Abstract
Collagen constitutes one-third of the human proteome, providing mechanical stability, elasticity, and strength to organisms. Normal type I collagen is a heterotrimer triple-helical molecule consisting of two α-1 chains and one α-2 chain. The homotrimeric isoform of type I collagen, which consists of three α-1 chains, is only found in fetal tissues, fibrosis, and cancer in humans. A mouse model of the genetic brittle bone disease, osteogenesis imperfect, oim, is characterized by a replacement of the α-2 chain by an α-1 chain, resulting also in a homotrimer collagen molecule. Experimental studies of oim mice tendon and bone have shown reduced mechanical strength compared to normal mice. The relationship between the molecular content and the decrease in strength is, however, still unknown. Here, fully atomistic simulations of a section of mouse type I heterotrimer and homotrimer collagen molecules are developed to explore the effect of the substitution of the α-2 chain. We calculate the persistence length and carry out a detailed analysis of the structure to determine differences in structural and mechanical behavior between hetero- and homotrimers. The results show that homotrimer persistence length is half of that of the heterotrimer (96 Å vs. 215 Å), indicating it is more flexible and confirmed by direct mechanical testing. Our structural analyses reveal that in contrast to the heterotrimer, the homotrimer easily forms kinks and freely rotates with angles much larger than heterotrimer. These local kinks may explain the larger lateral distance between collagen molecules seen in the fibrils of oim mice tendon and could have implications for reducing the intermolecular cross-linking, which is known to reduce the mechanical strength.
Introduction
Collagen constitutes one-third of the human proteome, providing mechanical stability, elasticity, and strength to organisms (1–6). Type I collagen is a heterotrimer consisting of two α-1 chains and one α-2 chain. Homotrimeric collagen, consisting of three α-1 chains has previously been studied to help define the role of the α-2 chain on collagen stability and mechanical integrity (7–9). In humans, homotrimeric collagen is found only in fetal tissues (10), fibrosis (11–13), and cancer (14–19). Studies have shown that homotrimers of type I collagen are resistant to mammalian collagenases (14,20). In a mouse model of the genetic brittle bone disease, osteogenesis imperfecta (OI), the oim mutation of type I collagen is characterized by complete replacement of the α-2 chain by an α-1 chain, resulting in homotrimeric collagen. The oim phenotype has extreme skeletal fragility, reduced stature, and bone deformities, mimicking moderate to severe OI in humans. Experimental studies have shown that the mechanical strength of oim bone and tail tendon is significantly less than that of the normal mice (9,21–25).
At the molecular level it has been proposed that the α-2 chain has a critical role in the integrity of the triple helix of collagen (9). X-ray diffraction patterns of oim tail tendon shows a loss of lateral packing in the collagen fibrils (9). Interestingly, the axial structure of the collagen molecule is unaltered in oim tissue and has the normal D-periodic banding (9,26). Using DSC, studies have shown that the denaturation temperature of oim homotrimeric collagen is higher than wild-type heterotrimeric collagen, which implies oim collagen is more thermally stable (27,28) and requires more energy to disrupt the integrity of the triple helix. At the same constant temperature, homotrimeric collagen denaturates 100 times slower than heterotrimeric collagen (27). Using DSC in both acetic acid and water, Miles et al. calculated that oim collagen has ∼5% greater volume fraction of water, which they propose increased the lateral distance between collagen molecules by 1.4 Å in oim fibers. The increase in water may be due in part to the lower hydrophobic content of the α-1 chain compared to the α-2 chain (8,29). The increased water content and therefore lateral distance between collagen molecules may be responsible for the loss of lateral packing and reduced cross-linking (28). Because cross-linking of molecules are critical for mechanical integrity of collagen (30,31), this could account for the weaker mechanical properties of the tissue at larger length scales. These studies provide insight into the role of the α-2 chain at the fibril level, however, it remains unclear how the loss of the α-2 chain in the homotrimer affects the behavior at the molecular level, which may lead to its altered fibril behavior.
