Abstract
We explored diverse alterations contributing to liposarcomagenesis by sequencing the genome, exome, transcriptome, and cytosine methylome of a primary and recurrent dedifferentiated liposarcoma (DLPS) from distinct chemotherapy/radiotherapy-naïve patients. The liposarcoma genomes had complex structural rearrangements, but in different patterns, and with varied effects on the structure and expression of affected genes. While the point mutation rate was modest, integrative analyses and additional screening identified somatic mutations in HDAC1 in 8.3% of DLPS. Liposarcoma methylomes revealed alterations in differentiation pathway genes, including CEBPA methylation in 24% of DLPS. Treatment with demethylating agents, which restored CEBPA expression in DLPS cells, was anti-proliferative and pro-apoptotic in vitro and reduced tumor growth in vivo. Both genetic and epigenetic abnormalities established a role for small RNAs in liposarcomagenesis, typified by methylation-induced silencing of microRNA-193b in DLPS but not its well-differentiated counterpart. These findings reveal an unanticipated role for epigenetic abnormalities in DLPS tumors and suggest demethylating agents as potential therapeutics.
Keywords: Dedifferentiated liposarcoma, DNA methylation, histone deacetylase, microRNA, adipocyte differentiation
Introduction
Liposarcoma is the most common soft-tissue sarcoma (~20% of all adult sarcomas). Dedifferentiated liposarcoma (DLPS) is a heterogeneous subtype that is particularly aggressive in the retroperitoneum (1). Excisional surgery remains the standard of care for localized DLPS, but ~80% of patients diagnosed with primary retroperitoneal DLPS will die from locally recurrent or metastatic disease within five years. As these tumors are largely resistant to conventional cytotoxic therapies, it is imperative to identify novel therapeutic targets for this disease.
DLPS is characterized by intermediate genomic complexity where ~80–90% of patients have amplifications of a 12q ring chromosome (2) spanning two canonical oncogenes, CDK4 and MDM2. Yet, recent large-scale genomic characterization of sarcomas has indicated the picture is more complex, both in this amplicon and in affiliated abnormalities throughout the liposarcoma genome (3).
Here, we performed 16 concurrent massively parallel sequencing experiments in four samples, a primary and a local recurrence of DLPS and their paired non-neoplastic adipose tissue controls from two unrelated male and female individuals (hereafter referred to as DLPS1 and DLPS2, respectively). These samples were selected from a large panel of molecularly characterized retroperitoneal DLPS of similar clinical and pathologic characteristics (Table S1). Initial array-based DNA copy number profiling was used to select tumors that harbored abnormalities typical of DLPS tumors, including 12q amplifications, among others (see Methods; Fig. S1A). A detailed histologic analysis indicated that both tumors, while heterogeneous, were predominantly dedifferentiated (see Supplementary Methods; Fig. S1B–H). Using high-grade areas of each sample, we performed long-insert mate-paired whole-genome sequencing (~5× coverage), exome sequencing (~30–70× coverage), RNA sequencing of the poly(A)+ transcriptome, and an enrichment of methylated DNA followed by sequencing (Fig. S2). These integrative analyses were complemented by analyses of selected mutations, methylation, and expression changes in larger sets of tumor and normal samples, and by assessing the functional significance of aberrant methylation of specific genes by pharmacological inhibition both in vitro and in an accompanying animal model of DLPS.
Results
Structural rearrangement in liposarcoma
We identified 355 and 543 somatically acquired genomic rearrangements (intra- and inter-chromosomal) in DLPS1 and DLPS2 respectively, from mate-paired sequencing reads that aligned atypically to the genome (Table S2, estimated false discovery rate (FDR) of 8.7%; see Supplementary Methods). In these genomes, genomic amplification at one or both breakpoints was the source of nearly all rearrangements, including 94.6% in the primary DLPS1, and all but three (99.4%) in the recurrent DLPS2 (all involving chromosome 9 sequence). These patterns are fundamentally different from those observed in breast or pancreatic cancers (4, 5), in which a greater fraction of rearrangements are associated with tandem duplication or deletion than genomic amplifications, though variability exists. Despite the common origin of most rearrangements found here, the patterns observed in each tumor were markedly different. Although chromosome 12 amplifications were prominent in both tumors and were the primary source of structural abnormality in DLPS1, the recurrent DLPS2 had a less complex 12q amplicon, but greater diversity of inter-chromosomal rearrangements (Fig. 1A–B).
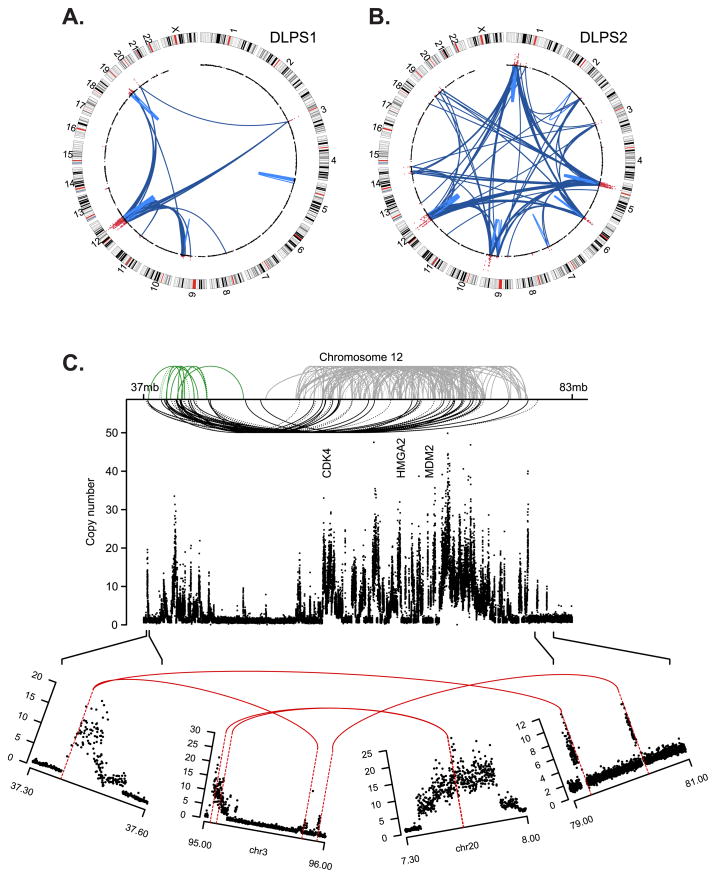
Figure 1. Somatic rearrangements in two sarcomas.
