Abstract
While genomically targeted therapies have improved outcomes for patients with lung adenocarcinoma, little is known about the genomic alterations which drive squamous cell lung cancer. Sanger sequencing of the tyrosine kinome identified mutations in the DDR2 kinase gene in 3.8% of squamous cell lung cancers and cell lines. Squamous lung cancer cell lines harboring DDR2 mutations were selectively killed by knock-down of DDR2 by RNAi or by treatment with the multi-targeted kinase inhibitor dasatinib. Tumors established from a DDR2 mutant cell line were sensitive to dasatinib in xenograft models. Expression of mutated DDR2 led to cellular transformation which was blocked by dasatinib. A squamous cell lung cancer patient with a response to dasatinib and erlotinib treatment harbored a DDR2 kinase domain mutation. These data suggest that gain-of-function mutations in DDR2 are important oncogenic events and are amenable to therapy with dasatinib. As dasatinib is already approved for use, these findings could be rapidly translated into clinical trials.
Keywords: Squamous cell lung cancer, DDR2, dasatinib, tyrosine kinase inhibitors, lung cancer genomics
INTRODUCTION
Lung cancer is the leading cause of cancer-related mortality in the United States with over 157,000 deaths projected in 2010 (1). The more common type of lung cancer, non-small cell lung cancer (NSCLC), accounts for 85% of cases and carries a grim prognosis with approximately 70% of patients presenting with advanced and often incurable disease at the time of diagnosis (2).
Despite these statistics a great deal of progress has been made in the targeted treatment of patients with NSCLC, largely due to the development of small molecule inhibitors of the epidermal growth factor receptor (EGFR) tyrosine kinase (3–5). Patients who respond to EGFR kinase inhibitors are much more likely to have the adenocarcinoma subtype of NSCLC (6). Patients with the other principal subtype of NSCLC, lung squamous cell cancer (lung SCC), very rarely respond to these agents and few advances have been made in the treatment of this type of lung cancer which comprises 25% of NSCLC. In addition to EGFR, several other promising therapeutic targets have been identified in the laboratory such as EML4-ALK, KRAS and MET; drugs directed against these proteins are being tested in clinical trials (7–10). However, it appears that these targets are likely limited to adenocarcinomas as well. A recent report has suggested that targeting FGFR1 amplifications in SCC of the lung may be a promising therapeutic strategy, though FGFR inhibitors are not currently in clinical use for the treatment of patients with lung cancer (11). Given the burden of disease from lung SCC we sought to identify new therapeutic targets for patients with lung SCCs by examining the tyrosine kinome of lung SCCs for novel mutated kinases.
Here, we report the identification of novel somatic mutations in the discoidin domain receptor 2 (DDR2) tyrosine kinase gene at a frequency of 3.8% (n=11) in a sample set of 290 squamous cell lung cancer samples. DDR2 is a receptor tyrosine kinase which binds collagen as its endogenous ligand and has been previously shown to promote cell migration, proliferation and survival when activated by ligand binding and phosphorylation (12–18). DDR1 and DDR2 mutations have been reported in several cancer specimens, including four DDR1 mutations (W385C, A496S, F866Y, F824W) and two DDR2 mutations in lung cancer (R105S and N456S), but these reports have not been confirmed in independent samples and functional characterization of the mutations has not been reported (19–21). We demonstrate that DDR2 mutation status is associated with sensitivity to the tyrosine kinase inhibitor dasatinib or to sh-RNA mediated depletion of DDR2. Additionally, we show that DDR2 mutations are oncogenic and that their ability to transform cells can be blocked by dasatinib treatment or by combination tyrosine kinase inhibitor treatment. Furthermore, we report a DDR2 kinase domain mutation in a clinical trial subject with SCC of the lung who had a radiographic response to combination therapy with erlotinib and dasatinib and who did not have an EGFR mutation. Together, these data suggest DDR2 may be an important therapeutic target in SCCs.
RESULTS
DDR2 is mutated in squamous cell lung cancer
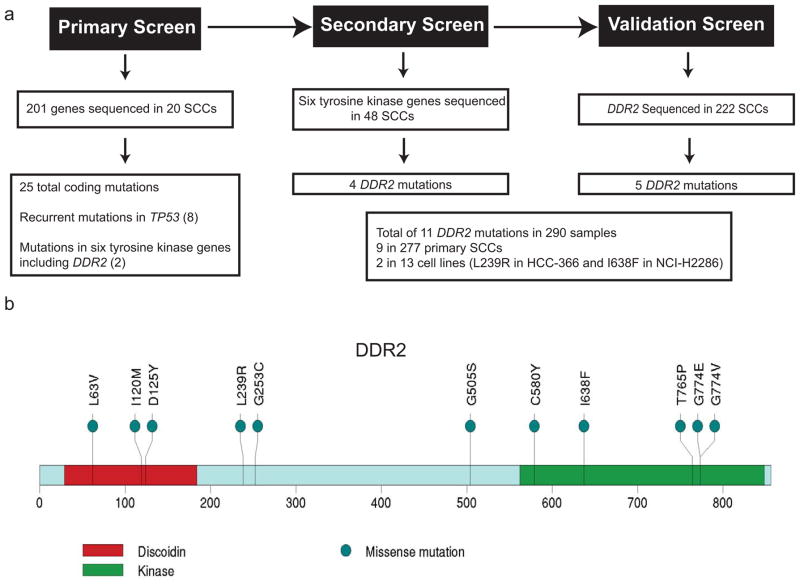
We performed Sanger sequencing of 201 genes including the entire tyrosine kinome in an initial set of 20 primary lung SCC samples and matched normal controls and identified somatic missense mutations in 25 genes in our discovery sample set including six in tyrosine kinase genes (Figure 1a). Recurrent somatic mutations were identified in TP53 (n=8), and in the tyrosine kinase genes: Discoidin Domain Receptor 2 (DDR2; n=2) and Kinase insert Domain Receptor (KDR; n=2) (Figure 1a). Subsequent sequencing of six of the mutated tyrosine kinase genes (DDR2, FGFR2, NTRK2, JAK2, FLT3 and CDK8), selected on the basis of being possible therapeutic targets, in a secondary screen of 48 squamous cell lung cancer samples including 13 cell lines revealed four additional DDR2 mutations (Figure 1a) as well as three FLT3 mutations, two NTRK2 and JAK2 mutations and one mutation in each of FGFR2 and CDK8.
Figure 1. Sequencing of squamous lung cancer samples identifies recurrent mutations in DDR2.
(a) Schema depicted for the primary, secondary and validation screens for DDR2 mutations in squamous lung cancer samples. (b) Amino acid sequence of DDR2 with the positions of the identified mutations shown in the context of the known domain structure of DDR2.