Here, we use a molecular simulation approach to study the mechanical and structural differences between type I heterotrimer and homotrimer of real sequence mouse collagen molecules (collagen, type I, α-1 and α-2 chain precursor (Mus musculus)). The persistence lengths are calculated and the structural analyses are given to provide a complete understanding of the differences between heterotrimer and homotrimer collagen molecules. The persistence length quantifies the stiffness of long polymer chains, making it an appropriate parameter for estimating collagen stiffness. However, persistence length calculations assume that the chain is homogeneous, which is not true for collagen molecules. Both experimental studies (32–35) and computational models (36–38) have shown that the triple helical domain of collagen molecules is not homogeneous and has varied structural and biological properties along its length. Multiscale modeling has also shown that the mechanical properties of collagen molecules depend on the variation of sequence repeat (39,40). We therefore use structural analyses (unit height, radius, distances between Cα atoms) to study differences in behavior along the heterotrimer and homotrimer. Atomistic structural analysis helps to explain the fundamental mechanisms that affect the overall behavior of collagen molecules.
Materials and Methods
Collagen molecule generation
The real sequences of type I α-1 and type I α-2 chains of Mus musculus (wild-type mouse) are used to generate the collagen molecules. The heterotrimer collagen molecule is built of two α-1 chains and one α-2 chain, whereas the homotrimer collagen molecule is built of three α-1 chains. The sequences are adopted from the National Center for Biotechnology Information protein database (http://www.ncbi.nlm.nih.gov/protein): AAH50014.1 for α-1 chain and NP_031769.2 for the α-2 chain. The entire α-1 and α-2 chains consist of 1014 residues with repeated G-X-Y triplets, excluding the C-terminal and N-terminal sequences. A specific section with a length of 57 residues of the 1,014 total residues (from the 403rd to 459th residues) is selected to generate the heterotrimer and homotrimer collagen molecules. The chosen sequences are
-
α-1:
GFPGPKGTAGEPGKAGERGLPGPPGAVGPAGKDGEAGAQGAPGPAGPAGERGEQGPA and
-
α-2:
GFPGPKGPSGDPGKPGERGHPGLAGARGAPGPDGNNGAQGPPGPQGVQGGKGEQGPA.
Note that these sequences of the α-1 chain and α-2 chain are chosen because the six residues of the two ends are the same for both α-1 and α-2 chains to avoid the possible boundary effects of the heterotrimer and homotrimer. The amino acid composition (by %) in the segment is similar to the composition of the complete collagen molecule.
The collagen molecules are created by inputting the sequences of three chains into the software THeBuScr (41), which enables a user to build a triple-helical molecule based on any specified amino acid sequence. The code uses derived conformations from statistical analyses of high-resolution x-ray crystal structures of triple-helical peptides to build collagen molecule structures. The snapshots of initial structures of the heterotrimer and homotrimer are shown in Fig. S2 in the Supporting Material. To neutralize the terminals, two ends of the heterotrimer and homotrimer are capped by assigning the first residue to ACE, the acetylated N-terminus, and the last residue to CT3, the N-methylamide C-terminus. The length of the collagen molecule is ∼160 Å and is solvated in a periodic water box composed of TIP3 water molecules. The VMD software (42) is used to solvate the collagen and also to neutralize the system by adding ions. The final solvated all-atom system contains ≈37,000 atoms, with a box size of 280 Å × 40 Å × 40 Å.
All-atom equilibration
Full atomistic simulations are performed using NAMD (43) and the CHARMM force field (44) that includes parameters for hydroxyproline amino acids based on a suggestion by Anderson (45). This force field has been widely validated for a variety of biochemical models of proteins including collagen (39,46–49). An energy minimization using a conjugate gradient scheme is performed before molecular dynamics (MD) simulations. Rigid bonds are used to constrain covalent bond lengths, thus allowing an integration time step of 2 fs. Nonbonding interactions are computed using a cutoff for the neighbor list at 13.5 Å, with a switching function between 10 and 12 Å for van der Waals interactions. The electrostatic interactions are modeled by the particle mesh Ewald summation method. After energy minimization, the collagen molecule is simulated through 50 ns at a constant temperature of 310 K and 1.013 bar pressure in MD simulations. The first 10 ns simulations are used to equilibrate the system and are excluded from analyses. During the last 40 ns simulations, the configurations of collagen molecules are recorded every 20 ps, which results in 2000 frames. Each frame of the 2,000 frames is used in the analyses. The total simulation takes ∼10,000 CPU hours. We also performed repeated simulations for both the heterotrimer and homotrimer, from a different starting point, to confirm the reliability of our results (additional information included in the Supporting Material).