Structural rearrangements and DNA copy number alterations detected in two retroperitoneal liposarcoma genomes, a primary tumor, DLPS1 (A) and a local recurrence, DLPS2 (B). Chromosomes are plotted in the outer ring with the centromeres indicated in red. DNA copy number data inferred from whole-genome sequencing is indicated in the inner ring with genomic amplifications highlighted (red). Structural rearrangements are edges between two indicated loci, either intra-chromosomal (light blue) or inter-chromosomal (dark blue). C. The remodeling of chromosome 12 in DLPS1 is shown across ~46Mb of the q-arm. Three clusters (green, gray, and black) of both inverted and non-inverted intra-chromosomal rearrangements (dotted and solid, respectively) are shown spanning the progressive amplicon (copy number as indicated, y-axis). Three landmark genes (CDK4, HMGA2, and MDM2) are annotated. In the lower panel, the amplicon structure and rearrangement pattern of the 5’-most and 3’-most breakpoints are shown, indicating a circular structure including interspersed sequences from chromosomes 3 and 20, on top of which the complex pattern of intrachromosomal rearrangements (top) were likely acquired through successive rounds of unstable replication.
The instability of the 12q locus in DLPS1 was noteworthy. The greater amplicon spanned ~44Mb of the q-arm and showed a complex network of back-and-forth intrachromosomal rearrangements in both inverted and non-inverted orientations (Fig. 1C). Three clusters of rearrangement were apparent and suggest a model of amplicon evolution that started with an initial co-amplification of CDK4 and MDM2 (albeit in distinct events) incorporating sequence from focal copy number amplifications of 3q11.2 and 20p12.3, respectively. This was cleaved out and replicated autonomously and unstably, likely acquiring the additional intra-chromosomal rearrangements sporadically over late cell cycles. While the clustering of rearrangements was distinct, and the systematic remodeling of chromosome 12 profound, its origin is unlikely to be a chromothripsis, a single cellular catastrophe posited to drive a subset of osteosarcomas or chordomas (6). The 12q structure in DLPS1 lacks many of the hallmarks of chromothripsis, such as cycling between discrete copy number states and retained heterozygosity (inferred from 250K SNP array data (3) and heterozygous SNPs in the whole-genome sequence data). The 12q structure in both DLPS1 and DLPS2 instead likely resulted from progressive rearrangement and amplification. Additionally, while retrotransposition of mobile elements (principally L1 and Alu) is a natural source for structural mutagenesis in human genomes (7, 8), none of the rearrangement breakpoints detected in either genome corresponds to known insertion polymorphisms (dbRIP release 2, ref. (9)) or those recently found in lung tumors (10). Finally, de novo assembly from reads that spanned the complex 12q rearrangement in DLPS1 failed to generate long contigs, confirming the complex interspersed pattern of the amplicon indicated by spectral karyotyping of DLPS cell lines (data not shown).
In total, 64% of rearrangements affected the sequence of protein-coding or non-coding genes (compared to ~56% expected by chance, p-value < 10−4; Supplementary Methods), and were diverse in pattern. Notable events included internal rearrangements and translocations involving HMGA2 in both DLPS genomes (Fig. 2A). High mobility group A2 (HMGA2) is a non-histone, chromatin-associated protein that lacks transcriptional activity, but regulates transcription in trans by altering chromatin architecture (11). Rearrangements involving HMGA2 on 12q are recurrent in DLPS, with whole-gene HMGA2 amplifications and a 5’ amplification with a breakpoint between exon 5 and the 3’ untranslated region (UTR) of the long isoform each affecting ~30% of tumors (3). The primary tumor (DLPS1) harbored the latter complex 5’ amplicon, so we examined its structure and transcriptional consequences in detail. This amplicon included the full HMGA2 open reading frame (ORF), which was fused to intergenic chromosome 12 sequence (Fig. 2A, top). This structure was confirmed by concurrent RNA sequencing with splice junction reads indicating that only the long isoform was expressed and that it lacked its 3’ UTR (Fig. 2A, bottom). RNA sequence data confirmed that HMGA2 was strongly over-expressed in DLPS1, though not in DLPS2 (Fig. 2A, inset), which had an internal intron 3 amplicon clustering in a position similar to fusion-associated rearrangements in benign lipomas (11) though without a fusion partner. In DLPS1, the structure and over-expression of HMGA2 suggest that the rearrangement eliminated microRNA repression of the gene mediated by let7 binding to the 3’ UTR (12). This combined with the genomic amplification of the full ORF, likely accounts for the potent over-expression of HMGA2 in this tumor.