Given that DDR2 was the most frequently mutated gene in the primary and secondary screen we sequenced DDR2 in a validation cohort of 222 primary lung SCC samples which yielded an additional five samples with mutation, resulting in an overall frequency of 3.8% (n=11) in 290 total samples and an overall frequency of 3.2% in primary lung SCC samples when cell lines were excluded (n=9/277) (Figure 1a). Mutations were found both in the kinase domain and in other regions of the protein sequence and two mutations were identified at G774 (Figure 1b). The L239R and I638F mutations were identified in the HCC-366 and NCI-H2286 SCC cell lines, respectively, and the remainder of the mutations were found in primary SCC samples. The majority of the mutations resided in regions of high degrees of amino acid conservation as compared to the murine, zebrafish and C. elegans homologs of DDR2 (Figure S1). Additional genomic analysis of previously reported copy number and gene expression datasets did not reveal any evidence of DDR2 overexpression in SCCs as compared to normal lung or lung adenocarcinoma nor did we identify copy number alterations in DDR2 (data not shown) (19, 22–24). A query of the limited clinical information accompanying the sequenced samples did not Identify any significant correlation of DDR2 mutation status with the age, sex or smoking status of the patients.
DDR2 mutant cell lines are selectively sensitive to tyrosine kinase inhibitors and to sh-RNA-mediated depletion of DDR2
To assess whether targeting DDR2 might be a promising therapeutic strategy in lung SCC, we analyzed several tyrosine kinase inhibitors reported to inhibit DDR2 including imatinib and dasatinib, drugs which are FDA-approved for clinical use for targeting BCR-Abl in chronic myelogenous leukemia and acute lymphoblastic leukemia, c-KIT in gastrointestinal stromal tumors and PDGFR in chronic myelomonocytic leukemia (21, 25–28). Fluorescence resonance energy transfer (FRET) measurements provided in vitro Kd values of dasatinib (5.4 nM) and imatinib (71.6 nM) for recombinant DDR2 (Table S1)
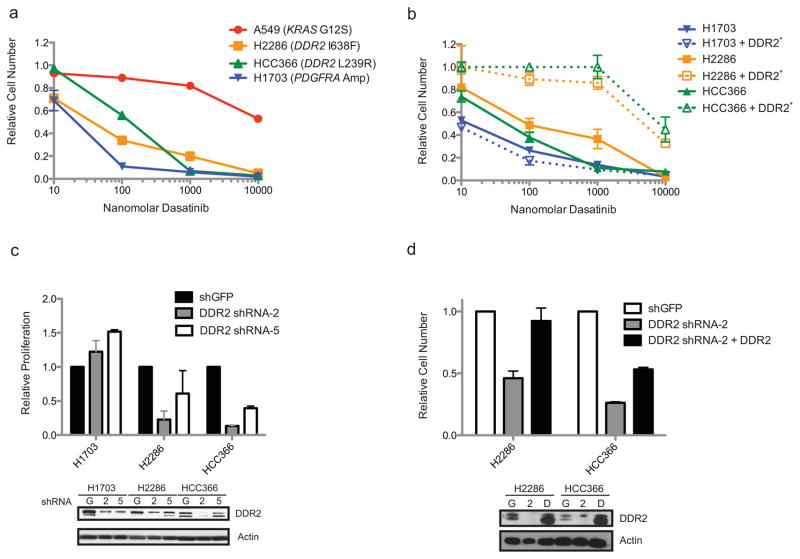
Dasatinib showed particular efficacy against SCC cell lines bearing DDR2 mutations, as dasatinib inhibited proliferation of the DDR2-mutant NCI-H2286 and HCC-366 cells with calculated IC50s of 139 and 140 nM respectively (Figure 2a). Of note, a recent pharmacokinetic analysis of dasatinib in lung cancer patients demonstrated that peak concentrations of dasatinib were in the range of 300 ng/ml (615 nM) at the maximum tolerated dose of 140 mg daily, a dose approved for use in leukemias (29). Imatinib was less potent when tested in the same cell lines with respective IC50s of 1.2 and 1.0 μM for the DDR2-mutant NCI-H2286 and HCC-366 cell lines (Figure S2a). Dasatinib and imatinib were less effective against the A549 cell line which is known to harbor a KRAS mutation and does not have any DDR2 mutations (calculated IC50 of 7.4 μM for dasatinib and 2.3 μM for imatinib). Consistent with previous reports, the NCI-H1703 SCC cell line, which contains a PDGFRA amplification, was sensitive to both drugs, serving as a positive control for our assay (30, 31). Notably, no other somatic mutations have been reported in the COSMIC database for NCI-H2286 or HCC-366 lines to suggest alternative dasatinib targets and a previous report examining the drug sensitivities of 83 NSCLC cell lines identified HCC-366 as the most sensitive squamous cell lung cancer line to dasatinib, though NCI-H2286 and NCI-H1703 were not assayed (32). Treatment of the DDR2 mutant cell lines with dasatinib appeared to lead to cell death as opposed to cell cycle arrest as measured by trypan blue exclusion (Figure S2b). Dasatinib treatment was associated with an increase in cellular annexin V staining, suggesting that the treated cells died by apoptosis (data not shown).
Figure 2. Lung cancer cell lines with DDR2 mutations are sensitive to drugs and RNAi targeting DDR2.
(a) Proliferation of A549, NCI-H2286, HCC-366 and NCI-H1703 grown for six days in the presence of various concentrations of dasatinib. Proliferation shown relative to untreated cells at the same time point. Standard errors are shown for triplicate samples. (b) Proliferation shown of NCI-H2286 and HCC-366 cell lines ectopically expressing the T654M gatekeeper mutation in DDR2, labeled as DDR2*. Six day proliferation in the presence of dasatinib is shown as above. For NCI-H2286 and HCC-366 the gatekeeper mutation is expressed in cis with the DDR2 mutation found in the cell line. (c) Proliferation measured as above for NCI-H2286, HCC-366 and NCI-H1703 cells stably expressing sh-RNA vectors targeting either GFP or the 3′ UTR of DDR2 (DDR2 sh-RNA-2) or the coding sequence of DDR2 (DDR2 sh-RNA-5). Proliferation is measured after four days in culture as compared to day 1. Standard errors are shown for triplicate samples. Immunoblot showing relative levels of DDR2 in the cell lines used in the experiment is shown in the inset. “G” indicates cells expressing shGFP, and “2” and “5” the numbered DDR2 targeted hairpins. (d) Four-day proliferation of the DDR2 mutant NCI-H2286 and HCC-366 cell lines stably expressing ectopic DDR2 following knock-down of DDR2 by a sh-RNA targeting the 3′ UTR of DDR2 (sh-RNA-2). Proliferation of triplicate samples is presented as above relative to cells transduced with a sh-RNA targeting GFP. Protein levels of DDR2 are shown in the immunoblot below, “G” indicates sh-GFP, “2” indicates expression of sh-RNA 2 and “D” indicates expression of sh-RNA2 and DDR2.