Mechanical testing
Steered MD is used to probe the mechanical properties of the heterotrimer and the homotrimer. This method is based on the concept of pulling a collection of chosen atoms via a spring along the pulling direction, while keeping another group of atoms fixed. After 40 ns NPT simulations, we fix the left ends of the collagen molecules and apply a constant velocity pulling method to stretch both the heterotrimer and the homotrimer. In both simulations, the spring constant is 7 kcal/mol/Å2 and the pulling velocity is set to 5 Å/ns.
Analysis of the collagen molecule structures
In collagen molecules, each chain is a repeating primary sequence of (Gly-X-Y)n and the three chains are staggered with respect to each other. The positions of the repeated Gly therefore capture the characteristic structures of collagen molecules. We use the Cα atom of each Gly residue as a measurement point to characterize molecular structure (Fig. 1, a and b). Five structural parameters of collagen molecule are analyzed: contour length, end-to-end distance, the distance between nearest Cα atoms, unit height, and radius (the distance from Cα atom to twisting axis).
Figure 1.
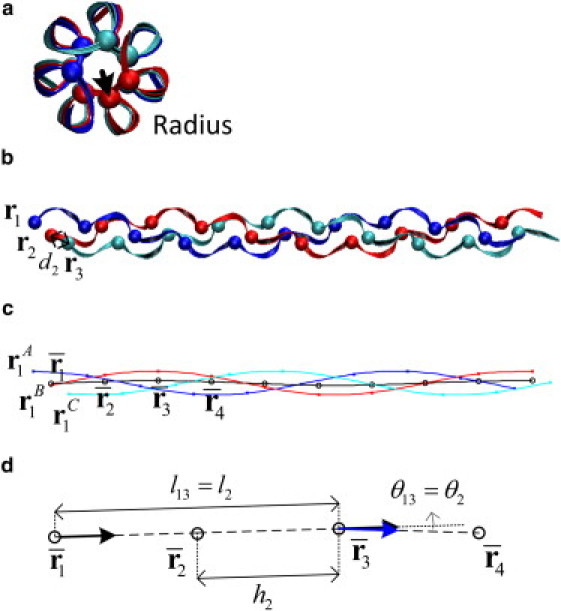
Structure of collagen protein materials. (a) The cross section of the collagen molecule. Each chain is plotted by ribbon in the same color. The spheres are the Cα atoms of Gly residues. The arrow indicates the radius, which is defined by the distance from the Cα atom to the twisting axis. (b) The side view of the collagen molecule. (c) The spline fit. The blue (red and cyan, respectively) line is the spline fit passing through the Cα atoms of chain A (B and C, respectively). x represents the positions of Cα atoms. The black line is the approximated twisting axis of the collagen molecule obtained by the spline fit. (d) Illustration on the unit height (h2), separated distance (l13) between two points on the collagen molecule and the angle (θ13) between the tangent vectors of these two points.
In our analysis, ri denotes the position of the Cα atom of i-th Gly in the collagen molecule. To estimate the contour length and the end-to-end distance of the collagen molecules, we compute the center of mass of all the nearest three Cα atoms (one from each chain because three chains in triple helix are staggered) by Here bi is the center of mass, and the contour length is then calculated by summing the distances between adjacent bi, i.e.,
| (1) |
where n is the number of the center of masses and Lc is the contour length. The end-to-end distance is estimated by the distance between the first and the last bi, i.e., ‖bn − b1‖. The distance between the i-th Cα atom to its nearest Cα atom can be simply computed by di = ‖ri + 1 − ri‖.
Furthermore, we use and to be the positions of the Cα atoms of the i-th Gly in chain A, B, and C, respectively, and to be the center of mass of the Cα atoms of the i-th Gly in three chains . The unit height is calculated by (see Fig. 1 d). The two unit heights of both ends are excluded from analyses to avoid the boundary effects. We calculate the radius as the distance between each Cα to the twisting axis of the collagen molecule, which is approximated by constructing a three-dimensional natural interpolating cubic spline curve to (Fig. 1 c). Once the spline curve is obtained, the i-th radius, i.e., the radius at is defined by averaging the radius at and . The radii at and are obtained by computing the distances between them to the twisting axis.