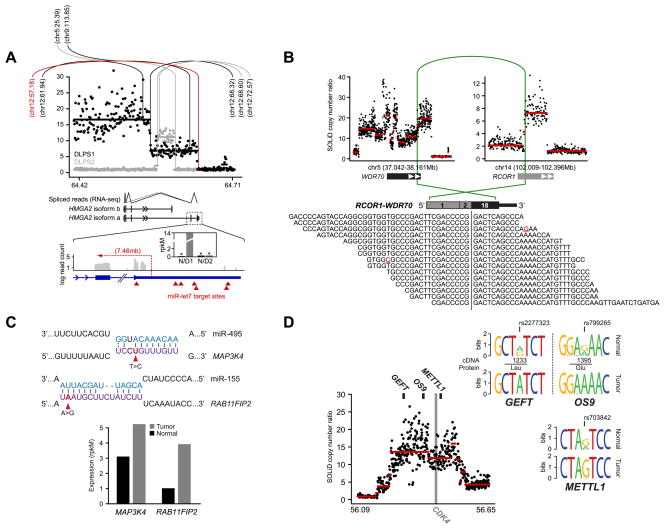
Figure 2. Diverse abnormalities manifest in RNA.
A. The segmented copy number profile, inferred from whole-genome sequencing, of the HMGA2 locus (12q14.3) is plotted for the both DLPS1 (black) and DLPS2 (gray). Rearrangements are indicated (vertical/arc lines) and the position of their second breakpoint is annotated as megabases on the recipient chromosome. In red is the structural rearrangement in DLPS1 affecting the boundary between HMGA2 exon 5 and the 3’ UTR. Splice junction reads from RNA sequencing of DLPS1 (bottom panel) confirm that the long isoform (a) is expressed. A high-resolution view of the final exons and 3’ UTR of the transcript is shown; transcriptome read coverage confirms truncation and loss of the 3’ UTR from the rearrangement and significant over-expression of HMGA2 in DLPS1 (inset; gray and black are normal and tumor samples respectively). B. Rearrangement in DLPS2 pairing a simple intragenic amplicon in RCOR1 with a complex amplicon spanning WDR70. The RCOR1 intron 2 breakpoint juxtaposed the 5’ region including exon 2 with exon 18 and the amplified 3’ end of WDR70 (green arc). In total, 15 RNA sequencing reads supported the predicted fusion junction of this out-of-frame chimera. C. Somatic mutations in the 3’ UTRs of MAP3K4 and RAB11FIP2 in DLPS2 that were detected from RNA but not exome sequencing fall in the seed regions of conserved target sites complementary to miR-495 and miR-155 respectively (purple, microRNA sequences are shown above in blue). Both genes have elevated expression in DLPS2 compared to their matched normal tissue (bottom, measured as rpkM), consonant with release from microRNA repression. D. Allele-specific expression of GEFT, OS9, and METTL1, three genes adjacent to the CDK4 oncogene on 12q13.3–14.1, was detected from heterozygous exonic variants from RNA sequencing in DLPS1 (reflected as sequence logos inferred from spanning reads) and is attributable to a single complex genomic amplification.
For those DNA rearrangements that juxtaposed two genes into a putative novel ORF, we used RNA sequencing data to explore the candidate fusion junctions and identified seven acquired chimeric fusion transcripts, of which five (71%) validated with RT-PCR of RNA across the rearranged exon-exon junction (Table S3 and Fig. S3; see Supplementary Methods). While in some cases highly expressed, as with the RCOR1-WDR70 out-of-frame fusion (Fig. 2B), studying additional cases is necessary to determine if these are actually passenger events, given the extensive genome remodeling in these two sarcomas. This large burden of rearrangements will, by chance, affect the coding regions of many genes, among which a minority will result in expressed gene fusions. Indeed, the scarcity of expressed fusions in these genomes on a background of systematic rearrangement may indicate instead that the primary driver of 12q amplification in DLPS tumors is the dosage increases of a large number of genes. Nevertheless, our current analysis confirms the value of cross-validating DNA rearrangements with concurrent RNA sequencing to identify expressed fusions.
The nucleotide mutational landscape
To determine somatic nucleotide substitutions in these tumors, we combined the results of RNA sequencing with those of exome capture and DNA sequencing in both tumors and matched normal samples. We respectively identified 8 and 13 confirmed somatic mutations in the two tumors after obtaining independent validation data with deep 454 sequencing on a subset of candidate mutations (Table S4; see Supplementary Methods). This is equivalent to ~0.53 tumor-specific mutations per million bases, a modest point mutation burden similar to our previous findings using limited Sanger sequencing of selected candidate genes (3). In fact, while the patterns of structural rearrangement between the primary and local recurrence were very different (Fig. 1A–B), the point mutation rate and the nucleotide context of the mutations were similar. Exome sequencing also captured a significant fraction of the whole-gene and intragenic copy-number alterations apparent in the whole-genome sequencing data (Fig. S4), as has previously been observed with targeted approaches (13).
Several mutated genes confirmed previously implicated dysregulated pathways in liposarcomagenesis, though by unexpected means, while others were novel (Supplementary Results). We investigated four of the mutant genes (HDAC1, MAPKAP1, PTPN9, and DAZAP2) by sequencing their coding exons in a validation set of 96 liposarcomas (80 tumors including DLPS1 and DLPS2 as controls, plus 16 cell lines). In total, somatic mutations in HDAC1, MAPKAP1, PTPN9, and DAZAP2 were identified in 8.3%, 3.1%, 3.1% and 2.1% of liposarcomas respectively (Table S4; in addition to rediscovering the four mutations in the originally sequenced samples). None of the mutations among the 94 additional liposarcomas were identical to those originally described. Nevertheless, the frequency, diversity, and predicted functional impact of the HDAC1 mutations (Table S4) identified here is noteworthy, and is consistent with the emergence of mutations in key epigenetic or chromatin machinery in multiple human cancer types.
