Abstract
The Gram-negative bacterium Sideroxydans lithotrophicus ES-1 (ES-1) grows on FeCO3 or FeS at oxic–anoxic interfaces at circumneutral pH, and the ES-1-mediated Fe(II) oxidation occurs extracellularly. However, the molecular mechanisms underlying ES-1’s ability to oxidize Fe(II) remain unknown. Survey of the ES-1 genome for candidate genes for microbial extracellular Fe(II) oxidation revealed that it contained a three-gene cluster encoding homologs of Shewanella oneidensis MR-1 (MR-1) MtrA, MtrB, and CymA that are involved in extracellular Fe(III) reduction. Homologs of MtrA and MtrB were also previously shown to be involved in extracellular Fe(II) oxidation by Rhodopseudomonas palustris TIE-1. To distinguish them from those found in MR-1, the identified homologs were named MtoAB and CymAES-1. Cloned mtoA partially complemented an MR-1 mutant without MtrA with regards to ferrihydrite reduction. Characterization of purified MtoA showed that it was a decaheme c-type cytochrome and oxidized soluble Fe(II). Oxidation of Fe(II) by MtoA was pH- and Fe(II)-complexing ligand-dependent. Under conditions tested, MtoA oxidized Fe(II) from pH 7 to pH 9 with the optimal rate at pH 9. MtoA oxidized Fe(II) complexed with different ligands at different rates. The reaction rates followed the order Fe(II)Cl2 > Fe(II)–citrate > Fe(II)–NTA > Fe(II)–EDTA with the second-order rate constants ranging from 6.3 × 10−3 μM−1 s−1 for oxidation of Fe(II)Cl2 to 1.0 × 10−3 μM−1 s−1 for oxidation of Fe(II)–EDTA. Thermodynamic modeling showed that redox reaction rates for the different Fe(II)-complexes correlated with their respective estimated reaction-free energies. Collectively, these results demonstrate that MtoA is a functional Fe(II)-oxidizing protein that, by working in concert with MtoB and CymAES-1, may oxidize Fe(II) at the bacterial surface and transfer released electrons across the bacterial cell envelope to the quinone pool in the inner membrane during extracellular Fe(II) oxidation by ES-1.
Keywords: Sideroxydans lithotrophicus ES-1, extracellular Fe(II) oxidation, decaheme c-type cytochrome MtoA, pH-dependent, ligand complexation
Introduction
The contribution of Fe(II)-oxidizing bacteria (FeOB) to iron cycling in freshwater, groundwater, and marine environments, as well as in most soils and sediments, has been well recognized (Emerson et al., 2010). A variety of neutrophilic and acidophilic Fe(II)-oxidizing microorganisms have the ability to derive energy for growth from the oxidation of dissolved or structural Fe(II) under either oxic or anoxic conditions. Unlike aerobic acidophilic or anaerobic neutrophilic FeOB, the geologic importance of aerobic neutrophilic FeOB has long been neglected because of the rapid auto-oxidation of Fe(II) by O2 at circumneutral pH. However, recent studies indicate that aerobic neutrophilic FeOB would play a key role in microoxic niches with low levels of O2 concentration, where microbial Fe(II)-oxidation can compete with the chemical oxidation of Fe(II). For example, the Gram-negative bacterium Sideroxydans lithotrophicus ES-1 (ES-1), originally isolated from the ground water with Fe(II) at neutral pH in MI, USA, grows on FeCO3 or FeS at oxic–anoxic interfaces (Emerson and Moyer, 1997; Emerson et al., 2007). ES-1 does not grow on Mn(II) oxides, sulfide, or organic carbon sources, such as acetate, pyruvate, and glucose, and does not reduce Fe(III) oxides. Fe(III) (oxy)(hydr)oxide precipitates are closely associated with the ES-1 cells, but do not form sheath- or stalk-like structures (Emerson and Moyer, 1997). Recently, the ES-1 genome was sequenced (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). However, the molecular mechanism by which ES-1 oxidizes Fe(II) remains unknown.
Because Fe(III) oxides produced from biotic Fe(II)-oxidation are usually sparingly soluble at circumneutral pH and in the absence of complexing ligands, bacteria oxidize Fe(II) extracellularly presumably to avoid accumulation of Fe(III) oxides inside their cells. To overcome the physical separation between the bacterial inner membrane where microbial oxidases are located and bacterial cell surface, FeOB have evolved different electron transfer pathways that link the inner membrane to the cell surface. The pathways identified to date include Cyc-2/Rus/Cyc-1 of Acidithiobacillus ferrooxidans, PioABC of Rhodopseudomonas palustris TIE-1 and FoxEYZ of Rhodobacter strain SW2 (Appia-Ayme et al., 1999; Yarzabal et al., 2002; Croal et al., 2007; Jiao and Newman, 2007; Castelle et al., 2008). Although these systems are phylogenetically unrelated, they all have at least one c-type cytochrome (c-Cyt) as a key electron transfer protein. These c-Cyts work in concert with other proteins, often in the form of protein–protein complexes that can span the entire microbial cell envelope to facilitate electron conductance between the inner membrane and Fe(II) external to the bacterial cell.
Notably, PioAB of R. palustris TIE-1 are homologs of MtrAB of the Fe(III)-reducing bacterium Shewanella oneidensis MR-1 (MR-1; Jiao and Newman, 2007). In MR-1, MtrA is a decaheme c-Cyt, while MtrB is a trans-outer membrane (OM), porin-like protein. They form a tight protein complex that transfers electrons across the OM to MtrC and OmcA, two OM decaheme c-Cyts that are localized on bacterial outermost surface (Ross et al., 2007; Shi et al., 2008; Hartshorne et al., 2009; Lower et al., 2009; Reardon et al., 2010). MtrABC and OmcA are key components of the MR-1 extracellular electron transfer pathway, which also includes a tetraheme c-Cyt CymA in the inner membrane. Together, they facilitate electron transfer from the quinone/quinol pool in the inner membrane across the periplasm, through the OM, to the surface of Fe(III) oxides (Richardson, 2000; Shi et al., 2007, 2009; Fredrickson et al., 2008). In addition to mediating electron transfer to and from Fe, MtrAB homologs are also involved in extracellular reduction of dimethylsulfoxide by MR-1 and are hypothesized to be the prototype of a model system for electron transfer across the bacterial OM (Gralnick et al., 2006; Hartshorne et al., 2009).
To investigate the molecular mechanism used by ES-1 for oxidizing Fe(II), we searched the ES-1 genome for the homologs of Cyc-2/Rus/Cyc-1 of A. ferrooxidans, FoxEYZ of Rhodobacter strain SW2 and PioAB/MtrAB. This search identified a three-gene cluster that encoded MtrA, MtrB, and CymA homologs. To distinguish them from those found in Fe(III)-reducing bacteria, we named the identified homologs MtoAB and CymAES-1. Cloned mtoA partially complemented an MR-1 mutant without MtrA in ferrihydrite (FH) reduction. Recombinant MtoA was purified following overexpression in MR-1 cells and characterized systematically. Purified MtoA was found to be a decaheme c-Cyt and able to oxidize soluble Fe(II) in vitro. Collectively, these results suggest that MtoA is a Fe(II)-oxidizing protein that works in concert with MtoB and CymAES-1 to mediate electron transfer reactions from the cell surface to the inner membrane during extracellular Fe(II) oxidation by ES-1.
Materials and Methods
Standard procedures
Protein concentrations were measured with a bicinchoninic acid (BCA) protein assay kit from Pierce (Rockford, IL, USA). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were conducted according to the instructions from Invitrogen (Carlsbad, CA, USA). To visualize proteins directly, gels were stained with GelCode blue stain from Pierce. Heme staining was carried out according the protocol described by Thomas et al. (1976). RGS-His antibody was used for detecting the recombinant protein tagged with RGS-His epitope by Western blot analysis (QIAGEN, Valencia, CA, USA). Kanamycin was used at 25 μg/ml.