To validate DDR2 as a relevant target of dasatinib in SCCs we ectopically expressed a DDR2 transgene with a threonine to methionine mutation at amino acid 654, a mutation site shown previously to render DDR2 dasatinib-insensitive in a manner similar to the ability of the T790M mutation in EGFR to confer acquired resistance to the tyrosine kinase inhibitors erlotinib and gefitinib (33). We introduced the dasatinib-insensitive DDR2 “gatekeeper” mutant in cis with the observed L239R and I638F mutations in the HCC-366 and NCI-H2286 cell lines respectively as well as alone in NCI-H1703. Expression of the gatekeeper mutation led to a decrease in dasatinib sensitivity in both DDR2 mutant cell lines and had a modest effect on NCI-H1703 (Figure 2b; transgene expression is shown in Figure S2e). While the calculated IC50 for NCI-H1703 did not change with ectopic expression of the gatekeeper, the IC50 increased by 35-fold for NCI-H2286 and 209-fold for HCC-366, respectively. Interestingly, a parallel sequencing project in our lab identified a T654I mutation in DDR2 in a primary endometrial carcinoma sample (data not shown).
Dasatinib was originally designed as an inhibitor of Src and is a multi-targeted tyrosine kinase inhibitor (34). Dasatinib treatment is associated with toxicity in patients including myelosuppression and the development of pleural and pericardial effusions (35, 36). In an attempt to identify additional agents which could potently inhibit DDR2 with less associated toxicity we screened a panel of 20 tyrosine kinase inhibitors which were predicted to have the potential to inhibit DDR2 based on their respective structures. We found that nilotinib, a second-generation BCR-Abl inhibitor, as well as with AP24534, a third generation BCR-Abl inhibitor which displays activity against BCR-Abl and imatinib-resistant BCR-Abl (36), inhibited the proliferation of SCC lines harboring DDR2 mutations (Figures S2c and S2d). We observed that AP24534 treatment resulted in a greater degree of inhibition than nilotinib which was agreement with calculated in vitro Kd values of 35.4 nM for nilotinib and 9.0 nM for AP24534 as compared to 5.4 nM for dasatinib (Table S1).
sh-RNAs targeting DDR2 kill DDR2-mutant SCC cell lines
As an independent measure of DDR2-dependency we expressed short-hairpin RNAs targeting DDR2 using lentiviral vectors in the NCI-H2286, HCC-366 and NCI-H1703 cell lines. We screened a set of sh-RNA-expressing plasmids for the ability to knock-down DDR2 mRNA expression by real-time PCR in NCI-H2286 cells and selected two hairpins for further analysis given their ability to reduce DDR2 mRNA levels by approximately 50% (data not shown). We observed that knock-down of DDR2 by these two sh-RNAs led to a reduction in proliferation of the two DDR2 mutant cell lines but not of PDGFRA-amplified NCI-H1703 cells which had been sensitive to imatinib and dasatinib in our proliferation assays (Figure 2c). The reduction in proliferation appeared to correlate with the degree of knock-down as the observed phenotype was greater with sh-RNA-2 than sh-RNA-5 (Figure 2c) and appeared to be caused by cell death and not cell cycle arrest (data not shown).
To assess the specificity of the observed knock-down phenotype we performed a similar experiment in NCI-H2286 and HCC-366 cells ectopically expressing their described mutated forms of DDR2 (I638F and L239R respectively) and then knocked-down endogenous DDR2 with sh-RNA-2 which targets the 3′ UTR of DDR2 and would not be expected to interfere with ectopic expression of DDR2. We observed for both NCI-H2286 and HCC-366 that ectopic expression of DDR2 attenuated the anti-proliferative effect of endogenous DDR2 knock-down and that the effect was of greater magnitude in NCI-H2286, perhaps due to a greater degree of off-target effects in HCC-366 (Figure 2d).
DDR2 mutations are associated with dasatinib sensitivity in vivo
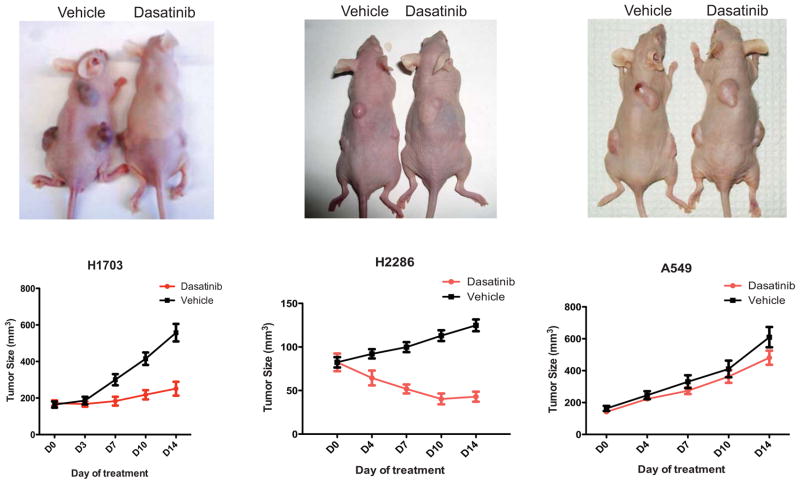
To analyze the effects of dasatinib treatment in a somewhat more physiological setting, we performed xenograft studies in athymic nude mice in which we injected cohorts of mice with NCI-H2286, HCC-366, NCI-H1703 and A549 cells. HCC-366 cells did not form tumors in the mice and could not be analyzed further. Following tumor formation of the three tested lines mice were treated with dasatinib at 50 mg/kg by oral gavage for two weeks or vehicle control. Dasatinib treatment led to a decrease in tumor size in the NCI-H1703 and NCI-H2286 lines but not in A549, consistent with our in vitro results (Figure 3).
Figure 3. Xenografts of squamous lung cancer cell lines demonstrate anti-tumor effects of dasatinib in vivo.
Athymic nu/nu mice were injected subcutaneously with A549, NCI-H1703, HCC-366 and NCI-H2286 cells (n=10) and treated with dasatinib or vehicle for two weeks following tumor formation. Depicted are representative images of mice from each cohort as well as measurements of tumor size. Tumors did not form in the mice injected with HCC-366 and these mice could not be analyzed further.