Persistence length calculations
The persistence length characterizes the stiffness of a long protein. Using θ to denote the angle between the tangent of two points of the protein separated by a distance L and Lp to denote the persistence length of the protein, the expectation value of the cosine angle exponentially decays with the distance (50), that is,
| (2) |
Alternatively, using r to denote the end-to-end distance of a long protein, the following relation can also be derived under the assumption that the protein is infinite long (51):
| (3) |
Both Eq. 2 and Eq. 3 are used to calculate the persistence length in this work. A brief derivation of Eq. 2 and Eq. 3 is provided in the Supporting Material. To use Eq. 2, we use the relation between θ and L. For each frame, we calculate lij(t), the distance between and and θij(t), the angle between the tangent of these two positions at time t (Fig. 1 d), thus we have
| (4) |
| (5) |
Note that here we approximate the tangent at by . The expectation value of the distance lm and the cosine angle cosθm between m-th neighboring centers of masses are then obtained by averaging through the simulation time:
| (6) |
| (7) |
The expectation value of the distance and the cosine angle are calculated by averaging the last 40 ns simulations. An exponential fitting, i.e., Eq. 2, is then applied to obtain the persistence length of collagen molecules. Note that the exponential decay in Eq. 2 holds only for long proteins and the length of the protein in the simulation is only 160 Å. Therefore, we only use the expected values up to the fifth neighboring center of masses.
An alternative way of calculating the persistence length is by using Eq. 3. The expectation value of the contour length and the end-to-end distance of collagen molecule are calculated by averaging the last 40 ns of the simulations. Then the persistence length is obtained directly by solving Eq. 3.
Analysis of the freely rotating behavior
We use , and to denote three center of masses on the collagen molecule. The rotating angle θ is the supplementary angle formed by these three center of masses (see Fig. 5 a). The expectation value of the rotating angle is obtained by averaging the rotating angles for last 40 ns simulations. To further reveal the freely rotating behavior, for each configuration, a rotation is performed such that is parallel to the x axis. Here, is the new coordinate of after rotation. Finally, we use to denote the unit vector of . The projection of on y-z plane is calculated for each recorded frame.
Figure 5.
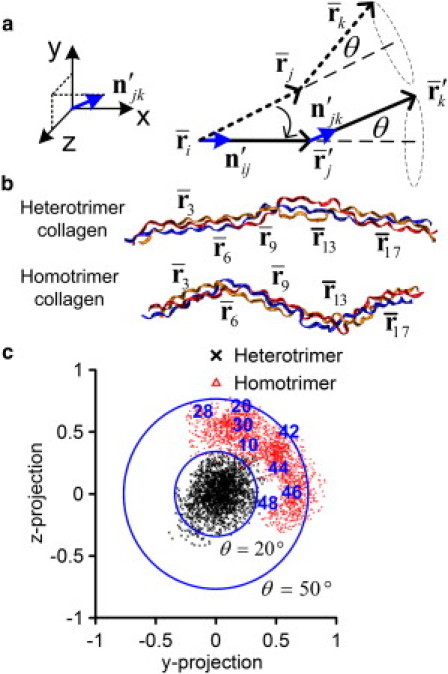
Comparison of the kink and freely rotating behaviors between heterotrimer and homotrimer collagen molecules. (a) Illustration of the analysis method. (b) Illustration of several centers of masses of collagen molecules. (c) The projection of the rotating vectors on the y-z plane for (i,j,k) = (3,6,9).
We analyze the freely rotating behavior at the center of masses of Cα atoms of the 2nd, 3rd, 4th, 5th, 6th, 9th, 13th, and 17th Gly residues. We chose these regions to show differences between local regions based on the structural parameters calculated previously.
Structures in PDB format
Geometries of initial and final (50 ns) structures of the heterotrimer and homotrimer molecules (without water molecules) in PDB format are available in the Supporting Material.
Results and Discussion
We calculate the averaged contour lengths and the end-to-end distances of heterotrimer and homotrimer collagen molecules of Mus musculus. The contour length of heterotrimer collagen is 166.6 ± 1.7 Å and homotrimer collagen is 162.5 ± 1 Å, a slightly smaller value. Interestingly, there is a striking difference between the end-to-end distance of the heterotrimer (147.6 ± 6.8 Å) and homotrimer (127.1 ± 5.5 Å). The persistence length, which characterizes the mechanical stiffness, can be obtained through the relation between the contour length and the end-to-end distance of the collagen molecule. The calculated persistence length is 215 Å for the heterotrimer and 96 Å for the homotrimer, suggesting that the homotrimer collagen molecule is more flexible (smaller overall bending stiffness).