Gene identification and cloning
The approach used to identify MtoAB from the ES-1 genome was similar to those described previously (Shi et al., 1998; Shi and Zhang, 2004). MR-1 MtrAB and PioAB of R. palustris TIE-1 were used as templates to search for open reading frames (ORFs) whose predicted peptide products displayed similarity to the MtrAB/PioAB by BLAST (E < 0.01). Following their identification, the polypeptide sequences of MtoA, MtoB, or CymAES-1 and MtrA homologs, MtrB homologs, or CymA homologs of 19 sequenced Shewanella strains described by Fredrickson et al. (2008) and PioA or PioB were used to construct phylogenetic trees with the neighbor-joining-based ALGNX program of Vector NTI (Invitrogen). Likewise, the ES-1 genome was also searched for the homologs of Cyc-2/Rus/Cyc-1 of A. ferrooxidans and FoxEYZ of Rhodobacter strain SW2. The ORF for the identified MtrA homolog was synthesized and then cloned into a protein expression vector pJexpress 401 to create pLS279 by DNA 2.0 (Menlo Park, CA, USA). pLS279 was introduced into MR-1 ΔmtrB–mtrD (locus tags SO_1776-SO_1782) or ΔmtrA (SO_1777) mutant by electroporation to create LS587 and LS597, respectively (Shi et al., 2005; Hartshorne et al., 2009; Clarke et al., 2011). ΔmtrA mutant was also transformed with pJexpress 401 to create LS620.
Ferrihydrite reduction
For ferrihydrite (FH) reduction, MR-1, LS597, and LS620 were grown in PIPES-buffered M1 medium with 20 mM sodium lactate aerobically at 30°C for 16 h with agitation (150 rpm) and harvested by centrifugation (5000 × g, 5 min). Harvested cells were washed once with the same medium, purged with N2, and transferred to Balch tubes at final concentration of 1 × 108 cells/ml. FH was added at final concentration of 10 mM. The tubes were incubated horizontally at 30°C with shaking (25 rpm; Shi et al., 2011). At predetermined time points, 0.5 N HCl extractable Fe(II) was determined by the ferrozine assay (Stookey, 1970).
Protein purification
MtoA was purified from a mutant without major MR-1 c-Cyts, such as MtrA, MtrC, and OmcA. Compared to that in wt, yields of purified c-Cyts from this mutant were often two to three times higher. LS587 was grown aerobically in Tryptic Broth at 30°C with agitation (150 rpm) until the culture reached an optical density at 600 nm of 0.6. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM. The LS587 cells were grown for another 17 h and then harvested by centrifugation at 6000 × g for 15 min. The harvested cells were washed once with buffer A (20 mM HEPES, pH 7.8, 150 mM NaCl) and stored at −20°C. Frozen cell pellets were resuspended in buffer B [buffer A + protease inhibitor (Roche Diagnostic, Indianapolis, IN, USA)] in a ratio of 5 ml/g wet weight cells. The cells were lysed by passage through a French press three times at 8000 lbf/in2. The unbroken cells and debris were removed by centrifugation at 15,000 × g for 30 min. The supernatant was transferred to ultracentrifugation tubes and further centrifuged at 150,000 × g for 1 h. The supernatant was loaded onto a Ni2+–nitrilotriacetic acid (NTA) agarose column pre-equilibrated with buffer B. The column was washed with following buffers in sequential order: buffer B, buffer C [buffer B + 10% (v/v) glycerol], and buffer D (buffer C + 40 mM imidazole). MtoA was eluted with buffer E (buffer C + 250 mM imidazole; Shi et al., 2005). The fractions containing MtoA were pooled and concentrated. The concentrated MtoA was loaded on a HiLoad 16/60 column of Superdex 200 and eluted with buffer B by means of an ÄKTA explorer fast protein liquid chromatography system (GE Healthcare, Piscataway, NJ, USA). The MtoA-containing fractions were pooled, concentrated, changed to buffer C, aliquoted, and stored at −20°C. All protein purification steps were performed at 4°C.
MALDI–TOF mass spectrometry
MALDI–TOF mass spectra were acquired using a Bruker UltraFlextreme (Billerica, MA, USA) mass spectrometer operated in linear mode. The instrument was calibrated with Protein Standard II (Bruker). MtoA in the final dialysis buffer was either desalted with a C4 OMIX pipette tip (Varian, Palo Alto, CA, USA) before spotting on the MALDI target, or spotted directly. Similar results were obtained with either method. The matrix solution was α-cyano-4-hydroxycinnamic acid in methanol. Laser power and number of lasers shots were adjusted and scans were averaged until the desired signal-to-noise ratio was obtained. Data from four experiments on three different preparations of MtoA were averaged.
Liquid chromatography–MS
Approximately 60 μg of MtoA in a volume of 200 μl was denatured by adding ∼96 mg of solid urea and incubating at room temperature for 1 h. The sample was diluted 12-fold with 100 mM ammonium bicarbonate buffer (pH 8) and then digested with sequencing-grade modified trypsin (Promega, Madison, WI, USA) overnight at 37°C at a protein-to-enzyme ratio of 20:1.The digest was desalted by solid phase extraction with a Supelco Discovery C18 cartridge. The desalted sample was then analyzed by LC–tandem mass spectrometry (MS/MS) on a custom-built LC system coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) as previously described (Livesay et al., 2007). The resulting MS/MS data were searched using TurboSequest v27.12 (Eng et al., 1994), with parent mass tolerance, 50 ppm; fragment mass tolerance, 0.5 Da; partially tryptic enzyme rules; a dynamic modification on cysteine (C) residues of 615.1694 (Yang et al., 2005), corresponding to the mass of a heme C group containing 56Fe and accounting for the charge on the heme iron. Because heme C peptides are known to be difficult to identify from database searches, we manually annotated tandem mass spectra of heme C peptides assigned to each observed heme motif.
Cyclic voltammetry
Cyclic voltammetry (CV) was performed inside a Faraday cage housed in a N2-filled chamber (atmospheric O2 < 2 ppm). The glass electrochemical cell contained three electrodes: a Ag/AgCl (saturated KCl) reference electrode, a basal plane pyrolytic graphite working electrode and a Pt wire counter electrode. The sample chamber was maintained at 4°C. Immediately prior to each experiment the working electrode surface was lightly abraded with “Wet and Dry Abrasive Paper” of fine grade (English Abrasives and Chemicals, Stafford, UK) and polished with an aqueous 0.3-μm Al2O3 slurry. After sonication, the electrode was rinsed, dried with a tissue and a few microliters of ice-cold solution containing 100 μM MtoA in 20 mM HEPES pH 7.6 + 100 mM NaCl + 10% (v/v) glycerol were placed on the electrode. After 30–60 s, excess solution was removed from the electrode, which was then immersed in the desired buffer-electrolyte: 25 mM HEPES pH 7.1 with 100 mM NaCl, or 20 mM Tris pH 7.8–9.2 with 100 mM NaCl. Baseline subtraction was carried out as previously described (Hartshorne et al., 2009). Midpoint potentials (Em) were determined by fitting oxidative and reductive scans to the sum of the theoretical response for 10 centers behaving as isolated n = 1 sites (Clarke et al., 2011). Potentials are quoted with respect to the standard hydrogen electrode (SHE) following addition of +0.197 V to the measured values.
Spectroscopic and stopped-flow kinetic measurement
All spectroscopic and kinetic measurements were conducted in an anoxic chamber filled with N2 from a liquid N2 boil-off supply (<1 ppm O2; Innovative Technologies, Port Washington, NY, USA). MtoA stock solution containing 4 μM protein, 150 mM NaCl, and 20 mM HEPES buffer (pH = 7.6) was purged with dry N2 gas for more than 1 h, then stored at 4°C in serum bottles capped with thick rubber stoppers and crimp sealed. All chemicals, plastic syringes, tubes, vials, and pipette tips were deoxygenated for at least 24 h inside the anoxic chamber prior to use. The UV–visible absorption spectra were collected using Agilent 8452 Diode Array Spectrophotometer (Santa Clara, CA, USA). Purified MtoA was in the oxidized form. To record the spectrum of fully reduced MtoA, 10 mM sodium dithionite solution was gradually added to 0.5 ml of protein stock solution until no changes in the UV–visible spectra were observed. In pH-dependent Fe(II) oxidation experiments, MtoA stock solution was diluted to 0.4 μM in Tris buffer (20 mM buffer with 150 mM NaCl) at the desired pH value. A small volume of FeCl2 stock solution (15 mM) was spiked into the diluted protein solution. Absorption spectra of the resulting solution mixture were observed at ∼5 min after the spiking, allowing the reactions to reach equilibrium.