DDR2 mutations are oncogenic and DDR2-driven transformation is dasatinib-sensitive
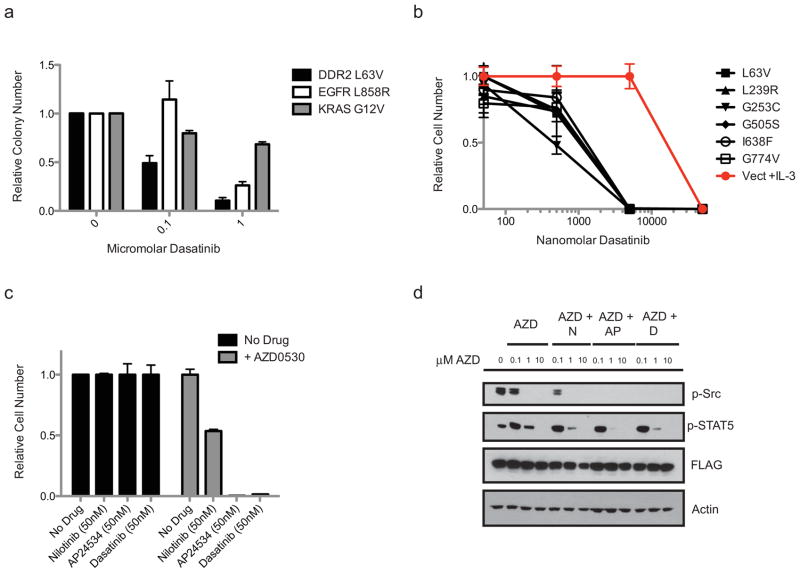
We examined whether DDR2 mutations could confer an oncogenic gain-of-function phenotype. Ectopic expression of a subset of the DDR2 mutants identified in our primary and secondary screens (n=2/6) promoted the formation of colonies in soft agar of NIH-3T3 cells (Figure S3a). Colony formation was greatest in the L63V and I638F mutants at a level comparable to that driven by expression of the gain-of-function L858R mutation in EGFR and modest in the remainder of the genotypes. Colony formation could be inhibited with a single application of dasatinib at the time of plating in the case of the L63V mutant, the mutant which reproducibly formed the most colonies in our assay (Figure 4a). Dasatinib treatment also inhibited the colony formation of NIH-3T3 cells expressing the L858R mutation in EGFR, consistent with previous reports, and did so to a lesser extent in NIH-3T3 cells stably expressing the activating G12V KRAS mutation (Figure 4a) (32, 37).
Figure 4. Ectopic expression of DDR2 mutants leads to cellular transformation which can be blocked by dasatinib or combination tyrosine kinase inhibitor treatment.
(a) Results from soft agar assay in which 3T3 fibroblasts expressing the L63V DDR2 mutation, the L858R EGFR mutation or the KRAS G12V mutation were plated in soft agar in the presence of various concentrations of dasatinib. Colony number of six independent samples with standard errors is shown. (b) Proliferation at four days of Ba/F3 cells expressing vector only or one of six DDR2 mutations shown in cells grown in the presence of dasatinib. For the vector control cells are grown in the presence of IL-3 to maintain viability and in the case of the DDR2 mutants all cells are IL-3 independent and cultured in the absence of IL-3. Proliferation is shown relative to untreated cells at the same time point for triplicate samples with standard errors. (c) Proliferation of Ba/F3 cells expressing DDR2 L63V co-cultured with 50 nM of nilotinib, AP24534 or dasatinib with or without 500 nM AZD0530. Proliferation is relative to untreated cells grown in parallel. (d) Immunoblots of DDR2 L63V transformed Ba/F3 cells treated for two days with the depicted concentrations of AZD0530 (AZD) in addition to 50 nM nilotinib (N), AP24534 (AP) or dasatinib (D). The first lane is an untreated sample. Shown are immunoblots probed with antibodies against phospho-Src Y416, phospho-STAT5 Y694, FLAG-DDR2 and actin.
As the observed gain-of-function phenotype was modest for many of the DDR2 mutants in NIH-3T3 cells, we evaluated the transforming potential of DDR2 in the interleukin-3 (IL-3)-dependent hematopoietic cell line Ba/F3. We observed that ectopic expression of all six DDR2 mutants identified in our primary and secondary screens led to IL-3-independent growth of Ba/F3 cells as did high levels of expression of wild-type DDR2 and no differences were observed in the time to transformation or the rate of IL-3 independent proliferation (Figure S3b). A kinase-dead DDR2 transgene (K608E) did not support the IL-3-independent growth of Ba/F3 cells (Figure S3b). While culture with the less potent DDR2 inhibitor imatinib did not lead to significant killing of Ba/F3 cells expressing DDR2 mutations as compared to cells grown in the presence of IL-3, culture with dasatinib led to cell death in all cell lines expressing DDR2 mutants with a mean calculated IC50 of 680 nM for the mutants and 30 μM for the control (Figures 4b and S3c). The third-generation BCR-Abl inhibitor AP24534 was also effective in killing the IL-3–independent Ba/F3 cells expressing mutant forms of DDR2, suggesting that this class of drugs may be effective against DDR2-driven neoplasms while the second generation BCR-Abl inhibitor nilotinib demonstrated modest activity against the DDR2-tranformed Ba/F3 cells (Figures S3d and S4a). Survival of Ba/F3 cells in the absence of IL-3 was associated with maintenance of STAT5 phosphorylation as has been previously shown (Figure S4b)(38).
DDR2 transformed cell lines maintain Src phosphorylation and are especially sensitive to dual inhibition of DDR2 and Src
Given that the type I kinase inhibitor dasatinib was more potent in DDR2-transformed Ba/F3 cells than the more target-specific type II inhibitors nilotinib and imatinib we sought to test whether the potency of dasatinib in this system might be due to effects of dasatinib on other kinases in addition to DDR2. DDR2 has previously been shown to require Src for maximal kinase activity (16) and we observed that levels of phosphorylated Src were maintained in Ba/F3 cells expressing DDR2 mutants in the absence of IL-3 (Figure S4b). To test whether the ability of DDR2 mutations to confer IL-3-independent proliferation in Ba/F3 cells might depend on both DDR2 and Src activity we treated Ba/F3 cells expressing DDR2 with AZD0530, a highly selective Src-family kinase inhibitor which displays minimal activity against DDR2 as compared to the other inhibitors described in this manuscript (in vitro Kd 291 nM, Table S1) (39). Similar to nilotinib treatment, AZD0530 had a modest effect on the proliferation of the IL-3-independent DDR2-expressing Ba/F3 cells (Figures S4a and S4c). However, when Ba/F3 cells expressing the L63V DDR2 mutation were grown in 50 nM nilotinib, a concentration associated with little effect on proliferation of wild-type Ba/F3 cells or Ba/F3 cells expressing DDR2 mutations (Figure S4a), the addition of AZD0530 led to a marked reduction in proliferation of Ba/F3 cells expressing DDR2 L63V, suggesting that the coordinated activity of DDR2 and Src-family kinases may be required for the ability of DDR2 mutated Ba/F3 cells to grow in the absence of IL-3 and thereby providing a possible explanation for the potency of dasatinib in this system (Figures 4c and S5a). A similar additive effect of AZD0530 was observed when the Ba/F3 cells were co-treated with AZD0530 and 50 nM of either AP24534 or dasatinib (Figure 4c for L63V; for a more detailed version of this experiment including additional DDR2 mutants see Figure S5). AZD0530 reduced Src and STAT5 phosphorylation in a dose-dependent fashion in the DDR2 L63V-expressing Ba/F3 cells when used as a single agent or in combination with nilotinib, AP24534 or dasatinib (Figures 4d and S4d).