Our calculation results of the persistent lengths are within the range of previously reported values. By fitting the force-extension curve from a mechanical test using optical tweezers to a worm-like chain elasticity model, the persistence length of type I heterotrimer collagen obtained by Sun et al. (52) is found to be 145 ± 73 Å and the persistence length of type II homotrimer collagen is found to be 112 ± 84 Å (53). These studies also suggest that the homotrimer has a smaller persistence length than the heterotrimer, but it should be noted that the homotrimer in the study cited previously is type II collagen. The persistence lengths obtained from earlier simulations are in the range from ∼150 to 250 Å (54), and coarse-grained modeling of collagen molecules using a MARTINI force field suggests a value of 515 ± 67 Å (likely higher due to the generally stiffer nature of protein structures in the MARTINI approach) (55).
As an alternative approach, we also calculate the persistence length as a measure of the direction of the tangent loss of a protein over a distance. The relation between the separated distance and the cosine of the angle between two tangent vectors at two points on the heterotrimer and homotrimer are shown in Fig. 2. The decrease of the cosine angles of homotrimer collagen molecule is larger than the heterotrimer collagen molecule as the distances increase. That is, the homotrimer collagen molecule has a shorter persistence length and is more flexible to change its direction. Exponential fits (see Eq. 2.) are fit to the results of heterotrimer and homotrimer collagen molecules shown in Fig. 2. The persistence lengths from the exponential fitting are 209 Å for the heterotrimer collagen molecule and 89 Å for the homotrimer collagen molecule. The results are consistent with the values obtained by the contour length and end-to-end distance approach used previously and again shows that the homotrimer collagen molecule has a shorter persistence length and is more flexible.
Figure 2.
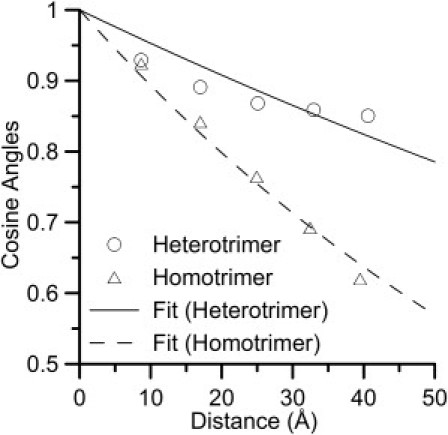
Relation between the separated distance and the loss of the direction of the tangent. The relation between averaged distance 〈lm〉 and cosine angles 〈cosθm〉 of heterotrimer and homotrimer collagen molecules. The line represents the exponential fit (Eq 2.) with persistence length of 209 Å for heterotrimer collagen molecule and 89 Å for homotrimer collagen molecule.
The persistence length calculations assume the collagen molecules to be homogeneous, which is an over-simplification of a heterogeneous molecular structure. We thus perform additional analyses to identify structural mechanisms that governed the apparent change in molecular stiffness. Fig. 3, a and b, show the averaged values of unit heights of heterotrimer and homotrimer collagen molecules, respectively. The results show that both the heterotrimer and homotrimer are not homogeneous along the length of the collagen section analyzed. To validate our results with experimental data, the average of all the unit heights (excluding two at each end) are calculated. The averaged unit height is 8.57 Å for the heterotrimer and 8.59 Å for the homotrimer collagen molecule. The results are similar to that obtained from a statistical analysis of high-resolution x-ray crystal structures of triple-helical peptides, which shows a varied unit height of 8.53 Å for imino-rich regions and 8.65 Å for amino-rich regions (56).
Figure 3.
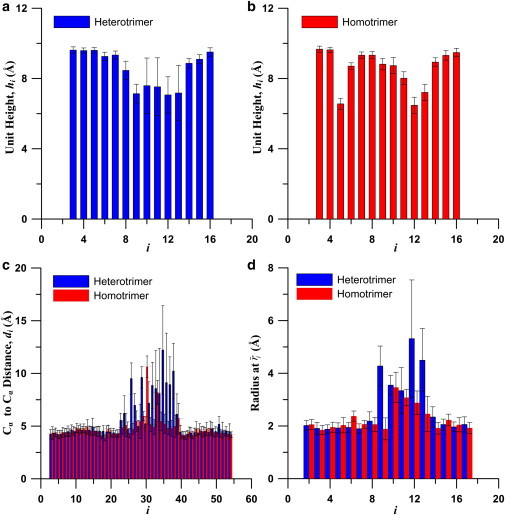
Statistics of structural parameters of collagen protein. (a) Averaged unit height of heterotrimer collagen molecule. (b) Averaged unit height of homotrimer collagen molecule. (c) Averaged nearest Cα distance of heterotrimer and homotrimer collagen molecules. (d) Averaged radius of heterotrimer and homotrimer collagen molecules.