Electron transfer in the Fe(II) oxidation by MtoA was measured using the reaction between 39.4 μM of FeCl2 and 4 μM of purified MtoA solution at pH 8. The amount of reduced MtoA after reaction was measured using the absorbance at the 552-nm band in the absorption spectrum. The Fe(II) concentration in the solution was measured by ferrozine assay. To minimize the spectral interference of MtoA, the solution was filtered by a 30-kDa centrifuge filter (Amicon Ultra-0.5, Millipore, Billerica, MA, USA) at 13,000 × g for 12 min, and then 0.4 ml of the filtrate into 1.6 ml ferrozine (1000 mg/l, pH 7) was added. A control experiment was conducted without adding MtoA solution, and the Fe(II) concentration after the same filtration process did not change substantially.
Ligand effects on oxidation of Fe(II)-complexes by MtoA were investigated using a stopped-flow system with a BioLogic MOS 250 spectrometer (Knoxville, TN, USA). The method was described in detail by Wang et al. (2008). Briefly, known volumes of MtoA and Fe(II)-complex solutions were rapidly mixed, and then the absorbance at 552 nm was tracked as a function of time. The concentration of oxidized MtoA at time t (Ct) can be calculated from the measured absorbance (At) using the following equation (Wang et al., 2008) after baseline correction:
| (1) |
where C0 is the initial concentration of oxidized MtoA, εox, and εred are the molar absorption coefficients of the oxidized and reduced MtoA, respectively. All stopped-flow kinetic experiments were conducted in Tris buffer (pH 8) containing 0.4 μM protein and 200 μM Fe(II) complexes. The ferrous iron-to-ligand ratio in Fe–citrate, Fe–NTA, and Fe–EDTA stock solutions was 1:10.
Analysis of kinetic data
The overall reaction between oxidized MtoA (ox-MtoA) and Fe(II)-complexes can be expressed as:
| (2) |
The rate of this reaction can then be expressed as follows:
| (3) |
where k is the rate constant, A and C are the Fe(II) and oxidized MtoA concentrations, respectively. Q is the ion activity product of the redox reaction, and K is the equilibrium constant. The concentration of oxidized MtoA (C) was calculated from the measured absorbance in the stopped-flow system and plotted against time. The residual Fe(II) concentration was calculated from the electron balance between the reacted Fe(II) and reduced MtoA. The calculated MtoA and Fe(II) concentrations as a function of time were then used to estimate rate parameters in Eq.3 Eq. 3. The second-order rate constant (k) and the equilibrium constant (K) were determined by the slope and the ending point of the fitted line, respectively.
Results
Identification of MtoAB and CymAES-1
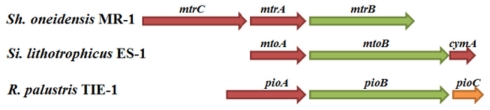
Search of the ES-1 genome did not reveal any homologs for Cyc-2/Rus/Cyc-1 of A. ferrooxidans or FoxEYZ of Rhodobacter strain SW2, but identified a gene cluster that encoded an MtrA/PioA homolog (Slit_2497) and an MtrB/PioB homolog (Slit_2496). Analysis of the genes adjacent to the mtrAB homologs also identified a cymA homolog (Slit_2495) that was next to an mtrB homolog. To distinguish them from those found in MR-1 and PioAB of R. palustris TIE-1, we named these genes and their encoding proteins mtoAB–cymA and MtoAB/CymAES-1, respectively (Figure 1). No pioC homolog was found in the ES-1 genome. The deduced MtoA polypeptide contains 317 amino acids with calculated molecular mass of 33556.2 Da. Ten CXXCH motifs (i.e., putative heme-binding sites) were found within the MtoA polypeptide, suggesting that matured MtoA contains up to 10 heme co-factors. In addition to the 10 histidine residues of the CX2CH motifs, the MtoA polypeptide contained 14 more histidine residues, most of which were probably used as the distal ligands for the heme Fe. Phylogenetic analysis of the deduced amino acid sequence of MtoA with the MtrA homologs of 19 sequenced Shewanella strains (Fredrickson et al., 2008) and PioA of R. palustris TIE-1 demonstrated that MtoA was 39% identical to PioA and 42–44% identical to the MtrA sequences analyzed. Furthermore, similar to Shewanella MtrA, MtoA lacked the N-terminal extension (∼200 amino acids) that was found in PioA polypeptides (Jiao and Newman, 2007). These results show that MtoA is almost equally related to PioA used for Fe(II) oxidation and MtrA for Fe(III) reduction, and the N-terminal extension of PioA is probably used for specific interaction with PioBC, but not directly for Fe(II) oxidation.
Figure 1.
Genetic organization of mtoA, mtoB, and cymA of Sideroxydans lithotrophicus ES-1. Shown are the relative positions of mtoA, mtoB, and cymA within the complete nucleotide sequence of S. lithotrophicus ES-1. The genes are labeled by arrows whose sizes and orientations indicate their relative lengths and directions in which they are presumed to be transcribed. For comparison, mtrCAB of Shewanella oneidensis MR-1 and pioABC of Rhodopseudomonas palustris TIE-1 are included. The genes encoding c-type cytochromes are labeled in red, while those encoding the β-barrel outer membrane proteins are labeled in green.
Likewise, MtoB was 17% identical to PioB and 19–21% identical to the MtrB sequences analyzed, while CymAES-1 was 56% identical to CymA of S. baltica OS223, 34% to MR-1 CymA and 32–36% identical to the other CymA sequences included in the alignment. Like PioB, MtoB is ∼110 amino acids longer than the MtrBs of Shewanella. Given that in MR-1, MtrAB, and CymA are the key components of the electron transfer pathway used for extracellular reduction of Fe(III) (Richardson, 2000; Shi et al., 2007, 2009), identification of an mtoAB–cymA cluster suggests that MtoAB and CymAES-1 may form a pathway for transferring electrons from the bacterial surface to the inner membrane during extracellular Fe(II) oxidation by ES-1.
Complementation of MR-1 mtrA deletion mutant in ferrihydrite (FH) reduction by MtoA
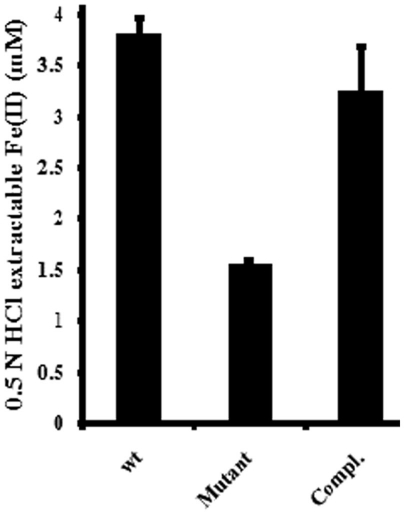
MtoA is a homolog of MR-1 MtrA that plays a critical role in FH reduction because deletion of mtrA impaired MR-1’s ability to reduce FH (Hartshorne et al., 2009). To determine whether MtoA can facilitate trans-membrane electron transfer, pLS279 with mtoA was introduced into an MR-1 ΔmtrA mutant and tested for FH reduction. The same mutant was also transformed with an empty vector, and the resulting strain was used as a negative control. Under the conditions tested, the MR-1 wt began to reduce FH within 24 h, while no FH reduction was detected in the reactions mediated by ΔmtrA mutants with either empty vector or mtoA at 48 h. After 120 h, FH reduction was detected in the reactions mediated by ΔmtrA mutant with either empty vector or mtoA, but no major difference was found between them. After 240 h, wt produced 3.82 ± 0.14 mM Fe(II) (n = 3), while mutants with empty vector or MtoA produced 1.56 ± 0.3 and 3.26 ± 0.42 mM Fe(II) (n = 3), respectively, which were 40 and 85% of that produced by wt (Figure 2). Thus, recombinant MtoA partially complements ΔmtrA mutant in FH reduction, demonstrating that it can be inserted into MtrB and transfer electron across the outer membrane to MtrC during FH reduction. Because deletion of mtrA up-regulates mtrD, an mtrA homolog (Coursolle and Gralnick, 2010), the up-regulated MtrD may contribute to the FH reduction observed in the mutant with empty vector.
Figure 2.
Complementation of Shewanella oneidensis MR-1 mutant without MtrA by MtoA. Fe(II) formation after reduction of 10 mM FH by S. oneidensis MR-1 wild type (wt), ΔmtrA mutant with empty vector, and the mutant complemented with MtoA (Compl.) for 240 h. The values reported are the means and SD of triplicate measurements.