Observation of a DDR2 kinase domain mutation in a clinical trial subject with a radiographic response to combination therapy with dasatinib and erlotinib
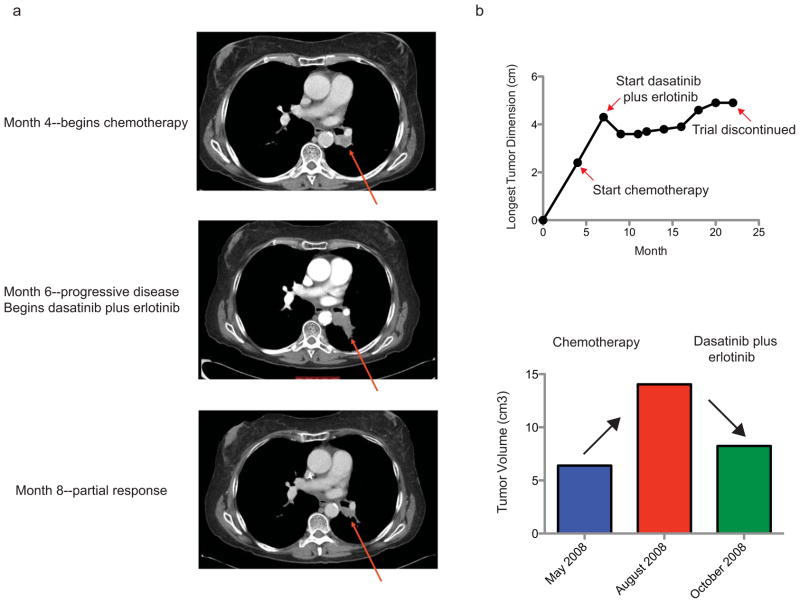
Two recent early-phase clinical trials of dasatinib have been reported in which subjects with advanced stage lung cancer were treated with either dasatinib or a combination of dasatinib and erlotinib (29, 40). One of seven subjects with a squamous cell lung cancer exhibited a significant shrinkage in tumor size while undergoing therapy with a combination of dasatinib and erlotinib, and unlike the other subject on study with lung adenocarcinoma who exhibited a response to treatment, there was no evidence of EGFR mutation in the subject with squamous cell lung cancer. The patient was a 59 year old Caucasian woman with a 1/3 pack per day smoking history for 38 years who quit one year before her diagnosis of lung cancer. She was found to have a left lower lobe stage I (T2N0M0) squamous cell lung cancer and received primary treatment with weekly carboplatin and paclitaxel with concomitant 70 Gy of radiation resulting in a complete response. However, approximately one year later she developed progression of disease within the radiation field and treatment was initiated with s standard dose carboplatin and paclitaxel without response. She then began combination dasatinib and erlotinib therapy on protocol. A restaging CT scan after nearly 2 months indicated tumor shrinkage and the patient experienced improved symptoms (resolved dyspnea and cough) (Figures 5A and 5B). She remained on treatment for 14 months until therapy had to be discontinued secondary to treatment-induced airspace disease and pleural effusions.
Figure 5. Radiographic response of a patient with a S768R DDR2 mutation treated with dasatinib plus erlotinib.
(a) CT scan images shown from a lung SCC patient who was treated with chemotherapy and later with dasatinib plus erlotinib. Serial CT scans are shown at the time of initiation of chemotherapy, initiation of study treatment with dasatinib and erlotinib and following two months of treatment with dasatinib plus erlotinib. (b) Top panel: Tumor dimension measurements from the subject above starting four months prior to chemotherapy treatment and extending to the time at which combination therapy with dasatinib and erlotinib was discontinued. Bottom panel: Bar graph depicting measured tumor volume by RECIST criteria of the subject prior to chemotherapy, following chemotherapy and following two months of dasatinib plus erlotinib therapy.
We performed directed sequencing of DDR2 in a pre-treatment tumor specimen derived from this individual and identified a novel DDR2 kinase domain mutation, S768R, which was present in 844 of 3020 (28%) of reads obtained by 454 sequencing (data not shown) and independently verified by Sanger sequencing (Figure S6). The mutation could not be verified as somatic as no normal DNA was available for this individual who is deceased. There were no other SCC subjects who responded to therapy on this study or a subsequent study of dasatinib alone (n=13 total) to further explore this correlation. We performed three-dimensional modeling of the S768R mutation in the context of the DDR2 kinase domain which suggested that the S768R substitution is likely to alter the kinase activity of DDR2 (Figure S7).
DISCUSSION
We report the identification of 11 novel mutations in DDR2 in a screen of 290 SCC samples, yielding an overall mutation rate of 3.8% in all samples and 3.2% in primary SCCs, a rate comparable to the fraction of lung adenocarcinoma patients bearing ALK fusions, a genomic event associated with dramatic responses to the ALK inhibitor crizotinib(10, 41). It remains an important question at this time whether DDR2 mutations will be primarily found in SCCs of the lung or whether they may be present in SCCs originating from other tissue types such as the head and neck or skin or in tumors of different histologies. Additionally, the rate of DDR2 mutations was lower in our validation screen than in our initial mutation screens, and it will likely take additional sequencing efforts, such as the upcoming Cancer Genome Atlas (TCGA), to further define the prevalence of DDR2 mutations. Interestingly, we have found two additional DDR2 mutations in a sample set of endometrial carcinomas (T654I and T685S) as well as a mutation in a colorectal cancer patient (T692N), supporting the possibility that DDR2 mutations may be present in multiple cancer types. A search of the COSMIC database is notable for DDR2 mutations in renal cell carcinoma, glioblastoma multiforme and the previously mentioned lung adenocarcinoma samples. Furthermore, our initial and secondary screens consisted largely of samples from the United States while the validation screen consisted of more samples from European patients, suggesting that demographics may also affect the rate of observed DDR2 mutations. A recent report in which over 1500 genes were sequenced in a cohort of 63 squamous cell lung cancers using mismatch repair technology did not identify any DDR2 mutations (42), though we calculate that sample size was not large enough to detect a statistically significant difference in the rates of DDR2 mutations as compared to our study assuming a power of 0.8 and alpha of 0.05 (43).
We evaluated the effects of ectopic expression of six mutant forms of DDR2 in NIH 3T3 cells and Ba/F3 cells and showed that mutated DDR2 could function as an oncogene in either context, though with differing potency. We did not complete an assessment of all identified DDR2 mutants nor did we evaluate the effects of expression of mutated DDR2 in the more appropriate context of primary squamous lung cells in a mouse or other model organism. The creation of these models is currently underway and will be critical to more fully characterize the function of mutated DDR2.
The precise mechanism by which mutated DDR2 promotes cellular transformation is unclear. While ectopic expression of DDR2 correlated with STAT5 and Src phosphorylation in transformed Ba/F3 cells and chemical inhibition of Src and DDR2 appeared to exhibit an additive if not synergistic effect in DDR2-transformed Ba/F3 cells, the mechanism by which mutations in DDR2 activate downstream signaling is not known. It is possible that the kinase domain mutations, in a manner similar to the modeled mutation at S768, alter the kinase activity of DDR2. The observation that ectopic expression of wild-type DDR2 was sufficient to transform Ba/F3 cells suggests that increased DDR2 signaling activity is a potential mechanism of transformation. It is also possible that the mutations in the discoidin domain or unclassified regions of DDR2 could impact upon the ligand binding or localization of DDR2, as prior reports have shown that DDR2 mutations in familial Spondylo-meta-epiphyseal dysplasia (SMED) alter the ligand binding and membrane trafficking of DDR2(44, 45).