In addition to the coding point mutations and small insertions and deletions described here, liposarcoma transcriptomes were altered in diverse ways. We confirmed two somatic mutations in DLPS2 in the 3’ UTRs of MAP3K4 (373T>C) and RAB11FIP2 (3879A>G) respectively (Table S4). These altered the seed sequence of predicted microRNA target sites (MAP3K4/miR-495 and RAB11FIP2/miR-155), and were consistent with elevated expression in the tumor (Fig. 2C, Supplementary Results). Differential allele-specific expression (ASE) between normal and tumor transcriptomes was also apparent (Table S5). Its origins could be traced with concurrent DNA sequencing, as in the case of three genes on or adjacent to the CDK4 amplicon on 12q13.3–14.1 (GEFT, OS9, and METTL1) with ASE resulting from a single allele-specific copy number amplification (Fig. 2D, see Supplementary Results). Finally, we explored ADAR-catalyzed adenosine-to-inosine [A>I(G)] RNA editing by searching for A>G transitions in cDNA data present as homozygous A alleles in DNA (Table S6; see Supplementary Methods). However, we found little evidence that A>I editing contributes to tumor transcriptome diversity in liposarcomas (Supplementary Results).
CEBPA is frequently methylated in DLPS
To investigate cell-type and tumor-specific DNA methylation patterns, we generated ~62 million uniquely aligned reads [3.1 gigabases (Gb)] in the matched tumor and normal samples with an MBD-based enrichment protocol (14) (see Methods). From peaks of methylation generated from read distributions, we found that global patterns of cytosine methylation were similar between each tumor and normal and between the two patients (Fig. S5–7). After normalizing methylation levels by the tumor's intrinsic copy number inferred from whole-genome sequencing (Fig. S8–9), we identified persistently differentially methylated regions (DMRs) between normal and tumor genomes (FDR <1%, see Supplementary Results and Methods). Between 2.5 and 3.4% of assessed regions were differentially methylated between a tumor and its matched normal genome, and these DMRs demonstrated significant overlap (p-value ~ 0). We identified 3,186 regions of recurrent differential methylation spanning 2.38 Mb of total sequence (143–1391bp; 10–90th percentile). Regions of statistically significant somatic gain or loss of methylation in the tumors appear to arise across diverse sequence features in a highly non-random fashion (Fig. 3A, Supplementary Results).
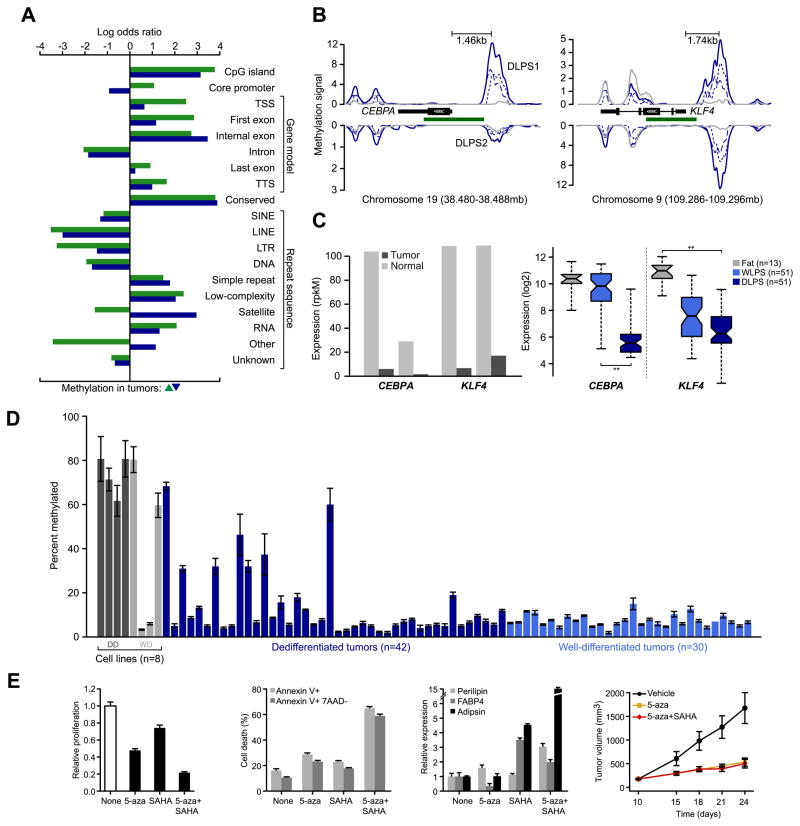
Figure 3. Core promoter methylation in mediators of adipogenesis.
A. Patterns of somatic methylation enriched or depleted (measured by log odds ratio relative to background sequence in the human genome) among 19 sequence contexts including the canonical gene cassette. Tumor-specific increases and decreases of methylation are green and blue, respectively. B. Methylation of CEBPA and KLF4 is increased in tumors (blue), but not their matched normal adipose tissues (gray) in upstream regions or in regions spanning adjacent CpG islands (green). Methylation signal for each tumor is plotted for both the positive and negative strands (dotted and dashed respectively) and combined into total signal (solid) for each sample. Also indicated is the distance between the peak of methylation in both promoters and their respective transcription starts sites. C. CEBPA and KLF4 expression, inferred from RNA sequencing in the methylated samples (normalized rpkM) indicates their reduction in DLPS1 and DLPS2 compared to their matched normal adipose tissues (left panel). On the right, expression measured by microarray in a cohort of 115 tissues (as shown; starred, p-values < 10−12, ANOVA). D. CEBPA promoter methylation status (average percent methylated, two biological replicates assessed by bisulfite pyrosequencing, error bars represent standard deviation) in 8 cell lines (dark and light gray are dedifferentiated and well-differentiated cell lines, respectively) and 72 tumors indicated that CEBPA methylation is high in cell lines and a subset of DLPS, but absent from WLPS tumors. E. Proliferation of DDLS8817 dedifferentiated liposarcoma cells after treatment with 5-aza, SAHA, or the combination of both (left; mean ± propagated error). Proliferation levels are shown relative to untreated cells. The percentage of apoptotic cells is shown (middle, left; measured by annexin V and 7-AAD staining, mean percent positive ± standard deviation) in untreated and drug-treated DDLPS8817 cells. Expression of early, intermediate, and late markers of differentiation (perilipin, FABP4, and adipsin respectively) were measured by RT-PCR (mean ± standard deviation) in the presence of drug and shown relative to the level in untreated cells (middle, right). The growth of DLPS tumors in mice (DDLS8817 xenografts) treated with 5-aza (decitabine), 5-aza plus SAHA, or vehicle (right; mean tumor volume ± standard error of the mean, n = 5 mice/group) was also analyzed.