Purification of recombinant MtoA
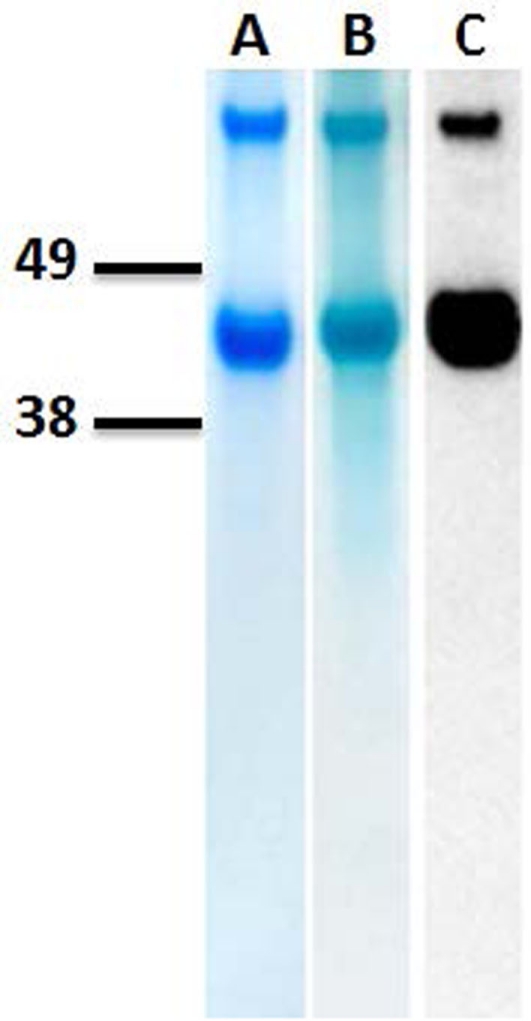
Following cell lysis and ultracentrifugation, recombinant MtoA was isolated from the soluble fraction by immobilized metal ion affinity chromatography followed by gel-filtration chromatography. The purified MtoA migrated as two bands on SDS-PAGE with apparent masses of ∼43 and 86 kDa, respectively. Heme staining and Western blot analyses of the same sample showed that both bands were the heme-containing MtoA, indicating that band with ∼86 kDa contained MtoA dimers (Figure 3). Measurement of purified MtoA with MALDI–TOF MS revealed that its molecular mass was 42,746 ± 114 Da (n = 3), which was very close to 42,639 Da, the calculated molecular mass for recombinant MtoA matured with 10 heme groups. In addition, liquid chromatography–mass spectrometry (LC–MS)/MS experiments following tryptic digest demonstrated heme modification at 8 out of 10 CXXCH heme-binding motifs in MtoA (all but motifs 9 and 10). Presumably, peptides from motifs 9 and 10 are not observed because of a lack of trypsin cleavage sites in their vicinity. Both motifs 9 and 10 would be in the same 56-residue, 6891-Da tryptic peptide, too large to be readily analyzed by LC–MS (data not shown). All these results clearly demonstrate that purified MtoA is a decaheme c-Cyt.
Figure 3.
Purification of MtoA. A total of 2 μg of isolated MtoA was run by SDS-PAGE and then visualized by GelCode staining (lane A) and heme staining (lane B) or probed with RGS-His-HRP antibody (lane C). Migration positions of protein standards with different molecular masses (kDa) are indicated at the left.
Cyclic voltammetry of purified MtoA
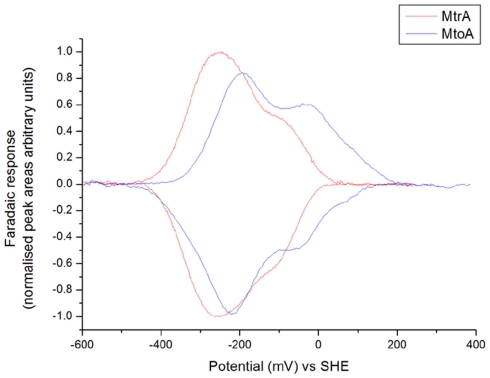
Cyclic voltammetry of basal plane graphite electrodes exposed to solutions of MtoA displayed clear peaks describing oxidative and reductive transformations that were not seen in the absence of MtoA (Figure 4). The oxidative and reductive peak areas were within error of each other consistent with reversible redox transformation of the adsorbed protein, and typically corresponded to 3 × 10−12 pmol of electroactive MtoA per square centimeter assuming all 10 hemes are redox active. This electroactive population is consistent with monolayer coverage of the electrode by MtoA, and peak currents were found to be directly proportional to the scan rate confirming that the response did indeed arise from an adsorbed protein film. As shown in Figure 4, MtoA is redox active over a more positive window of potential than MR-1 MtrA at pH 7.1, suggesting that MtoA is better poised to oxidize Fe(II) than MR-1 MtrA.
Figure 4.
Cyclic voltammograms of MtoA of Sideroxydans lithotrophicus ES-1 and MtrA of Shewanella oneidensis MR-1 adsorbed on graphite electrodes. Scan rate = 50 mV/s, buffer-electrolyte = 25 mM HEPES pH 7.1 with 100 mM NaCl, T = 277 K.
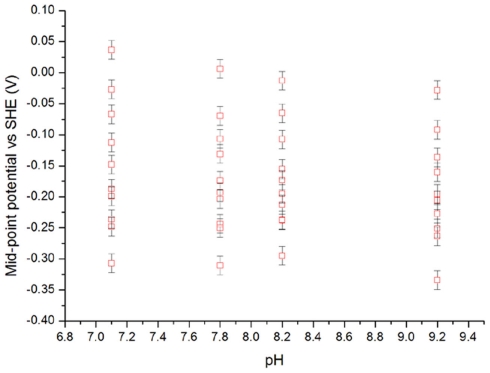
The redox properties of MtoA were measured by CV at pH 7.1, 7.8, 8.2, and 9.2. For each pH the protein was redox active between ∼+100 and −400 mV (vs. SHE) with the heme Em values clustering to define a low potential peak and a higher potential shoulder. The major impact of increased pH was to produce a greater proportion of hemes with lower Em. A smaller effect was a shift of the higher potential end of redox active window to more negative values. These effects were quantified by assuming that the oxidative and reductive peaks arose from the sum of contributions from 10 independent centers displaying reversible, n = 1 electron transfer. This assumption yielded a good fit to the data and averaging the Em values from oxidative and reductive peaks yielded 10 apparent Em values for each pH (Figure 5).
Figure 5.
Average Em values determined from cyclic voltammetry of MtoA at the indicated pH.
Fe(II) oxidation by MtoA
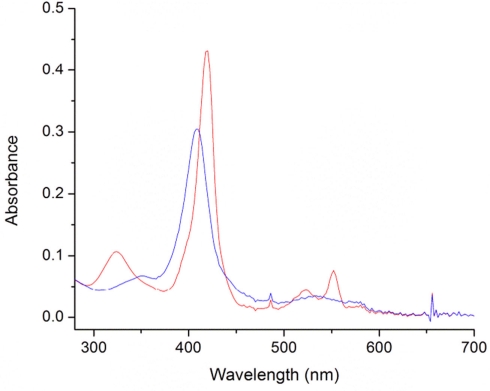
The absorption spectrum of purified, oxidized MtoA exhibited a characteristic peak at 408 nm (α peak) with a shoulder at ∼352 nm and a weak absorption band at ∼530 nm. After addition of sodium dithionite solution at pH 7, α peak shifted to 418 nm, and the shoulder shifted to 324 nm to form a distinct peak. In addition, two absorption peaks at 522 nm (β peak) and 552 nm (γ peak) appeared (Figure 6). A similar change was observed in absorption spectra of oxidized and reduced MtrC and OmcA (Wang et al., 2008). The fully reduced MtoA could be rapidly re-oxidized by addition of 1 mM of Fe(III)–NTA solution (data not shown), which is consistent with previous results that MR-1 MtrA is a bidirectional protein and can mediate electrons in and out of MR-1 cells as well as proteoliposomes (Hartshorne et al., 2009; Ross et al., 2011). To investigate the mass balance in FeCl2 oxidation by MtoA, the concentration changes of Fe(II) ions (Δ[Fe(II)]) and reduced MtoA ([MtoA]red) were investigated with 4 μM of oxidized MtoA reacted with ∼40 μM of FeCl2 at pH 9. At the end of the reaction, the ratio of Δ[Fe(II)] to [MtoA]red was 9.6 ± 1.5 (n = 3). Changing the initial concentration of MtoA to 2.3 and 3 μM, respectively, did not significantly change the ratio. Although not all MtoA was reduced by FeCl2 in all experiments, the amount of Fe(II) ions formed was close to 10 times more than the amount of MtoA reduced. It suggests that, under this condition, 10 hemes in MtoA participated in electron transfer, and no other electron acceptor or donor participated in this reaction.
