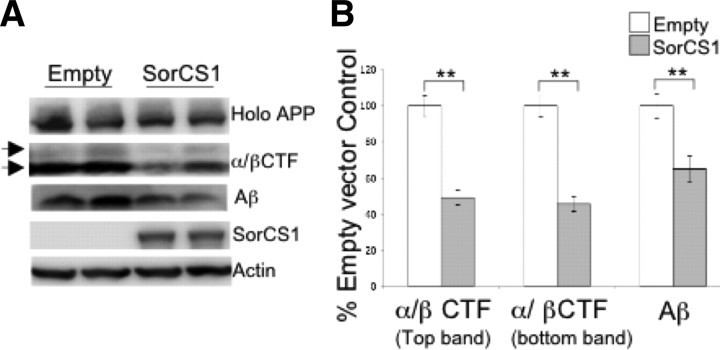
To determine whether SorCS1cβ modulates APP metabolism and Aβ formation, we analyzed levels of exogenously expressed human APP and its metabolites in HEK293t cells upon co-overexpression of SorCS1cb-myc. Upon co-overexpression of APP and SorCS1cb-myc, levels of cellular α/β-C-terminal fragment (α/βCTF) were decreased by 54% (top band; t(16) = 7.336, p ≤ 0.001) and 51% (bottom band; t(16) = 7.214, p ≤ 0.001) when compared to empty vector control (pCDNA4) (Fig. 1A,B). Total secreted Aβ was decreased by 35% (t(16) = 3.687, p = 0.002) following correction for holoAPP levels.
Figure 1.
Overexpression of SorCS1cβ-myc decreases Aβ generation. A, Western blot analysis of APP metabolites in HEK293t cells transiently transfected with APP695 and SORCS1cβ or APP695 and empty vector control. Lysates and conditioned media were probed for holoAPP, APPCTFs, and Aβ using pAb369 and 6E10, respectively. B, Protein levels were normalized to actin and expressed as percentage of empty vector control. APPCTFs and Aβ were additionally normalized to holoAPP levels to account for transfection variations. Data were collected in duplicate or triplicate from three independent experiments. Significant reductions (**p ≤ 0.01) in cellular α/βCTF (p ≤ 0.001) and secreted Aβ (p = 0.002) were observed upon overexpression of SorCS1cβ-myc, as compared to empty vector control.
Sorcs1 hypomorphic mouse brains accumulate APP metabolites, including Aβ42
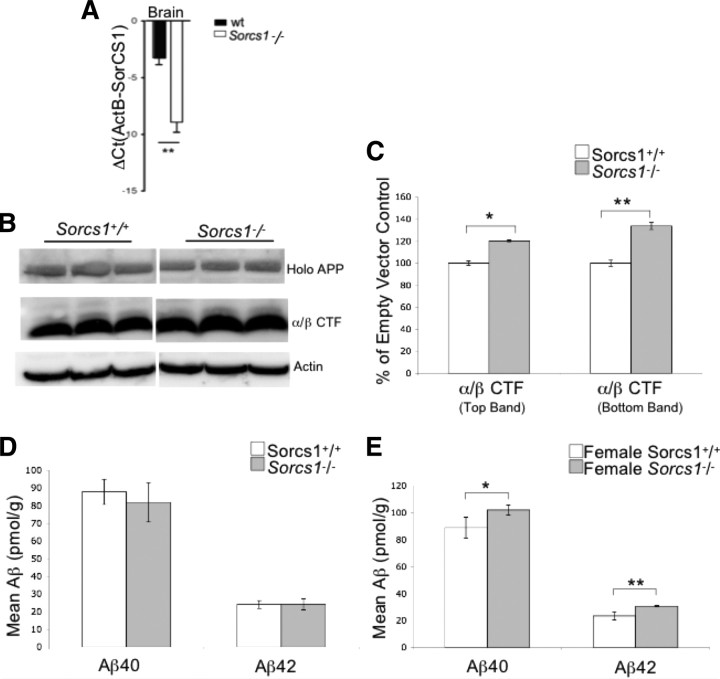
To determine whether SorCS1 has a direct effect on Aβ generation in vivo, we next analyzed the levels of endogenous murine APP and its metabolites in the brains of Sorcs1 hypomorphic mice. These mice exhibited a 50% decrease in Sorcs1 mRNA transcripts in the brain (Fig. 2A). The cross-reaction of our anti-SorCS1 antibody (anti-SorCS1/3) with SorCS3, however, precludes a more precise statement regarding the level of SorCS1 protein. When compared to wild-type littermates, the levels of α/βCTF in the brain of Sorcs1 hypomorphs were increased by 20% (top band t(10) = −2.612, p = 0.026) and 30% (bottom band t(10) = −3.201, p = 0.009) (Fig. 2B,C). Analysis of total Aβ40 and Aβ42 levels by sandwich ELISA revealed that the levels of Aβ40 (t(4) = −2.912, p = 0.044) and Aβ42 (t(4) = −5.113, p = 0.007) were increased in the brains of female Sorcs1 hypomorphs in comparison to wild-type females (Fig. 2E). No differences in Aβ40 or Aβ42 levels were observed when Sorcs1 hypomorphs were compared to wild type (male and female grouped) (Fig. 2D).