In total, 833 DMRs originated from the canonical promoters of 677 genes. RNA sequencing indicated that 175 of these genes were expressed in the normal adipose samples and differentially expressed (≥ 2-fold) in the affected tumor. Noteworthy among these were multiple effectors of adipocyte differentiation: GATA2, KLF4, and CEBPA. While GATA2 was among the most hypomethylated genes, GATA2 mRNA levels were not induced despite its proven role as an anti-adipogenic factor. KLF4 plays a vital role in multiple differentiated pathways, including adipogenesis, through its early regulation of C/EBPβ, and is among a set of genes whose expression reprograms somatic cells to acquire pluripotency. Promoter methylation reduced KLF4 expression in both tumors (Fig. 3B–C), and in a larger cohort of 115 normal adipose samples, DLPS tumors, and well-differentiated liposarcomas (WLPS), a precursor of DLPS, KLF4 expression was reduced in WLPS compared to normal fat and reduced further in DLPS.
C/EBPα (encoded by CEBPA) is a master transcriptional regulatorof adipocyte differentiation, along with PPARγ (15). The gene is mutated and methylated in a subset of acute myeloid leukemias, and in DLPS, CEBPA under-expression is associated with greater risk of distant recurrence (16). Nevertheless, our previous studies did not identify genomic deletions or point mutations that could account for the lower CEBPA transcript levels in DLPS (3). Here, we identified a peak of methylation centered ~1.5 kb upstream of the CEBPA locus in both tumors (q-value = 2×10−10, Fig. 3B), although the relative levels of methylation differed (2- and 12-fold greater methylation than the matched normal samples after copy number normalization). We confirmed methylation at this site with pyrosequencing of bisulfite-treated DNA targeting the boundaries of the consensus peak (see Supplementary Methods). We also observed an accompanying loss of CEBPA expression in both methylated tumors (18- to 25-fold reduction, normalized rpkM, Fig. 3C). In the larger cohort of normal adipose tissue, DLPS, and WLPS tumors, loss of CEBPA expression was evident in DLPS, but not in the less-advanced WLPS tumors (Fig. 3C). To determine the frequency of CEBPA methylation in liposarcomas, we performed similar bisulfite sequencing in a panel of 80 WLPS and DLPS tumors and cell lines. CEBPA methylation was common, but only in the DLPS tumors (~24% versus 0% of WLPS tumors, p-value = 0.004, Fig. 3D). Additionally, we characterized CEBPA methylation in heterogeneous tumors containing both well-differentiated and dedifferentiated regions and identified CEBPA methylation in only the dedifferentiated cell populations (data not shown). In DLPS1 and DLPS2, the similar reduction of CEBPA mRNA levels despite the order-of-magnitude difference in methylation (Figs. 3B–C) suggests that CEBPA expression is sensitive to low levels of promoter methylation, and that additional methylation above the level observed in DLPS2 has little additional effect on expression.
Treating DLPS cells (the DDLS8817 cell line) with the DNA methyltransferase inhibitor 5-Aza-2’-deoxycytidine (5-aza; decitabine) reduced CEBPA promoter methylation levels by 30–50% (Fig. S10A). While no further reduction was observed after the addition of the histone deacetylase inhibitor SAHA, expression levels were induced 3- and 19-fold with 5-aza alone and in combination with SAHA, respectively, confirming a synergistic effect of demethylation and the release of transcriptionally repressive chromatin (Fig. S10B). More so than either compound alone, the combination of 5-aza and SAHA also substantially induced anti-proliferative and pro-apoptotic effects in DLPS cells in vitro and led to increased expression of early, intermediate, and late differentiation markers (Fig. 3E), confirming reactivation of the differentiation pathway. Finally, the combination of 5-aza and SAHA produced a significant reduction in DLPS tumor volume in vivo (Fig. 3E). We therefore propose that CEBPA methylation is common in DLPS, contributes in part to the adipogenic block in tumors that lack JUN amplification-driven C/EBPβ disruption (17), and is an important step in the progression of well-differentiated to dedifferentiated liposarcomas.
Epigenetic regulation of small RNAs in high-grade DLPS
While conventional methylation profiling focuses on CpG-rich regulatory regions of the genome, unbiased genome-wide methylation sequencing can identify epigenetic control of additional genomic features. Indeed, the regulation of small RNAs by methylation has emerged as a mechanism in cancer, but has been investigated predominantly in a microRNA-specific manner (18) or has been measured indirectly (19). Here, we identified 13 microRNAs whose putative promoters were differentially methylated (data not shown). Notably, a peak of methylation spanned the putative promoter of miR-193b on 16p13.12 in the 3’ shore of an adjacent CpG island in DLPS1, but not its matched normal sample (q-value = 1.1×10−13, Fig. 4A–B). While microRNA promoters are less well characterized than their protein-coding counterparts, this locus has significant H3K4me3 enrichment in multiple human lineages, a histone modification that marks active promoters(20, 21) (Fig. 4B). To test whether increased promoter methylation silenced miR-193b in this tumor, we explored miR-193b expression with small RNA sequencing in this sample and a larger cohort of normal adipose tissue, WLPS, and DLPS (n = 58, ref. (22)). This confirmed that miR-193b expression was selectively lower in DLPS (p-value < 10−8, Fig. 4C). Among WLPS, miR-193b expression levels were similar to those of normal adipose tissue, indicating that loss of miR-193b expression is a late event in liposarcoma progression.