Figure 6.
Absorption spectra of MtoA (4 μM) in 50 mM NaCl and 100 mM HEPES buffer (pH = 7) in the oxidized (blue line) and the reduced form (red line).
pH-dependent Fe(II) oxidation by MtoA
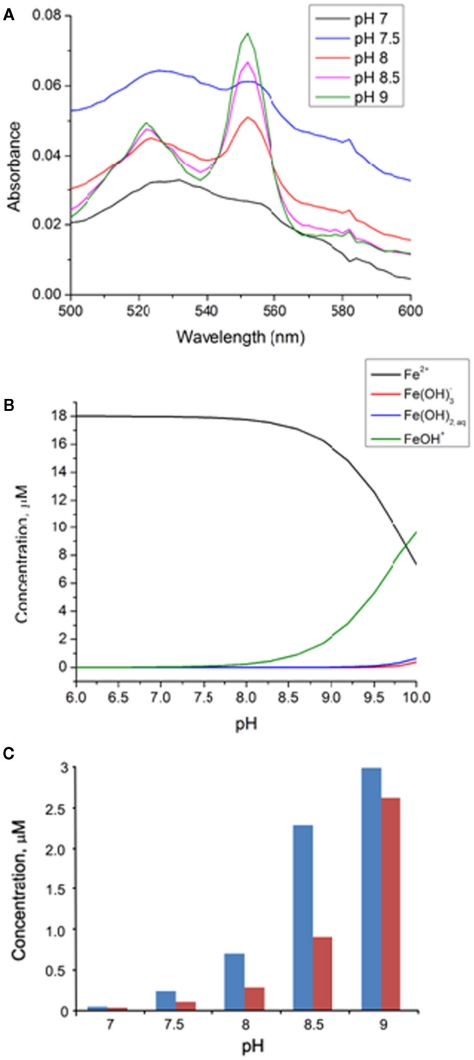
Absorption spectra of 0.4 μM MtoA after reaction with 18 μM FeCl2 at pH 7, 7.5, 8, 8.5, and 9, respectively, were compared (Figure 7). The comparison showed that, with the same amount of FeCl2, the absorbance at 552 nm increased as pH increased. At pH 7, only 1.0 ± 1.1% (n = 3) of MtoA was reduced by 18 μM FeCl2, but this percentage increased to 4.9 ± 0.3% (n = 3) at pH 7.5, 14.7 ± 3.3% (n = 3) at pH 8, 48.2 ± 1.7% (n = 3) at pH 8.5, and 63.0 ± 9.8% (n = 3) at pH 9. The maximum fraction of total MtoA reduced by FeCl2 was therefore systematically larger at higher pH. The same trend was also observed in HEPES buffer at pH 7 and 8 (data not shown). It should be noted that in all of these experiments, MtoA was not totally reduced, even though there was an excess of electron donor, FeCl2. At pH 9, increasing the concentration of FeCl2 up to 400 μM did result in increasingly reduced MtoA. At pH 8, no additional MtoA was reduced as [FeCl2] was increased to 840 μM. Thus, MtoA has the ability to oxidize FeCl2 in the pH range of 7–9, but reduction of MtoA is incomplete even in the presence of excess FeCl2. Similar incomplete redox reaction was observed in the oxidation of reduced MtrF by flavin mononucleotide (FMN). For that system it was proposed that only a subgroup of the 10 hemes in each MtrF molecule participated in FMN reduction based on reduction potential difference estimated between individual hemes and the midpoint potential of FMN/FMNH2 (Clarke et al., 2011). However, the mass balance experiment in this study showed that the molar ratio of the consumed Fe(II) to reduced MtoA was close to 10:1, consistent in principle with participation of 10 hemes in each MtoA molecule per 10 Fe(II) oxidized. If only a subgroup of MtoA hemes reacted with FeCl2, this ratio should be <10.This suggests that at the end of reaction a subpopulation of MtoA molecules are fully reduced, and incomplete reduction of MtoA is accounted for by a fully oxidized subgroup of MtoA. Hence, in contrast to MtrF, MtoA appears to undergo redox reactions via a cascade whereby once electron transfer into an MtoA molecule begins, it subsequently becomes more energetically favorable to continue reduction of that molecule to completion. This suggests that for any individual MtoA, redox potentials for individual hemes, in CV measurements for example, progressively shift to more oxidizing potentials as reduction proceeds, and vice versa. As Fe(II), Fe(III), and oxidized MtoA concentrations change during the course of reaction, the redox potential difference between ferrous species and MtoA may decrease to zero, at which point the reaction reaches equilibrium. Involvement of all of its 10 hemes during Fe(II) oxidation implies the ability of MtoA for transferring electrons from extracellular Fe(II), across the outer membrane, and into the periplasm, which is consistent with the complementation results described above.
Figure 7.
(A) Absorption spectra of MtoA (0.4 μM) after the oxidation of 18 μM FeCl2 at pH 7 (black), pH 7.5 (blue), pH 8 (red), pH 8.5 (pink), and pH 9 (green). (B) Speciation of Fe(II; 18 μM) as a function of pH in 150 mM NaCl solution (Allison et al., 1991). (C) Variation of Fe(II; blue columns) oxidized by MtoA at the end of reactions and initial [Fe(OH)+] (red columns) as a function of pH.
Ligand-dependent Fe(II) oxidation by MtoA
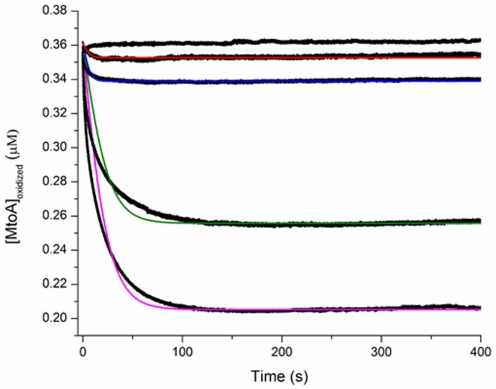
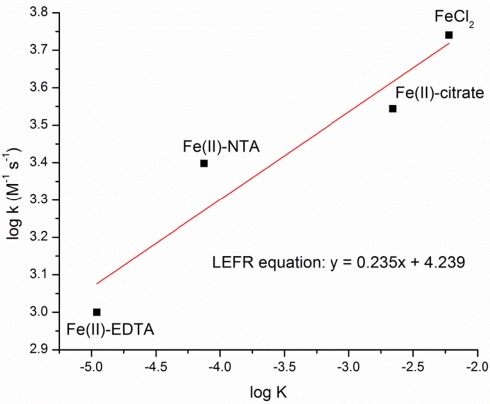
Rates of Fe(II)-complex oxidation by MtoA at pH 8 were investigated using a stopped-flow system. In all experiments, the molar ratio of Fe(II) to MtoA was 500. The change of MtoA concentration with time was calculated according to the absorbance change at the γ peak. A control experiment was included by mixing MtoA and Tris buffer (pH 8) in which no significant change was observed in the concentration of oxidized MtoA during the time course of study (Figure 8). In all Fe(II)-complex oxidation reactions, the concentration of oxidized MtoA changed in a similar way. It initially decreased very fast, and then the rate of change decreased to reach redox equilibrium. All reactions reached equilibrium within 2 min. The fraction of MtoA reduced by Fe(II)-complexes at the end of each reaction was in the order Fe(II)Cl2 > Fe(II)–citrate > Fe(II)–NTA > Fe(II)–EDTA (Figure 8).
Figure 8.