Figure 2.
Sorcs1 hypomorphic mouse brains accumulate APP metabolites, including Aβ42. A, Sorcs1 gene expression was quantified using real-time PCR. Sorcs1 mRNA was normalized to Actb mRNA. Significant reductions in Sorcs1 mRNA were observed in the brain in Sorcs1 hypomorphic mice (Sorcs1−/− n = 3) compared to wild-type mice (Sorcs1+/+ n = 3). B, C, Western blot analysis of APP metabolites in Sorcs1+/+ (n = 6) and Sorcs1−/− (n = 6) hemibrains. Membrane proteins were fractionated by differential solubilization and analyzed by SDS-PAGE and Western blotting. Endogenous holo-APP and APP α/βCTFs were visualized with pAb369. Protein levels were normalized to actin and presented as expression percentage of control. Sorcs1−/− mice exhibited significant increases (*p < 0.05, **p < 0.01) in α/βCTF compared to wild-type littermates (top band p = 0.026, bottom band p = 0.009). D, E, Aβ40 and Aβ42 levels in Sorcs1+/+ and Sorcs1−/− hemibrains were determined by sandwich ELISA (Wako). Aβ40 and Aβ42 levels were normalized to brain weight and presented as picomoles per gram. No difference was observed in total Aβ40 or Aβ42 levels when male and female mice were grouped for comparison of Sorcs1+/+ (n = 6) to Sorcs1−/− (n = 6) (D); however, when Aβ40 and Aβ42 were compared in female Sorcs1+/+ (n = 3) versus Sorcs1−/− (n = 3), increases in Aβ40 (p = 0.044) and Aβ42 (p = 0.007) were observed (E).
SorCS1 associates with and colocalizes with APP
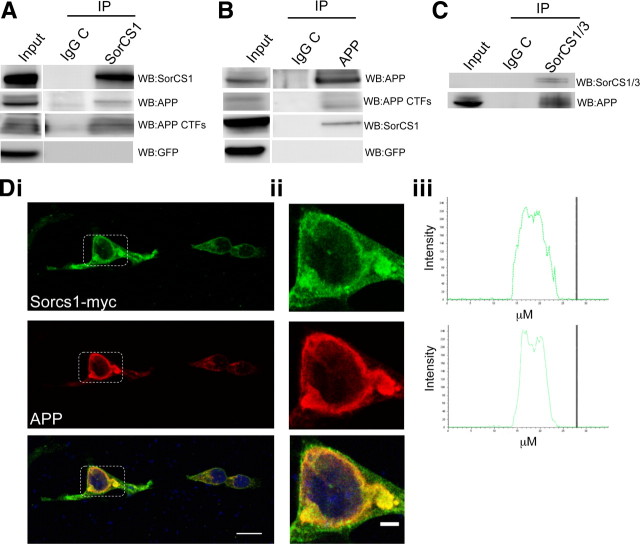
SorL1 and SorCS1 are both members of the VPS10-domain-containing family of proteins. SorL1 directly interacts with APP and modulates APP processing (Andersen et al., 2006; Spoelgen et al., 2007), raising the question of whether a similar interaction might occur involving APP and SorCS1. In immunoprecipitation/immunoblotting experiments using transfected 293t cell lines, we observed co-recovery of holo-SorCS1cβ-myc together with holoAPP and the α/βCTFs (Fig. 3A,B). To further validate this interaction, we sought to determine whether APP coimmunoprecipitated with SorCS1/3 in nontransgenic C57BL/6J brain tissue. Again, immunoprecipitation of endogenous SorCS1/3 resulted in coimmunoprecipitation of holoAPP (Fig. 3C). Furthermore indirect immunofluorescence microscopy for SorCS1cβ-myc and APP revealed substantial physical codistribution throughout the cell, most especially in the perinuclear region (Fig. 3D). Together, these data suggest the existence of a physiological complex that includes APP and SorCS1/3.