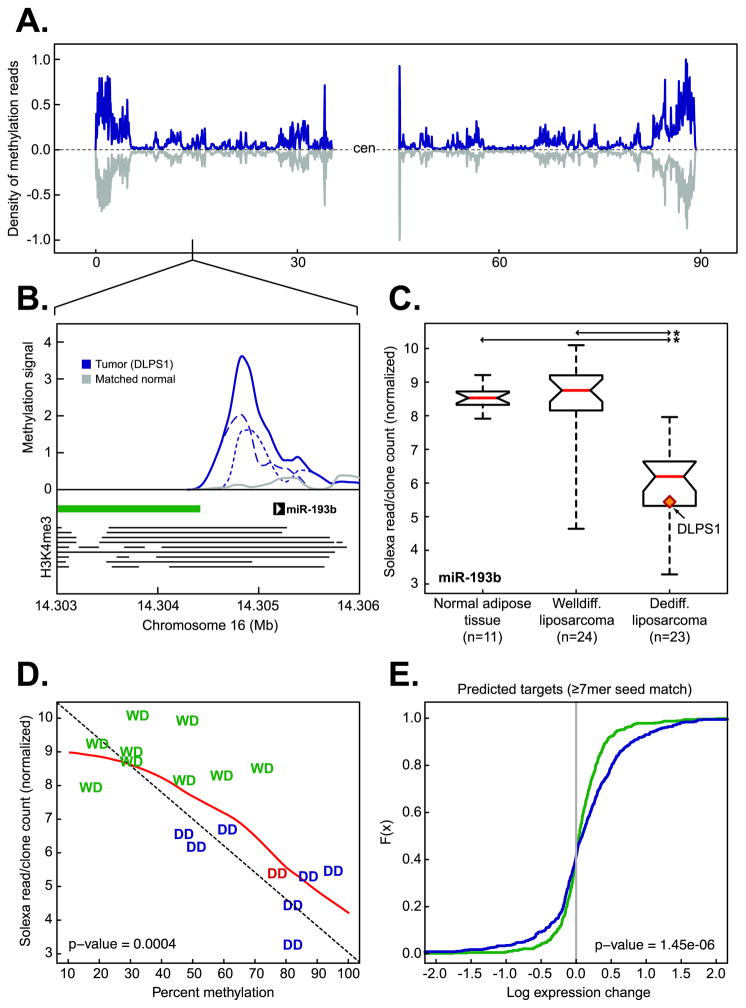
Figure 4. Epigenetic regulation of miR-193b in liposarcomagenesis.
A. The density of methylation on chromosome 16 in DLPS1 (blue) and its matched normal adipose tissue (gray). B. Methylation in the primary tumor (solid blue, positive and negative strands are dotted and dashed respectively) and matched normal adipose tissue (gray) in the region of the putative promoter [the position of enrichment of histone H3K4me3 in nine human cell types (20) is indicated by horizontal lines] of miR-193b overlapping the shore of a CpG island (green bar). C. Expression of miR-193b in normal adipose tissue samples, WLPS, and DLPS tumors determined by small RNA sequencing. The tumor in which methylation was observed (panel B) is highlighted (starred; p-value < 10−9, Student’s t-test). D. Expression of miR-193b as a function of percent methylation in the putative miR-193b promoter in a panel of both well-differentiated (WD, green) and dedifferentiated (DD, blue; DLPS1 is in red ) liposarcomas [n = 17; the diagonal is indicated by dotted line; the red line is the regression (loess)]. E. Cumulative distributions of expression changes of predicted miR-193b target genes (≥ 7 bp seed match length, n = 547 genes with expression data) between DLPS tumors and normal adipose tissues (green) or WLPS tumors and normal adipose tissues (blue) indicate that a greater number of predicted targets had increased expression in DLPS tumors with methylated miR-193b than expected by chance (p-value as indicated, Kolmogorov-Smirnov).
To explore the possibility that methylation was responsible for the reduction of miR-193b in this larger cohort, we performed pyrosequencing of bisulfite-converted DNA in the region representing the peak of methylation from DLPS1. In a panel of WLPS and DLPS tumors for which small RNA sequencing data were available, we found that as the percent of methylation increased in this region, miR-193b expression was reduced, and this was specific to DLPS (R = −0.76, p-value = 0.0004; Fig. 4D). To further explore the effect of epigenetic silencing of miR-193b, we characterized the expression changes of its predicted targets. Predicted gene targets of miR-193b were determined with miRanda-mirSVR (23) and included only those highly conserved target sites with a seed match length of ≥7bp. We examined their global expression changes in WLPS and DLPS compared to normal adipose tissue samples. In DLPS tumors in which miR-193b is silenced by methylation, genes with predicted targets sites for miR-193b were significantly upregulated compared to normal adipose tissue, an effect we did not observe in well-differentiated tumors with normal miR-193b levels (one sided Kolmogorov-Smirnov p-value = 1.45 × 10−6; Fig. 4E). These data indicate that miR-193b is selectively silenced by methylation in aggressive disease, which broadly affects the expression of its putative targets. This finding hints at a contribution of epigenetic alteration of small RNAs to liposarcomagenesis and a broader role of systematic epigenetic deregulation of small RNAs in cancer genomes.