Stopped-flow spectrophotometry results of the oxidation of 0.2 mM Fe(II)-complexes by 0.4 μM MtoA at pH 8 [black dots; from the top to the bottom is MtoA solution without Fe(II)-complexes (control), Fe(II)–EDTA, Fe(II)–NTA, Fe(II)–citrate, and Fe(II)Cl2]. The fitted curves for the kinetic data of MtoA reduction with Fe(II)Cl2 (purple), Fe(II)–citrate (green), Fe(II)–NTA (blue), and Fe(II)–EDTA (red).
The stopped-flow kinetic data were fitted to a second-order rate model using Eq.3. The rate constant, k, was derived from the slope of fitted curves in the fast stage, and the equilibrium constant, K, was determined from the equilibrium stage (Table 1). Rate and equilibrium constants were significantly affected by ligand type. The order for the oxidation rates are similar to the final oxidation fraction, which was Fe(II)Cl2 > Fe(II)–citrate > Fe(II)–NTA > Fe(II)–EDTA. These experiments showed that the type of ligand significantly affected the oxidation kinetics of Fe(II)-complexes by MtoA. Compared to the reduction rates of the analogous Fe(III)-complexes by MtrC and OmcA, which ranged from 0.872 μM−1 s−1 for the reaction between MtrC and Fe(III)–EDTA to 0.012 μM−1 s−1 for the reaction between OmcA and the Fe(III)–citrate (Wang et al., 2008), the oxidation rates of Fe(II)-complexes by MtoA were two orders of magnitude slower. Moreover, in the Fe(III)-complex reduction reactions by MtrC and OmcA, there was enough free energy driving the redox reaction to consume all heme groups. However, consistent with the discussion above regarding reaction incompleteness, the free energy in the present system was not sufficient for all MtoA to participate in the redox reaction.
Table 1.
Kinetic parameters for Fe-complexes oxidation by MtoA at pH 8.
| Complex | Rate constants (k, μm−1 s−1) | Equilibrium constant (K) | Log Koba | Log Kcalcb |
|---|---|---|---|---|
| FeCl2 | (6.3 ± 0.3) × 10−3 | (6.5 ± 0.4) × 10−3 | −2.22 | 7.04c |
| Fe–citrate | (4.1 ± 0.8) × 10−3 | (2.4 ± 0.4) × 10−3 | −2.66 | −5.46d |
| Fe–NTA | (2.5 ± 0.3) × 10−3 | (8.3 ± 1.5) × 10−5 | −4.125 | −0.82 |
| Fe–EDTA | (1.0 ± 0.3) × 10−3 | (2.1 ± 1.5) × 10−5 | −4.959 | −1.35 |
aLog Kob was calculated from the K values estimated from the fitted curve.
bLog Kcalc was calculated from theoretical Log K using experimental condition.
cCalculated assuming in equilibrium with ferrihydrite.
dCalculated by using thermodynamic data of Fe(III)citrate in Minteqa database.
Discussion
The increased oxidation of Fe(II)Cl2 by MtoA from pH 7 to pH 9 may be attributed mainly to the expected increased concentration of hydroxylated species of Fe(II), such as Fe(OH)+ and Fe(OH)2. Equilibrium speciation calculations based on Minteqa2 thermodynamic database (Allison et al., 1991) as plotted in Figure 7B show the speciation of Fe(II; 18 μM) in 150 mM NaCl solution as a function of pH. Hexaquo ferrous ion is the dominant species in the pH range used in this study, but the amount of Fe(OH)+ increases with increasing pH, especially when pH is greater than 7.5. Both thermodynamic calculations and experimental data indicate that hydroxylated species of Fe(II) have higher reactivity relative to hexaquo Fe(II) (e.g., Wehrli, 1990; Sedlak and Chan, 1997). OH− ligands in the inner coordination shell of Fe(II) increase its reducing potential and increase its oxidation rate, and therefore could increase reaction to product Fe(III) phases. The concentration change of Fe(II), including hexaquo Fe(II) and Fe(OH)+, at the end of pH-dependent experiments was compared to the initial Fe(OH)+ concentration (Figure 7C). It shows that the amount of Fe(II) oxidized by MtoA appears to correlate positively with the initial concentration of Fe(OH)+ from pH 7 to pH 9. Notably, at pH 9, the initial concentration of Fe(OH)+ is expected to be significantly higher than that at pH 8.5, but the amount of Fe(II) oxidized by MtoA only increases slightly. This discrepancy is caused, at least in part, by the fact that the average redox potential of MtoA decreases as pH increases. This decrease is particularly accelerated from pH 8.2 to pH 9.2. The decrease of the redox potential of MtoA from pH 8.2 to pH 9.2 could negatively offset the increase of Fe(OH)+ concentration in the same pH range, which may have the net result of only a slight increase of the Fe(II) oxidized by MtoA at pH 9. These results imply that change of pH in the environments may also have significant influence on Fe(II) speciation, which in turn will strongly affect microorganism-mediated Fe(II) oxidation in their natural settings.
Ligand types impact both reaction rates and equilibrium constants of redox reactions between Fe(II)-complexes and MtoA. The equilibrium speciation calculations showed that the dominant Fe(II) species in the four ligand systems is hexaquo Fe(II), Fe(II)–citrate−, FeOH–NTA−, and Fe–EDTA−, respectively, at a ferrous ion-to-ligand ratio of 1:10. The equilibrium constant for the half electron transfer reactions between Fe(II)–ligand and Fe(III)–ligand (Table 2) was calculated using the thermodynamic cycle that involves ligand detachment from Fe(II)–ligand complex, Fe(II) oxidation to Fe(III) and Fe(III)–ligand complexation:
Table 2.
Relevant speciation reactions for calculating reaction-free energy at 25°C.
| Reaction | Log K (I = 0) | Reference |
|---|---|---|
| Fe(III) + e− → Fe(II) | 13.00 | Martell and Smith (1995) |
| Fe(III)–3H+ + 3H2O = Fe(OH)3 | −3.96 | Cornell and Schwertmann (2003) |
| Fe(II) + citrate3− → Fe(II)–citrate− | 5.89 | Martell and Smith (1995) |
| Fe(III) + citrate3− → Fe(III)–citrate | 13.43 | Allison et al. (1991), Timberlake (1964) |
| Fe(III)OH–NTA− + e− → Fe(II)OH–NTA2− | 0.82 | Wang et al. (2008) |
| Fe(III)–EDTA− + e− → Fe(II)–EDTA2− | 1.35 | Wang et al. (2008) |
I, ionic strength.
| (4) |
Reaction equilibrium constants in this electron transfer pathway are provided in Table 2. For the case of FeCl2 at pH 8, it is reasonable to assume that the concentration of produced Fe(III) was controlled by the solubility of FH [Fe(OH)3] because organic ligand was not provided. The equilibrium constant for electron transfer from Fe(II) to FH (log K = 7.04; Table 1) was then calculated by combining the redox reaction of Fe(II)/Fe(III) and the formation reaction of FH (Table 2). The trend of calculated log K is consistent with that of the log K values fitted from the experimental data except for the Fe–citrate case (Table 1). The exception is likely because of incomplete understanding of the speciation in the Fe(III)–citrate system; two Fe(III)–citrate speciation models were assembled previously based on literature data (Liu et al., 2001) that involve completely different Fe(III) speciation. In this study, we used the speciation model 1 in Liu et al. (2001), which is also used in the Minteqa2 database (Allison et al., 1991). In this model, the log K for the equation:
| (5) |
is 13.43 Speciation calculations in our Fe(III)–citrate system using Eq.5, however, suggested that Fe(III)–citrate is not a stable species if Fe(III) is allowed to precipitate as FH. The alternative Fe(III)–citrate speciation model produced the same result. If the FH is used as the end product, then the overall reaction constant (log K) from Fe(II)–citrate to FH at pH 8 is 1.15. With this value, the trend of the calculated equilibrium reaction constants was consistent with the value estimated from the experimental data.