Figure 3.
SorCS1cβ-myc interacts with and colocalizes with APP. A, Transfected SorCS1cβ-myc was immunoprecipitated from HEK293t cells using α-myc antibody or control IgG. Immunoprecipitates were subjected to SDS-PAGE, transferred to PVDF membranes, and probed with antibodies as indicated. Coprecipitating APP and APPCTFs were detected by Western blot using pAb369. GFP was not detected in the immunoprecipitate, providing additional evidence for specificity of the SorCS1cβ-myc:APP complex. B, Immunoprecipitation of APP and APPCTFs using pAb369 resulted in coimmunoprecipitation of SorCS1cβ-myc as detected by α-myc antibody. GFP was not detected in the coprecipitate. C, Immunoprecipitation of endogenous SorCS1/3 from C57BL/6J brain tissue resulted in coimmunoprecipitation of APP. Control immunoprecipitations using an irrelevant IgG were included to provide evidence for specificity of the coprecipitation of SorCS1/3 and APP. Di, Detection of APP and SorCS1cβ-myc in transiently transfected HEK293t cells using sequential scanning confocal microscopy demonstrates colocalization of SorCS1cβ-myc (red) and APP (green) in the merged image (colocalization yellow) to the perinuclear region (nucleus blue) (scale bar, 20 μm). Dii, Enlarged image of the reference region (white box, scale bar 5 μm). Diii, Graphical representation of intensity of fluorescence across the perinuclear region demonstrates a strong colocalization for SorCS1cβ-myc and APP fluorescence.
Brains from female Sorcs1 hypomorphs contain decreased levels of Vps35
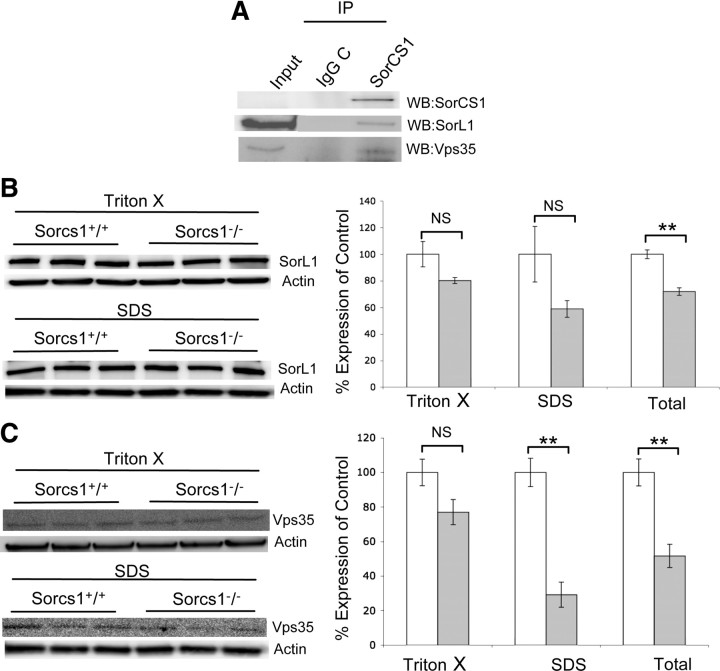
Andersen et al. (2010) recently reported that the retromer component Vps35 is required for SorL1 to modulate APP metabolism (Andersen et al., 2005; Offe et al., 2006; Nielsen et al., 2007; Schmidt et al., 2007). We therefore tested the possibility that the Vps35 and/or SorL1 forms complexes with SorCS1/3 and measured SorL1 and Vps35 levels in Sorcs1 hypomorphs. We were able to demonstrate coimmunoprecipitation of SorL1 and Vps35 with SorCS1/3 from brain tissue (Fig. 4A). Differential solubilization of brain tissue in Triton X-100 and SDS was subsequently used to study the cellular localization of these proteins in Sorcs1 hypomorphs. While we found that both total SorL1 (t(4) = 2.020, p = 0.003, 29% decrease) and Vps35 (t(4) = 4.708, p=0.009) protein levels were decreased in female Sorcs1 hypomorphs, differential solubilization revealed that only Vps35 protein levels in the SDS fraction (t(4) = 6.400, p = 0.003, 71% decrease) were reduced (Fig. 4C), indicating drastically reduced levels of Vps35 in subcellular fractions/compartments insoluble in Triton X-100 but soluble in SDS (Ali et al., 1989; Messier et al., 1993).