Discussion
Here we report the concurrent sequencing of human tumor genomes, exomes, transcriptomes, and cytosine methylomes, the results of which were integrated to yield a picture of genomic alterations in a primary and a locally recurrent liposarcoma. While these tumors harbored abnormalities at all levels, the structural remodeling of liposarcoma genomes was profound and had varied effects on their transcriptomes. Exome sequencing revealed a modest point mutation rate in these tumors, yet uncovered for the first time genes recurrently mutated in this subtype of soft-tissue sarcoma (HDAC1, MAPKAP1, PTPN9, and DAZAP2). Also, this first portrait of liposarcoma methylomes indicates that aberrant methylation has broad effects tied closely to differentiation phenotypes and suggests a role for demethylating agents in the treatment of DLPS tumors.
The prevalence of CEBPA methylation found here, as well as its specificity and clonality in DLPS, raises interesting questions about the dysregulation of adipogenesis in these tumors. The C/EBPβ-C/EBPα transcriptional network is complex, regulating multiple stages of the commitment of cells to terminal differentiation. C/EBPβ directly induces the expression of CEBPA, a process disrupted by JUN amplification in a subset of undifferentiated tumors (17). While the adipogenic block is common to all DLPS tumors, we and others observed JUN genomic amplification and over-expression in only ~24% of cases (3) (though in the absence of amplification, JUN may also be over-expressed via upstream kinase-driven activation). Moreover, recent evidence indicates that activated JUN, while oncogenic in DLPS cells, is not sufficient to block adipocytic differentiation, implying alternative abnormalities are necessary (24). We speculate that the CEBPA promoter methylation identified here may uncouple CEBPA expression from C/EBPβ-mediated induction and therefore may at least partially contribute to the unexplained adipogenic block in DLPS.
The relationship between CEBPA methylation and the HDAC1 mutations discovered here in adipogenesis is unknown. Generally, the histone deacetylase activity of HDAC1 targets promoters and blocks transcription, and although the loss of HDACs can enhance adipogenesis (25, 26), this is not true in all contexts (27). HDAC1 specifically sequesters and represses the C/EBPα promoter, and PPARγ is necessary to release C/EBPα from HDAC1-mediated repression (28). These data, in combination with our observation of KLF4 and CIDEA methylation (data not shown), suggests the possibility of a series of alterations necessary to mediate different aspects of the adipogenic block in these tumors.
Other pathways and non-adipogenic phenotypes are likely also involved in the progression of well-differentiated liposarcomas to dedifferentiated disease, and our finding of specific miR-193b methylation and silencing in aggressive disease implies an important role for aberrant expression of small RNAs in liposarcomagenesis. A role for miR-193b in cancer has been proposed based on its reduced expression in both hepatocellular carcinoma and melanoma (29, 30), and recent evidence suggests miR-193b is epigenetically silenced in prostate cancer (31). While the validated miR-193b targets CCND1 and ETS1 were over-expressed in some of these malignancies (29, 30), expression analysis of liposarcomas with and without miR-193b methylation indicated that neither gene is elevated in a DLPS-specific manner, as might be expected for miR-193b loss. However, because CCND1 regulates CDK4 activity, and CDK4 is amplified in ~90% of DLPSs, upregulation of CCND1 due to miR-193b loss might be functionally redundant. This is confirmed by its lack of induction in DLPS tumors harboring miR-193b loss. Nevertheless, other predicted miR-193b targets, including KRAS, ETV1, STMN1, and RAD51, were preferentially upregulated in miR-193b-silenced tumors, in not only the original tumor in which we identified miR-193b methylation (DLPS1), but also in the larger panel of dedifferentiated tumors. These data raise the possibility that the effects of miR-193b silencing are lineage-and context-specific.
This work represents an initial integrated sequence analysis of liposarcoma, and while most non-coding sequence is insufficiently covered by these experiments, the depth and breadth of alterations revealed here indicates the somatic complexity of cancers is still under-appreciated. These data also reveal an unanticipated role for epigenetic abnormalities in dedifferentiated liposarcoma and the potential therapeutic importance of demethylating agents for treating liposarcomas.
Materials and Methods
Patient specimens
Tumor and normal tissue were procured at Memorial Sloan-Kettering Cancer Center (MSKCC) and obtained with both patient consent and institutional review board approval. Representative samples were selected for sequencing from a large panel of dedifferentiated liposarcomas of typical clinical and histopathological characteristics in the Clinical Sarcoma Database (MSKCC). This panel was profiled on Agilent 244K aCGH arrays and hierarchically clustered (Supplementary Methods, Fig. S1). Samples selected for sequencing were required to (a) harbor canonical amplification of the 12q ring chromosome (2), (b) have additional lesions similar to those detected in our previous study of this and six additional sarcoma subtypes (3), (c) have moderate-to-low additional non-12q genome complexity, and (d) have available normal subcutaneous fat culled at the time of surgery. Among the tumors meeting these criteria, two were chosen: a primary tumor (DLPS1) from a male patient and a locally recurrent tumor (DLPS2) from an unrelated female patient. Following histological review by a pathologist, the tumor samples were cryomold macro-dissected to remove regions of necrosis.
Sequencing
Genomic DNA for all DNA-based sequencing was isolated using the DNeasy Tissue Kit (Qiagen). For genome sequencing, mate-pair libraries were constructed with an insert size range of 1–2kb. Library preparation, emulsion PCR, bead deposition, and sequencing were all performed according to the Applied Biosystems SOLiD Library Preparation Protocol, and a full slide of 2 × 50 bp sequence was produced per sample. For exome sequencing, pre-capture sequencing libraries were hybridized in solution with the Agilent SureSelect platform per the manufacturer’s instructions. For each capture library, a quad of 50-bp fragment sequence was generated from an upgraded SOLiD 4 instrument. For transcriptome sequencing, total RNA was isolated with the RNeasy Lipid Tissue Mini Kit (Qiagen) and subjected to both poly(A)+ selection and ribosomal RNA depletion. A 50-bp fragment library was generated and sequenced in a quad per sample according to the manufacturer’s instructions (SOLiD 3). Methylation libraries were generated after enrichment of methylated DNA with the Sequence Tag Analysis of Methylation Patterns (STAMP) protocol (14). This MBD affinity-purification assay is selective for methylated CpGs and suitable for massively parallel sequencing. MBD-enriched fragment libraries were prepared and run in either quad or octet format on a SOLiD 3 instrument. The small RNA sequencing discussed here was performed on an Illumina Genome Analyzer II, and is described in greater detail elsewhere (22). In all experiments, sequencing generated 50-mer dinucleotide reads in color space, and data were obtained from concurrent sequencing runs on the SOLiD 3 and 4 platforms. Sequencing data are available from the NCBI Short Read Archive (SRA) with accession number SRA046057.