We report the novel identification of recurrent somatic mutations in the DDR2 kinase gene and show that dasatinib can efficiently inhibit the proliferation of DDR2-mutated SCC cell lines in vitro and in vivo as well as cells ectopically expressing mutant DDR2. Together, these data identify a potential first therapeutic target in lung SCC for which clinically approved drugs already exist, thereby providing a rationale for clinical trials of tyrosine kinase inhibitors in this disease. We additionally report a DDR2 kinase domain mutation in a patient with squamous cell lung cancer who exhibited a radiographic response to the combination of dasatinib and erlotinib who did not harbor an EGFR mutation. While this is an interesting result we feel that caution should be exercised in its interpretation given that only one response was reported in a SCC patient in the trial, thus precluding a thorough assessment of this correlation. Additionally, the subject discontinued therapy in the context of unacceptable toxicity after 14 months of treatment, suggesting that investigation of tyrosine kinase inhibitors other than dasatinib as anti-DDR2 agents is warranted. While it is not possible to definitively conclude that this patient responded specifically to dasatinib treatment due to a DDR2 mutation, we feel it is unlikely that the response was due to erlotinib given the absence of an EGFR kinase domain mutation and the previously reported GI50 of 990 nM for erlotinib in the DDR2 mutant HCC-366 cell line(32). Additionally, a search of the literature did not identify any prior reports of erlotinib as a potent inhibitor of DDR2 and initial experiments we have done in the DDR2-mutant cells lines HCC-366 and NCI-H2286 indicate a sensitivity to erlotinib which is at least ten-fold less than dasatinib (data not shown).
In conclusion, we hope that our data may stimulate the initiation of larger clinical trials of dasatinib or other tyrosine kinase inhibitors in lung SCC patients and testing of these patients for DDR2 mutations, potentially leading to a less toxic and more effective treatment for a subset of patients with this deadly disease.
MATERIALS AND METHODS
Ethics Statement
All animal experiments were performed under an IACUC-approved animal protocol at the Dana Farber Cancer Institute and all experiments performed with human DNA were performed in accordance with Institutional Review Board (IRB) approved protocols as is described below.
Collection of Squamous Cell Lung Cancer Samples
For the primary and secondary screens tumor samples were obtained from the Dana Farber Cancer Institute/Brigham and Women’s Hospital/Harvard Cancer Center under institutional protocol 02-180, approved by the Dana Farber/Harvard Cancer Center IRB in September 2002 and renewed yearly thereafter. This is a general tissue collection protocol for patients with lung cancer who are consented to tissue collection for research, including DNA sequencing, prior to surgery. All patients with resectable biopsy-proven squamous cell lung cancers (as diagnosed by a board-certified Anatomic Pathologist) were eligible and eligible subjects underwent a detailed informed consent procedure prior to enrollment on the protocol which included a discussion of the use of tissue samples for DNA sequencing studies and written documentation of consent. In addition, DNA samples from de-identified squamous cell lung cancer patients were obtained from the Ontario Cancer Institute for the primary and secondary screens as part of a Dana Farber/Harvard Cancer Center IRB-approved collection of de-identified tumor samples for DNA sequencing studies. IRB approval for collection of de-identified samples was subject to a review of the local IRB-approved protocols for all external sites to ensure an adequate informed consent process had taken place.
The validation screen was performed at the University of Cologne and Max Planck Institute for Neurological Research. Samples were collected in accordance with a tissue collection protocol approved by the University of Cologne Ethics Committee which involved a detailed informed consent process including a discussion of genetic testing prior to the subject’s surgery with written documentation of consent. Again, all patients with biopsy-proven squamous cell lung cancer were eligible regardless of disease stage as long as their tumors were considered resectable. De-identified samples were also collected from additional European sites including Haukeland University Hospital, University Hospital Zurich, Université Joseph Fourier, Oslo University Hospital, Jena University Hospital and University Medical Centre Groningen. At all sites samples were obtained in accordance with an IRB approved tissue collection protocol and the collection of de-identified samples from these sites was approved by the University of Cologne Ethics Committee after a review of local collection protocols.
For the single patient sample obtained from the recent clinical trial of combination therapy with dasatinib and erlotinib for advanced lung cancer DNA was provided to the University of Cologne by the H. Lee Moffitt Cancer Center. DNA was obtained under an IRB-approved protocol at the H. Lee Moffitt Cancer Center and the sequencing of the de-identified sample by both 454 and Sanger sequencing was performed with the approval of the University of Cologne Ethics Committee after a review of the collection protocol.
In all instances specimens were continuously selected at the site of surgery in order to avoid sampling bias and all samples were de-identified prior to processing for DDR2 sequencing. When available, de-identified correlative clinical data was provided with the samples, though this data was not available to the investigators prior to sample genotyping. Patients with a prior history of tumors involving a visceral organ site were excluded to avoid the inclusion of metastases.
DDR2 Sequencing
DDR2 was sequenced from genomic DNA obtained from squamous lung cancer cell lines and patient samples by conventional Sanger sequencing. In the discovery set 20 patient samples and matched normal DNA were used for sequencing 201 genes including 90 kinases. All mutations were verified as somatic. Mutations were identified using an automated mutation caller and then verified manually with comparison made to the matched normal sequence in the case of all primary tumor samples. In the secondary screen 35 additional patient samples and 13 SCC cell lines were used for sequencing the six mutated tyrosine kinases identified in the primary screen (DDR2, FGFR2, NTRK2, JAK2, CDK8 and FLT3). In the validation screen 222 total samples underwent sequencing of the DDR2 gene. In all cases except D125Y matched normal DNA was available to verify the mutation as somatic.
Cell Culture
A549, NCI-H2286, HCC-366 and NCI-H1703 cells were obtained from the core collection at the Dana Farber Cancer Institute, having previously been purchased from the ATCC and used to establish a collection of early passage lung cancer cell lines which were analyzed by fingerprinting and SNP arrays (32). All cells used for the experiments described in this manuscript were obtained from freezes made at that time and no further validation was performed. Lung cancer cell lines were grown in RPMI (Invitrogen) with 10% fetal calf serum, NIH-3T3 cells were grown in DMEM (Mediatech) with 10% serum and Ba/F3 cells in RPMI supplemented with 10% serum and IL-3 (BD Biosciences) at 10 ng/ml. For IL-3 withdrawal experiments Ba/F3 cells were collected via centrifugation, washed once in sterile PBS and then resuspended in media without IL-3. Colony formation assays in NIH-3T3 cells were performed in six-well plates in which 25,000 NIH-3T3 cells were plated in triplicate in 1 ml of 0.33% top agar on top of 2 ml of 0.5% bottom agar. After three weeks colonies were counted using the NIH ImageJ software.