Figure 4.
SorCS1/3 coprecipitates with SorL1 and Vps35 in vivo and SorL1 and Vps35 expression is decreased in Sorcs1 hypomorphic mice. A, Immunoprecipitation of endogenous SorCS1/3 resulted in coimmunoprecipitation of SorL1 and Vps35 in brain tissue of C57BL/6J mice. Control immunoprecipitations using an irrelevant IgG were included to provide evidence for specificity of the coprecipitation of SorCS1/3, SorL1, and APP. B, C, Western blot analysis of SorL1 and Vps35 expression in female Sorcs1+/+ (n = 3) and Sorcs1−/− (n = 3) hemibrains. Membrane proteins were fractionated by differential solubilization (Triton X-100 and SDS fractions) and analyzed by SDS-PAGE and Western blotting. Endogenous SorL1 and Vps35 were visualized with α-SorL1 and α-Vps35 antibodies respectively. Protein levels were normalized to actin and presented as expression percentage of control. B, Female Sorcs1−/− mice exhibited a decrease in SorL1 protein levels in the Triton X-100 and SDS fractions that was not statistically significant. However, a significant decrease in SorL1 protein levels was detected in total SorL1 protein levels (Triton X-100+SDS) (p = 0.002). **p < 0.01. C, Female Sorcs1−/− exhibited a significant decrease in Vps35 protein levels (p = 0.003) in the SDS fraction. No significant change was detected in the Triton X-100 fraction. Total Vps35 protein levels were decreased in female Sorcs1−/− mice (p = 0.009).
Discussion
SorCS1 is the most recent member of the Vps10 family of proteins (Hermey et al., 2003) to be associated with AD (Liang et al., 2009). This has prompted us to hypothesize that SorCS1 might play some of the same roles already established for SorL1 in the modulation of APP metabolism. To investigate that possibility, we performed cell-based assays that clearly demonstrated that APP α/βCTF (p < 0.001, ∼50% decrease) and total Aβ (p = 0.002, 35% decrease) were decreased upon overexpression of SorCS1cβ. To validate this observation in vivo, we turned to the Sorcs1 hypomorphic mouse, where, in the brains of Sorcs1 hypomorphs, we observed a 25–30% increase in APP α/βCTF (p = 0.026, p = 0.009 respectively), a 14% increase in Aβ40 (p = 0.004), and a 24% increase in Aβ42 (p = 0.007) in the brains of female, but not male, Sorcs1 hypomorphic mice. These changes in APP metabolism are highly reminiscent of those observed in Sorl1 knockdown mice (Andersen et al., 2005; Dodson et al., 2008), except that sexual dimorphism in Aβ levels has not been reported for Sorl1 knockdown mice. The sexual dimorphism is especially interesting in light of the observation that the genetic linkage to SORCS1 is stronger for women in both T2DM (Goodarzi et al., 2007) and AD (Liang et al., 2009) populations. Similar observations were recently reported in abstract form by Reitz et al. (2010), although those investigators used siRNA in cultured cells as their SorCS1 knockdown model and so were unable to assess the possibility of sexual dimorphism.
We next assessed the possibility that SorCS1, like SorL1, directly influences APP metabolism and Aβ generation through molecules known to modulate APP metabolism. Specifically, as suggested by Small and Gandy (2006), SorL1 and other Vps10-containing proteins might modulate APP processing by mediating the interaction between the retromer complex and APP. We were able to detect APP, APP α/βCTFs, SorL1, and Vps35 in the anti-SorCS1 immunoprecipitates from both APP/SorCS1cβ-myc-doubly transfected cells and nontransgenic mouse brain. Interestingly, both SorL1 and Vps35 total protein levels were also decreased in the brains of Sorcs1 hypomorphs. Further study will be required to determine whether the protein–protein complexes and/or decreased expression of SorL1 and Vps35 play roles in the elevation of brain Aβ42 that we have observed in female Sorcs1 hypomorphs. Based on these coimmunoprecipitation data, SorCS1 is well positioned to modulate one or more steps in APP metabolism. While the focus of SorL1-related and, in this study, SorCS1-related effects on APP has been on protein trafficking, it is important to remember that many Vps10-domain proteins are γ-secretase substrates, and competition for access to the catalytic site of γ-secretase may also contribute to the mechanism (Nyborg et al., 2006). Further studies are required (1) to confirm the pathogenic importance of the Aβ42 changes in vivo by crossing Sorcs1 hypomorphic mice with human APP-overexpressing mice capable of forming Aβ oligomers and plaques; and (2) to elucidate the detailed mechanism through which SorCS1 regulates APP/Aβ metabolism. We propose that probing the molecular consequences of SorCS1 dysfunction will lead to pathways that elucidate the link between DM and AD.