Analysis and validation experiments
The details of read alignment and visualization, as well as the analysis of DNA copy number alterations, structural rearrangements, mutations, gene fusions, transcript and microRNA expression, methylation, mutation validation and recurrence testing, and CEBPA validation experiments, are available in the Supplementary Data. The primers used for methylation and mutation validation are available in Table S7. Liposarcoma cell lines used here including DDLS8817, ALT9070, DD5590p0, DD6960-1A, RWD8000-2, RWD8000-3, RDD6960-2, RDD8107-2, RWD5700-1, RWD3051, and WD0082 were established from WLPS and DLPS tissue samples obtained from consenting patients at MSKCC. LPS141 was established by J. Fletcher and S. Singer from a patient with DLPS resected at the Brigham and Women's Hospital. Molecular cytogenetics and/or array comparative genomic hybridization studies confirmed all cell lines contained the characteristic 12q amplification.
Supplementary Material
Significance.
Multi-modality sequence analysis of dedifferentiated liposarcomas (DLPS) revealed recurrent mutations and epigenetic abnormalities critical to liposarcomagenesis and to the suppression of adipocyte differentiation. Pharmacological inhibition of DNA methylation promoted apoptosis and differentiated DLPS cells in vitro and inhibited tumor growth in vivo, providing a rationale for investigating methylation inhibitors in this disease.
Acknowledgments
For advice and discussion we thank D. Betel and A. Olshen. We also thank M. Ladanyi for critical reading of the manuscript, members of the MSKCC Genomics Core Laboratory for support, and M. Pirun for assistance with the mutation validation pipeline to process 454 data.
Grant Support
This study was supported by the National Cancer Institute and the National Institutes of Health through the Soft Tissue Sarcoma Program Project grant P01 CA047179 (S.S, N.D.S, and B.S.T). B.S.T. is the David H. Koch Fellow in cancer genomics at MSKCC. This work was also partially supported by a generous donation from Richard Hanlon.
Footnotes
Conflict of interest: No potential conflicts of interest to disclose.
References
- 1.Fletcher C, Unni K, Mertens F. Pathology and genetics of tumors of soft tissue and bone. International Agency for Research on Cancer Press; Lyon: 2002. [Google Scholar]
- 2.Pedeutour F, Forus A, Coindre JM, Berner JM, Nicolo G, Michiels JF, et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer. 1999;24:30–41. [PubMed] [Google Scholar]
- 3.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–70. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–82. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27:323–9. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7:899–910. doi: 10.1038/nrc2271. [DOI] [PubMed] [Google Scholar]
- 12.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman DS, Hovingh GK, Iartchouk O, Rehm HL, Kucherlapati R, Seidman JG, et al. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods. 2009;6:507–10. doi: 10.1038/nmeth.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One. 2011;6:e14524. doi: 10.1371/journal.pone.0014524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–96. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 16.Gobble RM, Qin LX, Brill ER, Angeles CV, Ugras S, O'Connor RB, et al. Expression Profiling of Liposarcoma Yields a Multigene Predictor of Patient Outcome and Identifies Genes that Contribute to Liposarcomagenesis. Cancer Res. 2011;71:2697–705. doi: 10.1158/0008-5472.CAN-10-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariani O, Brennetot C, Coindre JM, Gruel N, Ganem C, Delattre O, et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell. 2007;11:361–74. doi: 10.1016/j.ccr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 19.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, et al. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ugras S, Brill ER, Jacobsen A, Hafner M, Socci N, Decarolis PL, et al. Small RNA sequencing and functional characterization reveals microRNA-143 tumor suppressor activity in liposarcoma. Cancer Res. 2011;71:5659–69. doi: 10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder EL, Sandstrom DJ, Law K, Fiore C, Sicinska E, Brito J, et al. c-Jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol. 2009;218:292–300. doi: 10.1002/path.2564. [DOI] [PubMed] [Google Scholar]
- 25.Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. J Biol Chem. 2006;281:6608–15. doi: 10.1074/jbc.M508982200. [DOI] [PubMed] [Google Scholar]
- 26.Wiper-Bergeron N, Wu D, Pope L, Schild-Poulter C, Hache RJ. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J. 2003;22:2135–45. doi: 10.1093/emboj/cdg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagace DC, Nachtigal MW. Inhibition of histone deacetylase activity by valproic acid blocks adipogenesis. J Biol Chem. 2004;279:18851–60. doi: 10.1074/jbc.M312795200. [DOI] [PubMed] [Google Scholar]
- 28.Zuo Y, Qiang L, Farmer SR. Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter. J Biol Chem. 2006;281:7960–7. doi: 10.1074/jbc.M510682200. [DOI] [PubMed] [Google Scholar]
- 29.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, et al. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol. 2010;176:2520–9. doi: 10.2353/ajpath.2010.091061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, et al. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur J Cancer. 2010;46:2828–36. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 31.Rauhala HE, Jalava SE, Isotalo J, Bracken H, Lehmusvaara S, Tammela TL, et al. miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int J Cancer. 2010;127:1363–72. doi: 10.1002/ijc.25162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.