The estimated reaction rate constants (k) and equilibrium constants (K) were positively correlated, establishing a linear free-energy relationship for this system (Figure 9). This result implies that it is the reaction-free energy that mainly determined the observed initial reaction rates when it was far away from equilibrium. It is interesting to note that the rate constant order of Fe(II)-complex oxidation, Fe–citrate > Fe–NTA > Fe–EDTA, is the inverse order observed in the reduction of Fe(III) complexes by MtrC and OmcA, where the Fe(III)–ligand reduction rate was Fe(III)–EDTA > Fe(III)–NTA > Fe(III)–citrate (Wang et al., 2008). This is expected from the trend of the activation-free energies for the redox reactions between Fe-complex and proteins (Wang et al., 2008). It is also expected from the relationship of K = kf/kb, where kf and kb are the forward [Fe(II) oxidation] and backward [Fe(III) reduction] reaction rate constants, respectively, and K is the equilibrium constant. Using the estimated K and k, which is kf here (Table 1), the calculated kb = kf/K, is on the order of Fe–citrate < Fe–NTA < Fe–EDTA, which is consistent with those observed by Wang et al. (2008). These two studies therefore demonstrate the same effect of ligand complexation on the reaction rates for both reduction and oxidation reactions, indicating that complexing ligands will have a significant impact on the reaction rates of microorganism-mediated Fe(II) oxidation in the environments.
Figure 9.
Linear free-energy relationship in Fe(II)-complexes oxidation by MtoA.
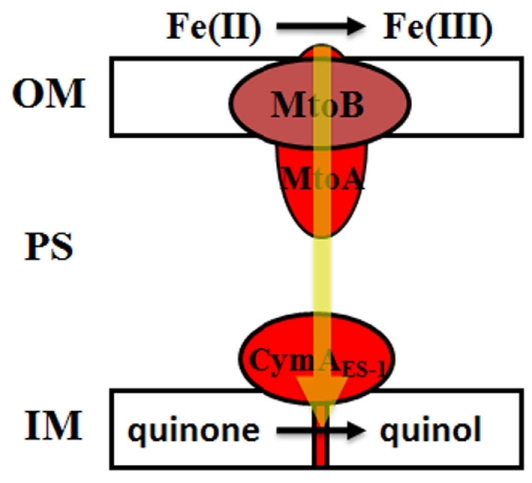
In MR-1, MtrAB form a tight protein complex on the OM where MtrB is proposed to serve as a sheath for embedding MtrA and MtrA mediates electron conductance across the OM, while CymA is located in the inner membrane where it recycles quinol back to quinone during extracellular Fe(III) oxide reduction (Hartshorne et al., 2009). In this study, we showed that MtoA was a functional Fe(II)-oxidizing protein with broad redox potential that was more positive than that of MR-1 MtrA, indicating that, like MR-1 MtrA, MtoA is also capable of mediating electron conductance across the OM, but with the direction opposite to that of MR-1 MtrA during metal-reducing conditions. In addition, ES-1 mtoAB–cymA is, to the best of our knowledge, the first example that the genes encoding MtrAB and CymA homologs are clustered together, which suggests that they may belong to the same operon and that their protein products may work together for mediating electron transfer reactions. Based on previous observations and the results from this study, we propose that, similar to MtrAB and CymA in MR-1 cells, MtoAB and CymAES-1 are also located in the OM and inner membrane, respectively, where MtoA is embedded inside MtoB. However, the direction of MtoAB/CymAES-1-mediated electron transfer during Fe(II) oxidation (i.e., outside-in) by ES-1 is opposite to that of MtrABC/CymA-mediated reactions during Fe(III) reduction (i.e., inside-out) by MR-1. We propose that through its heme group(s) exposed to the extracellular environment, MtoA oxidizes Fe(II) directly and then transfers the released electrons across the OM to periplasmic proteins that have yet to be identified, which in turn relay the electrons to CymAES-1. CymAES-1 uses the received electrons to reduce quinone to quinol in the inner membrane (Figure 10). Quinol then shuttles the electrons to the redox proteins in the inner membrane for reducing O2 and/or NAD+.
Figure 10.
The proposed roles of MtoAB and CymAES-1 in Sideroxydans lithotrophicus ES-1-mediated extracellular Fe(II) oxidation. Decaheme c-Cyt MtoA, which is inserted into the porin-like, outer membrane (OM) protein MtoB, oxidizes Fe(II) directly on the bacterial surface and transfers the released electrons across the OM to the periplasmic proteins that have yet to be identified. The periplasmic proteins relay the electrons through the periplasm (PS) to the tetraheme c-Cyt CymAES-1. CymAES-1, a homolog of NapC/NrfH family of quinol dehydrogenase that is located in the cytoplasmic or inner membrane (IM), reduces quinone to quinol. c-Cyts are labeled in red and direction of electron transfer is indicated by a yellow arrow.
PioABC homologs are also found in the Fe(II)-oxidizing bacterium Gallionella capsiferriformans (Bonnefoy and Holmes, 2011), indicating the broad involvements of MtoAB/PioAB homologs in microbial Fe(II) oxidation. It should be noted that a group of proteins that show no homologous to MtoAB/CymAES-1 have recently been proposed to be involved in Fe(II) oxidation by the bacterium Mariprofundus ferrooxydans PV-1 and homologs of these proteins are also present in ES-1 (Singer et al., 2011). These results emphasize the uncertainty that remains regarding the electron transfer pathway(s) utilized by ES-1 for Fe(II) oxidation and the need for additional research.
In summary, an mtoAB–cymA gene cluster is found in the genome of the Fe(II)-oxidizing bacterium S. lithotrophicus ES-1. Protein purification and characterization results confirm that MtoA is a decaheme c-type cytochrome and oxidizes soluble Fe(II). MtoA-mediated Fe(II) oxidation is pH- and Fe(II)-complexing ligand-dependent. It is proposed that, together, MtoAB and CymAES-1 form a pathway for electron conductance from extracellular Fe(II) to the quinone pool in the bacterial inner membrane.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Subsurface Biogeochemical Research program (SBR)/Office of Biological and Environmental Research (BER), U.S. Department of Energy (DOE), and is a contribution of the Pacific Northwest National Laboratory (PNNL) Scientific Focus Area. A portion of the research was performed using EMSL, a national scientific user facility sponsored by DOE-BER and located at PNNL. PNNL is operated for DOE by Battelle under contract DE-AC05-76RLO 1830.
References
- Allison J. D., Brown D. S., Novo-Gradac K. J. (1991). MINITEQ2//PRODEFA2, A Geochemical Assessment Model for Environmental Systems, 3 Edn Washington, DC: US Environmental Protection Agency [Google Scholar]
- Appia-Ayme C., Guiliani N., Ratouchniak J., Bonnefoy V. (1999). Characterization of an operon encoding two c-type cytochromes, an aa(3)-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl. Environ. Microbiol. 65, 4781–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher W. B., Bren K. L. (2010). A heme fusion tag for protein affinity purification and quantification. Protein Sci. 19, 1830–1839 10.1002/pro.460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefoy V., Holmes D. S. (2011). Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol. 10.1111/j.1462-2920.2011.02626.x [DOI] [PubMed] [Google Scholar]
- Castelle C., Guiral M., Malarte G., Ledgham F., Leroy G., Brugna M., Giudici-Orticoni M. T. (2008). A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 283, 25803–25811 10.1074/jbc.M802496200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T. A., Edwards M. J., Gates A. J., Hall A., White G. F., Bradley J., Reardon C. L., Shi L., Beliaev A. S., Marshall M. J., Wang Z. M., Watmough N. J., Fredrickson J. K., Zachara J. M., Butt J. N., Richardson D. J. (2011). Structure of a bacterial cell surface decaheme electron conduit. Proc. Natl. Acad. Sci. U.S.A. 108, 9384–9389 10.1073/pnas.1017200108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R. M., Schwertmann U. (2003). The Iron Oxides: Structure, Properties, Reactions, Occurrences and Uses. Weinheim: Wiley-VCH [Google Scholar]
- Coursolle D., Gralnick J. A. (2010). Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 77, 995–1008 [DOI] [PubMed] [Google Scholar]
- Croal L. R., Jiao Y., Newman D. K. (2007). The fox operon from Rhodobacter strain SW2 promotes phototrophic Fe(II) oxidation in Rhodobacter capsulatus SB1003. J. Bacteriol. 189, 1774–1782 10.1128/JB.01395-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D., Fleming E. J., Mcbeth J. M. (2010). Iron-oxidizing bacteria: an environmental and genomic perspective. Annu. Rev. Microbiol. 64, 561–583 10.1146/annurev.micro.112408.134208 [DOI] [PubMed] [Google Scholar]