Footnotes
J.W.S. is a trainee in the Integrated Pharmacological Sciences Training Program supported by Grant T32GM06754 from the National Institute of General Medical Sciences. This work was supported by National Institute on Aging Grant P01AG10491 (to S.G.), Veterans Affairs Merit Review Grant 1I01BX000348 (S.G.), and National Institute of Diabetes and Digestive and Kidney Diseases Grant DK58037 (A.D.A.). S.G. and R.E.T. are members of the Cure Alzheimer's Fund (CAF) Research Consortium and acknowledge the support of CAF. Confocal laser scanning microscopy was performed at the Mount Sinai School of Medicine-Microscopy Shared Resource Facility, supported with funding from a National Institutes of Health (NIH)–National Cancer Institute Shared Resources Grant (5R24 CA095823-04), a National Science Foundation Major Research Instrumentation Grant (DBI-9724504), and an NIH Shared Instrumentation Grant (1 S10 RR0 9145-01). We thank Efrat Levy (New York University), Gopal Thinakaran (University of Chicago), and Nabil Seidah (Clinical Research Institute, Montreal) for cDNA constructs.
References
- Ali N, Miligan G, Evans WH. G-proteins of rat-liver membranes: subcellular compartmentation and disposition in the plasma membrane. Mol Cell Biochem. 1989;91:75–84. doi: 10.1007/BF00228081. [DOI] [PubMed] [Google Scholar]
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- Andersen O, Fjorback A, Seaman M, Ilsoe E, Gustafsen C, Willnow T, Madsen P, Nykjaer A. The role of retromer in SorLA dependent APP transport and processing. Alzheimers Dement. 2010;6:S101. [Google Scholar]
- Buxbaum JD, Gandy SE, Cicchetti P, Ehrlich ME, Czernik AJ, Fracasso RP, Ramabhadran TV, Unterbeck AJ, Greengard P. Processing of Alzheimer beta/A4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc Natl Acad Sci U S A. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clee SM, Yandell BS, Schueler KM, Rabaglia ME, Richards OC, Raines SM, Kabara EA, Klass DM, Mui ET, Stapleton DS, Gray-Keller MP, Young MB, Stoehr JP, Lan H, Boronenkov I, Raess PW, Flowers MT, Attie AD. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet. 2006;38:688–693. doi: 10.1038/ng1796. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, Peng J, Levey AI, Willnow TE, Lah JJ. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer's disease. J Neurosci. 2008;28:12877–12886. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandy S. The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. J Clin Invest. 2005;115:1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi MO, Lehman DM, Taylor KD, Guo X, Cui J, Quiñones MJ, Clee SM, Yandell BS, Blangero J, Hsueh WA, Attie AD, Stern MP, Rotter JI. SORCS1: A novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes. 2007;56:1922–1929. doi: 10.2337/db06-1677. [DOI] [PubMed] [Google Scholar]
- Granhall C, Park HB, Fakhrai-Rad H, Luthman H. High-resolution quantitative trait locus analysis reveals multiple diabetes susceptibility loci mapped to intervals<800 kb in the species-conserved Niddm1i of the GK rat. Genetics. 2006;174:1565–1572. doi: 10.1534/genetics.106.062208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermey G, Keat SJ, Madsen P, Jacobsen C, Petersen CM, Gliemann J. Characterization of sorCS1, an alternatively spliced receptor with completely different cytoplasmic domains that mediate different trafficking in cells. J Biol Chem. 2003;278:7390–7396. doi: 10.1074/jbc.M210851200. [DOI] [PubMed] [Google Scholar]
- Kang DE, Saitoh T, Chen X, Xia Y, Masliah E, Hansen LA, Thomas RG, Thal LJ, Katzman R. Genetic association of the low-density lipoprotein receptor-related protein gene (LRP), an apolipoprotein E receptor, with late-onset Alzheimer's disease. Neurology. 1997;49:56–61. doi: 10.1212/wnl.49.1.56. [DOI] [PubMed] [Google Scholar]