Vectors
The full-length DDR2 cDNA was obtained from Origene and cloned into the EcoRI site of the retroviral vector p-Wzl-Blast and p-Babe-puro following the addition of a c-terminal FLAG tag by PCR. Mutants were generated by site-directed mutagenesis using the Quickchange site directed mutagenesis kit (Strategene). All mutations were verified by sequencing. sh-RNA lentiviral vectors for DDR2 were obtained from The RNAi Consortium at the Broad Institute (46, 47). DDR2 sh-RNA-2 corresponds to TRC clone TRCN0000121117 with hairpin sequence 5′-CCGG-CCCATGCCTATGCCACTCCAT-CTCGAG-ATGGAGTGGCATAGGCATGGG-TTTTTG-3′ targeting the 3′ UTR of DDR2. DDR2 sh-RNA-5 corresponds to TRC clone TRCN0000121121 with hairpin sequence 5′-CCGG-CCCTGGAGGTTCCATCATTTA-CTCGAG-TAAATGATGGAACCTCCAGGG-TTTTTG-3′ targeting the coding sequence of DDR2. Both hairpins were provided in the pLKO vector. A hairpin targeting GFP (shGFP) was obtained from TRC as well and used as a control.
Viral Infections
The DDR2 transgene was expressed in the lung cancer cell lines, NIH-3T3 cells and Ba/F3 cells using retroviral transduction with the pWzl vector, as has been previously described. Briefly, 293T cells were used to generate the virus with the appropriate pWzl or pBabe vector and packaging vector transfected using Fugene (Roche). Cells were subjected to two rounds of overnight infection in the presence of polybrene and stable cells generated using blasticidin selection at 10 μg/ml for 3T3, Ba/F3 and A549, 2 μg/ml NCI-H2286 and for NCI-H1703 and 1 μg/ml for HCC-366. Lentiviral infections were performed per the on-line TRC protocol (48) with 293T cells transfected with the suggested three vector combination of pLKO, VSVG and delta 8.9. Virus was collected and used to infect the lung cancer cell lines for six hours in the presence of polybrene. Stable cell lines were generated using puromycin selection at a concentration of 2 μg/ml for NCI-H2286 and 4 μg/ml for NCI-H1703, A549 and HCC-366.
Cell Proliferation and Viability Assays
Cell proliferation was measured with the Cell-Titer-Glo reagent (Promega) per the manufacturer’s instructions. For experiments with the SCC cell lines cells were plated in clear-bottomed 96 well plates at a density of 1500 cells per well. The following day the drug was added and cell proliferation was measured six days later for the SCC cell lines. For Ba/F3 cells were plated at 5000 cells per well and the drug added the same day. Proliferation was measured four days later. Proliferation measurements were made using a standard 96 well plate luminometer/plate reader. Data are shown as relative values in which the luminescence at a given drug concentration is compared to untreated cells of the same cell type. Kinase inhibitors were purchased from LC Labs or were synthesized by Nathanael Gray’s laboratory at Harvard Medical School. In vitro IC50s for DDR2 were determined for all compounds by LanthaScreen TR-FRET kinase activity assays performed by Invitrogen. Cell viability was measured using a vi-Cell reader to stain cells with trypan blue and to generate 50 independent images for each measured sample. Annexin V (BD Biosciences) analysis was performed on dasatinib treated cells 48 hours after addition of drug per the manufacturer’s protocol. For sh-RNA experiments cells were plated at a density of 1500 cells per well in 96 well plates following puromycin selection. Proliferation was measured four days later as compared to cells expressing a hairpin targeting GFP.
Immunoblots
Immunoblots were performed using the Nupage system (Invitrogen) per the manufacturer’s protocol. Cells were lysed in 1% NP-40 with protease (Roche) and phosphatase inhibitors (Calbiochem) and protein concentration assayed with the Bradford reagent (Bio-Rad). Primary antibodies used were Flag-M2 (Sigma), phospho-Y417-Src (Cell Signaling Technologies), phospho-Y694-STAT5 (Cell Signaling) and Actin (Santa Cruz). A DDR2 antibody was generated for this project by Bethyl Labs. Secondary HRP-conjugated antibodies were all obtained from Pierce and proteins detected by pico-ECL (Thermo Scientific). Images were imported into Adobe Illustrator using an Epson 4490 scanner. In some cases brightness and/or contrast of the scanned images was adjusted for clarity and blots were cropped to display the area of interest in the displayed figures. In all cases adjustment of brightness or contrast the adjustment was applied uniformly to the image as a whole.
Xenografts
All animal experiments were performed according to institutional guidelines regarding animal safety. Nude mice were injected with the lung cancer cell lines at a density of 2.5 (A549), 3.0 (NCI-H1703) or 5.0 (NCI-H2286 and HCC-366) million cells per injection to try to control for the variable rates of tumor growth in the animals. Cohorts of ten mice were injected at three sites for each cell type and the mice were observed until the tumor volume approached 150 cubic milliliters for A549 and NCI-H1703 or 100 cubic milliliters for NCI-H1703. At that time mice were treated with dasatinib at 50 mg/kg or vehicle control daily for two weeks and tumor size measured during the treatment period.
Statistics
For proliferation and colony formation assays mean values from a minimum of triplicate samples are reported as well as standard errors as calculated by Microsoft Excel. IC50 values were obtained using Graph Pad Prism software. Power and sample size calculations were performed using the Interactive Statistics Web Resource(43).
Supplementary Material
SIGNIFICANCE.
DDR2 mutations are present in approximately 4% of squamous cell lung cancers and DDR2 mutations are associated with sensitivity to dasatinib. These findings provide a rationale for the design of clinical trials of the FDA-approved drug dasatinib in patients with squamous cell lung cancer.
Acknowledgments
We would like to thank Eric McIntush of Bethyl Laboratories for his efforts in producing DDR2 antibodies.
Footnotes
Financial Disclosure: P.S.H. is the recipient of a Department of Defense Lung Cancer Research Fellowship # LC090577. C.B. and E.B. are supported by PNES INCA grant 2008. E.B.H. is supported by the SPORE grant (P50-CA119997). RKT is supported by the Deutsche Krebshlife (grant 107954), by the German Ministry of Science and Education (BMBF) as part of the NGFNplus program (grant 01GS08100), by the Max Planck Society (M.IF.A.NEUR8061) and by the Deutsche Forschungsgemeinschaft (DFG) through SFB832 (TP6). This work was supported by grants to M.M. from the American Lung Association, United Against Lung Cancer, the Sarah Thomas Monopoli Fund, the Seaman Foundation and Genentech.
Conflicts of Interest: M.M. is a consultant to Novartis and receives research support from Novartis, receives research support from Genentech, is a founding advisor and consultant to, and an equity holder in, Foundation Medicine and patent holder for EGFR mutation testing, licensed to Genzyme Genetics. N.S.G. receives research support from Novartis. E B.H. was the principal investigator of an industry-sponsored clinical trial of dasatinib and erlotinib in lung cancer funded in part by Bristol-Myers-Squibb and the American Society for Clinical Oncology. R.K.T. reports consulting and lecture fees (Sequenom, Sanofi-Aventis, Merck, Roche, Infinity, Boehringer, Astra-Zeneca, Johnson&Johnson, Atlas-Biolabs) and research support (Novartis, AstraZeneca).