- Emerson D., Moyer C. (1997). Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63, 4784–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D., Rentz J. A., Lilburn T. G., Davis R. E., Aldrich H., Chan C., Moyer C. L. (2007). A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2, e667. 10.1371/journal.pone.0000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J. K., McCormack A. L., Yates Iii J. R. (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K., Romine M. F., Beliaev A. S., Auchtung J. M., Driscoll M. E., Gardner T. S., Nealson K. H., Osterman A. L., Pinchuk G., Reed J. L., Rodionov D. A., Rodrigues J. L., Saffarini D. A., Serres M. H., Spormann A. M., Zhulin I. B., Tiedje J. M. (2008). Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603 10.1038/nrmicro1947 [DOI] [PubMed] [Google Scholar]
- Gralnick J. A., Vali H., Lies D. P., Newman D. K. (2006). Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U.S.A. 103, 4669–4674 10.1073/pnas.0505959103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne R. S., Reardon C. L., Ross D., Nuester J., Clarke T. A., Gates A. J., Mills P. C., Fredrickson J. K., Zachara J. M., Shi L., Beliaev A. S., Marshall M. J., Tien M., Brantley S., Butt J. N., Richardson D. J. (2009). Characterization of an electron conduit between bacteria and the extracellular environment. Proc. Natl. Acad. Sci. U.S.A. 106, 22169–22174 10.1073/pnas.0900086106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Newman D. K. (2007). The pio operon is essential for phototrophic Fe(II) oxidation in Rhodopseudomonas palustris TIE-1. J. Bacteriol. 189, 1765–1773 10.1128/JB.00776-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. G., Zachara J. M., Gorby Y. A., Szecsody J. E., Brown C. F. (2001). Microbial reduction of Fe(III) and sorption/precipitation of Fe(II) on Shewanella putrefaciens strain CN32. Environ. Sci. Technol. 35, 1385–1393 10.1021/es001261b [DOI] [PubMed] [Google Scholar]
- Livesay E. A., Tang K., Taylor B. K., Buschbach M. A., Hopkins D. F., Lamarche B. L., Zhao R., Shen Y., Orton D. J., Moore R. J., Kelly R. T., Udseth H. R., Smith R. D. (2007). Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem. 80, 294–302 10.1021/ac701727r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower B. H., Yongsunthon R., Shi L., Wildling L., Gruber H. J., Wigginton N. S., Reardon C. L., Pinchuk G. E., Droubay T. C., Boily J. F., Lower S. K. (2009). Antibody recognition force microscopy shows that outer membrane cytochromes OmcA and MtrC are expressed on the exterior surface of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 75, 2931–2935 10.1128/AEM.02108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell R. E., Smith R. M. (1995). Critical Stability Constants of Metal Complexes Database. Gaithersburg, MD: NIST Standard Reference Database [Google Scholar]
- Reardon C. L., Dohnalkova A. C., Nachimuthu P., Kennedy D. W., Saffarini D. A., Arey B. W., Shi L., Wang Z., Moore D., Mclean J. S., Moyles D., Marshall M. J., Zachara J. M., Fredrickson J. K., Beliaev A. S. (2010). Role of outer-membrane cytochromes MtrC and OmcA in the biomineralization of ferrihydrite by Shewanella oneidensis MR-1. Geobiology 8, 56–68 10.1111/j.1472-4669.2009.00226.x [DOI] [PubMed] [Google Scholar]
- Richardson D. J. (2000). Bacterial respiration: a flexible process for a changing environment. Microbiology 146(Pt 3), 551–571 [DOI] [PubMed] [Google Scholar]
- Ross D. E., Flynn J. M., Baron D. B., Gralnick J. A., Bond D. R. (2011). Towards electrosynthesis in Shewanella: energetics of reversing the mtr pathway for reductive metabolism. PLoS ONE 6, e16649. 10.1371/journal.pone.0016649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross D. E., Ruebush S. S., Brantley S. L., Hartshorne R. S., Clarke T. A., Richardson D. J., Tien M. (2007). Characterization of protein-protein interactions involved in iron reduction by Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 73, 5797–5808 10.1128/AEM.00146-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak D. L., Chan P. G. (1997). Reduction of hexavalent chromium by ferrous iron. Geochim. Cosmochim. Acta 61, 2185–2192 10.1016/S0016-7037(97)00077-X [DOI] [Google Scholar]
- Shi L., Belchik S. M., Wang Z., Kennedy D. W., Dohnalkova A. C., Marshall M. J., Zachara J. M., Fredrickson J. K. (2011). Identification and characterization of UndAHRCR-6, an outer membrane endecaheme c-type cytochrome of Shewanella sp. strain HRCR-6. Appl. Environ. Microbiol. 77, 5521–5523 10.1128/AEM.00260-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Deng S., Marshall M. J., Wang Z., Kennedy D. W., Dohnalkova A. C., Mottaz H. M., Hill E. A., Gorby Y. A., Beliaev A. S., Richardson D. J., Zachara J. M., Fredrickson J. K. (2008). Direct involvement of type II secretion system in extracellular translocation of Shewanella oneidensis outer membrane cytochromes MtrC and OmcA. J. Bacteriol. 190, 5512–5516 10.1128/JB.01049-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Lin J. T., Markillie L. M., Squier T. C., Hooker B. S. (2005). Overexpression of multi-heme C-type cytochromes. BioTechniques 38, 297–299 10.2144/05382PT01 [DOI] [PubMed] [Google Scholar]
- Shi L., Potts M., Kennelly P. J. (1998). The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22, 229–253 10.1111/j.1574-6976.1998.tb00369.x [DOI] [PubMed] [Google Scholar]
- Shi L., Richardson D. J., Wang Z., Kerisit S. N., Rosso K. M., Zachara J. M., Fredrickson J. K. (2009). The roles of outer membrane cytochromes of Shewanella and Geobacter in extracellular electron transfer. Environ. Microbiol. Rep. 1, 220–227 10.1111/j.1758-2229.2009.00035.x [DOI] [PubMed] [Google Scholar]
- Shi L., Squier T. C., Zachara J. M., Fredrickson J. K. (2007). Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20 10.1111/j.1365-2958.2007.05783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Zhang W. (2004). Comparative analysis of eukaryotic-type protein phosphatases in two streptomycete genomes. Microbiology 150, 2247–2256 10.1099/mic.0.27057-0 [DOI] [PubMed] [Google Scholar]
- Singer E., Emerson D., Webb E. A., Barco R. A., Kuenen J. G., Nelson W. C., Chan C. S., Comolli L. R., Ferriera S., Johnson J., Heidelberg J. F., Edwards K. J. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS ONE 6, e25386. 10.1371/journal.pone.0025386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stookey L. (1970). Ferrozine-a new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781 10.1021/ac60289a016 [DOI] [Google Scholar]
- Thomas P. E., Ryan D., Levin W. (1976). An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176 10.1016/0003-2697(76)90067-1 [DOI] [PubMed] [Google Scholar]
- Timberlake C. F. (1964). Iron-malate + iron-citrate complexes. J. Chem. Soc. 5078–5085 10.1039/jr9640005078 [DOI] [Google Scholar]
- Wang Z. M., Liu C. X., Wang X. L., Marshall M. J., Zachara J. M., Rosso K. M., Dupuis M., Fredrickson J. K., Heald S., Shi L. (2008). Kinetics of reduction of Fe(III) complexes by outer membrane cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 74, 6746–6755 10.1128/AEM.02800-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli B. (1990). “Redox reactions of metal ions at mineral surfaces,” in Aquatic Chemcial Kinetics: Reaction Rates of Processes in Natural Waters, ed. Stumm (Wiley) W. 311–336 [Google Scholar]
- Yang F., Bogdanov B., Strittmatter E. F., Vilkov A. N., Gritsenko M., Shi L., Elias D. A., Ni S., Romine M., Paša-Tolic L., Lipton M. S., Smith R. D. (2005). Characterization of purified c-type heme-containing peptides and identification of c-type heme-attachment sites in Shewanella oneidensis cytochromes using mass spectrometry. J. Proteome Res. 4, 846–854 10.1021/pr049774v [DOI] [PubMed] [Google Scholar]
- Yarzabal A., Brasseur G., Ratouchniak J., Lund K., Lemesle-Meunier D., DeMoss J. A., Bonnefoy V. (2002). The high-molecular-weight cytochrome c Cyc2 of Acidithiobacillus ferrooxidans is an outer membrane protein. J. Bacteriol. 184, 313–317 10.1128/JB.184.1.313-317.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]