- Kawarabayashi T, Younkin LH, Saido TC, Shoji M, Ashe KH, Younkin SG. Age-dependent changes in brain, CSF, and plasma amyloid β protein in the Tg2576 transgenic mouse model of Alzheimer's disease. J Neurosci. 2001;21:372–381. doi: 10.1523/JNEUROSCI.21-02-00372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendon CL, Talbot CJ, Craddock NJ, Han SW, Wragg M, Morris JC, Goate AM. Genetic association studies between dementia of the Alzheimer's type and three receptors for apolipoprotein E in a Caucasian population. Neurosci Lett. 1997;222:187–190. doi: 10.1016/s0304-3940(97)13381-x. [DOI] [PubMed] [Google Scholar]
- Liang X, Slifer M, Martin ER, Schnetz-Boutaud N, Bartlett J, Anderson B, Züchner S, Gwirtsman H, Gilbert JR, Pericak-Vance MA, Haines JL. Genomic convergence to identify candidate genes for Alzheimer disease on chromosome 10. Hum Mutat. 2009;30:463–471. doi: 10.1002/humu.20953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lublin AL, Gandy S. Amyloid-beta oligomers: possible roles as key neurotoxins in Alzheimer's disease. Mt Sinai J Med. 2010;77:43–49. doi: 10.1002/msj.20160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier JM, Shaw LM, Chafel M, Matsudaira P, Mercurio AM. Fimbrin localized to an insoluble cytoskeletal fraction is constitutively phosphorylated on its headpiece domain in adherent macrophages. Cell Motil Cytoskeleton. 1993;25:223–233. doi: 10.1002/cm.970250303. [DOI] [PubMed] [Google Scholar]
- Muhammad A, Flores I, Zhang H, Yu R, Staniszewski A, Planel E, Herman M, Ho L, Kreber R, Honig LS, Ganetzky B, Duff K, Arancio O, Small SA. Retromer deficiency observed in Alzheimer's disease causes hippocampal dysfunction, neurodegeneration, and Abeta accumulation. Proc Natl Acad Sci U S A. 2008;105:7327–7332. doi: 10.1073/pnas.0802545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Gustafsen C, Madsen P, Nyengaard JR, Hermey G, Bakke O, Mari M, Schu P, Pohlmann R, Dennes A, Petersen CM. Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor sorLA. Mol Cell Biol. 2007;27:6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MS, Keat SJ, Hamati JW, Madsen P, Gutzmann JJ, Engelsberg A, Pedersen KM, Gustafsen C, Nykjaer A, Gliemann J, Hermans-Borgmeyer I, Kuhl D, Petersen CM, Hermey G. Different motifs regulate trafficking of SorCS1 isoforms. Traffic. 2008;9:980–994. doi: 10.1111/j.1600-0854.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Nyborg AC, Ladd TB, Zwizinski CW, Lah JJ, Golde TE. Sortilin, SorCS1b, and SorLA Vps10p sorting receptors, are novel γ-secretase substrates. Mol Neurodegener. 2006;1:3. doi: 10.1186/1750-1326-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid β production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Waggott D, Boright AP, Hosseini SM, Shen E, Sylvestre MP, Wong I, Bharaj B, Cleary PA, Lachin JM, Below JE, Nicolae D, Cox NJ, Canty AJ, Sun L, Bull SB, MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium) Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes. Diabetes. 2010;59:539–549. doi: 10.2337/db09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Tokuhiro S, Clark L, Conrad C, Vonsattel J-P, Lantigua R, Medrano M, Simkin I, Haines J, Pericak-Vance M, Farrer L, Lee J, Rogaeva E, St George Hyslop P, Mayeux R. SorCS1 alters APP processing and variants may increase Alzheimer's disease risk. Alzheimers Dement. 2010 doi: 10.1002/ana.22308. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager KL, Wuu J, Leurgans SE, Rees HD, Gearing M, Mufson EJ, Levey AI, Lah JJ. Neuronal LR11/sorLA expression is reduced in mild cognitive impairment. Ann Neurol. 2007;62:640–647. doi: 10.1002/ana.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. sorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- Small SA, Gandy S. Sorting through the cell biology of Alzheimer's disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, Honig L, Vonsattel JP, Kim TW. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- Spoelgen R, von Arnim CA, Thomas AV, Peltan ID, Koker M, Deng A, Irizarry MC, Andersen OM, Willnow TE, Hyman BT. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and the β-secretase β-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]