References
- 1.Lennes IT, Lynch TJ. Quality indicators in cancer care: development and implementation for improved health outcomes in non-small-cell lung cancer. Clin Lung Cancer. 2009;10(5):341–6. doi: 10.3816/CLC.2009.n.046. [DOI] [PubMed] [Google Scholar]
- 2.West H, Harpole D, Travis W. Histologic considerations for individualized systemic therapy approaches for the management of non-small cell lung cancer. Chest. 2009;136(4):1112–8. doi: 10.1378/chest.08-2484. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97(5):339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki T, Rodig SJ, Chirieac LR, Janne PA. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer. 2010 doi: 10.1016/j.ejca.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammerman PS, Janne PA, Johnson BE. Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. Clin Cancer Res. 2009;15(24):7502–9. doi: 10.1158/1078-0432.CCR-09-0189. [DOI] [PubMed] [Google Scholar]
- 9.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell. 1997;1(1):13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 13.Vogel W. Discoidin domain receptors: structural relations and functional implications. FASEB J. 1999;13 (Suppl):S77–82. doi: 10.1096/fasebj.13.9001.s77. [DOI] [PubMed] [Google Scholar]
- 14.Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, et al. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol Cell. 1997;1(1):25–34. doi: 10.1016/s1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- 15.Leitinger B. Molecular analysis of collagen binding by the human discoidin domain receptors, DDR1 and DDR2. Identification of collagen binding sites in DDR2. J Biol Chem. 2003;278(19):16761–9. doi: 10.1074/jbc.M301370200. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda K, Wang LH, Torres R, Zhao H, Olaso E, Eng FJ, et al. Discoidin domain receptor 2 interacts with Src and Shc following its activation by type I collagen. J Biol Chem. 2002;277(21):19206–12. doi: 10.1074/jbc.M201078200. [DOI] [PubMed] [Google Scholar]
- 17.Olaso E, Labrador JP, Wang L, Ikeda K, Eng FJ, Klein R, et al. Discoidin domain receptor 2 regulates fibroblast proliferation and migration through the extracellular matrix in association with transcriptional activation of matrix metalloproteinase-2. J Biol Chem. 2002;277(5):3606–13. doi: 10.1074/jbc.M107571200. [DOI] [PubMed] [Google Scholar]
- 18.Labrador JP, Azcoitia V, Tuckermann J, Lin C, Olaso E, Manes S, et al. The collagen receptor DDR2 regulates proliferation and its elimination leads to dwarfism. EMBO Rep. 2001;2(5):446–52. doi: 10.1093/embo-reports/kve094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford CE, Lau SK, Zhu CQ, Andersson T, Tsao MS, Vogel WF. Expression and mutation analysis of the discoidin domain receptors 1 and 2 in non-small cell lung carcinoma. Br J Cancer. 2007;96(5):808–14. doi: 10.1038/sj.bjc.6603614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies H, Hunter C, Smith R, Stephens P, Greenman C, Bignell G, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65(17):7591–5. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 21.Day E, Waters B, Spiegel K, Alnadaf T, Manley PW, Buchdunger E, et al. Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur J Pharmacol. 2008;599(1–3):44–53. doi: 10.1016/j.ejphar.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Raponi M, Zhang Y, Yu J, Chen G, Lee G, Taylor JM, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66(15):7466–72. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 23.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41(11):1238–42. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stegmeier F, Warmuth M, Sellers WR, Dorsch M. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin Pharmacol Ther. 2010;87(5):543–52. doi: 10.1038/clpt.2009.297. [DOI] [PubMed] [Google Scholar]
- 26.Manley PW, Drueckes P, Fendrich G, Furet P, Liebetanz J, Martiny-Baron G, et al. Extended kinase profile and properties of the protein kinase inhibitor nilotinib. Biochim Biophys Acta. 2010;1804(3):445–53. doi: 10.1016/j.bbapap.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Rix U, Fang B, Bai Y, Edwards A, Colinge J, et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6(4):291–9. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26(1):127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 29.Haura EB, Tanvetyanon T, Chiappori A, Williams C, Simon G, Antonia S, et al. Phase I/II study of the Src inhibitor dasatinib in combination with erlotinib in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(8):1387–94. doi: 10.1200/JCO.2009.25.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102(27):9625–30. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos AH, Dutt A, Mermel C, Perner S, Cho J, Lafargue CJ, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther. 2009;8(21):2042–50. doi: 10.4161/cbt.8.21.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119(6):1727–40. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nat Biotechnol. 2009;27(1):77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quintas-Cardama A, Kantarjian H, Cortes J. Imatinib and beyond--exploring the full potential of targeted therapy for CML. Nat Rev Clin Oncol. 2009;6(9):535–43. doi: 10.1038/nrclinonc.2009.112. [DOI] [PubMed] [Google Scholar]
- 35.Kim LC, Rix U, Haura EB. Dasatinib in solid tumors. Expert Opin Investig Drugs. 2010;19(3):415–25. doi: 10.1517/13543781003592097. [DOI] [PubMed] [Google Scholar]
- 36.O’Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16(5):401–12. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song L, Morris M, Bagui T, Lee FY, Jove R, Haura EB. Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006;66(11):5542–8. doi: 10.1158/0008-5472.CAN-05-4620. [DOI] [PubMed] [Google Scholar]
- 38.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18(17):4754–65. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hennequin LF, Ballard P, Boyle FT, Delouvrie B, Ellston RP, Halsall CT, et al. Novel 4-anilinoquinazolines with C-6 carbon-linked side chains: synthesis and structure-activity relationship of a series of potent, orally active, EGF receptor tyrosine kinase inhibitors. Bioorg Med Chem Lett. 2006;16(10):2672–6. doi: 10.1016/j.bmcl.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 40.Johnson FM, Bekele BN, Feng L, Wistuba I, Tang XM, Tran HT, et al. Phase II study of dasatinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(30):4609–15. doi: 10.1200/JCO.2010.30.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDermott U, Iafrate AJ, Gray NS, Shioda T, Classon M, Maheswaran S, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68(9):3389–95. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 42.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 43.Available from: http://statpages.org/proppowr.html
- 44.Bargal R, Cormier-Daire V, Ben-Neriah Z, Le Merrer M, Sosna J, Melki J, et al. Mutations in DDR2 gene cause SMED with short limbs and abnormal calcifications. Am J Hum Genet. 2009;84(1):80–4. doi: 10.1016/j.ajhg.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali BR, Xu H, Akawi NA, John A, Karuvantevida NS, Langer R, et al. Trafficking defects and loss of ligand binding are the underlying causes of all reported DDR2 missense mutations found in SMED-SL patients. Hum Mol Genet. 2010;19(11):2239–50. doi: 10.1093/hmg/ddq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 47.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(51):20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Available from: http://www.broadinstitute.org/rnai/public/resources/protocols
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.