Fig. 3, a and b, show that both the heterotrimer and homotrimer collagen molecules have abnormal shorter unit heights at the region around the 12th triplet (corresponding to the 436th residues in terms of the entire type I collagen molecule). The standard deviation of unit heights at this region of the heterotrimer collagen molecule is larger than other regions of the molecule, which indicates that thermal energy easily varies the conformations of heterotrimer. This can be also confirmed by unit height distributions through simulation time (Fig. S3), time history of unit heights (Fig. S4), and the root mean-square deviation (Fig. S5). This finding is in accordance with experimental studies that have shown lower denaturation temperature of heterotrimers (27,28). There is an additional region with a shorter unit height for the homotrimer collagen molecules around the 5th triplet (corresponding to the 418th residues of type I collagen molecules).
The distances between the nearest Cα atoms of the Gly residues are shown in Fig. 3 c. The results show that at the region around the 34th to 39th carbon atoms in Fig. 3 c, the distance between the nearest Cα atoms of both heterotrimer and homotrimer collagen are larger than other regions. Note that each triplet has three Gly residues (one from each chain), and therefore the 12th triplet corresponds to the 34th to 36th Gly residues. Similarly, the radii (Fig. 3 d) at this region are larger than other regions; that is, the distances from the Cα atoms of Gly to the twisting axis are larger. Therefore, we refer to this region as the microunfolding region (Fig. 4). This finding is consistent with previous simulation results that show microunfolding at the 436th Gly residue of type I collagen (as can be confirmed in Table 1 in (36)). In a region of microunfolding, the H-bond between Gly on adjacent chains might not form easily because the distances between Gly are large. There are only two nearest Cα distances (d30 and d33) abnormally larger in homotrimer collagen molecules, whereas many abnormally larger nearest Cα distances are found in heterotrimer collagen molecule. This also supports our finding that the heterotrimer collagen molecule is more thermally unstable and the microunfolding is more severe. In contrast to the microunfolding region, at the region near the 5th triplet of the homotrimer, where unit height decreases, there is no increase in the radius or the distances between the nearest Cα atoms. These decreases of the unit height do not result in thermal instability in this region. It is also shown directly in the snapshot in Fig. 4 that this region does not feature microunfolding.
Figure 4.
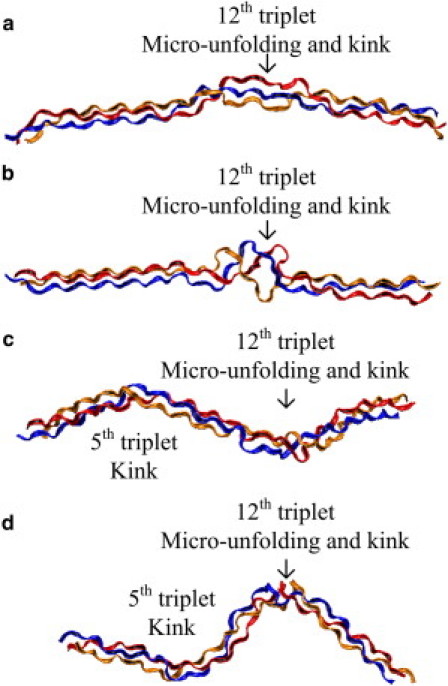
Snapshots of collagen protein. Each chain is plotted in ribbon with the same color. (a) 20 ns, heterotrimer collagen molecule. (b) 40 ns, heterotrimer collagen molecule. (c) 20 ns, homotrimer collagen molecule. (d) 40 ns, homotrimer collagen molecule.
Table 1.
Comparison of kink angles at different regions of heterotrimer and homotrimer collagen molecules
| (i,j,k) | Heterotrimer | Homotrimer |
|---|---|---|
| (2,3,4) | 7.6° ± 4.5° | 7.7° ± 4.2° |
| (3,4,5) | 6.3° ± 3.3° | 7.2° ± 3.7° |
| (3,6,9) | 10.1° ± 5.3° | 37.3° ± 7.6° |
| (9,13,17) | 22.4° ± 9.6° | 63.9° ± 10.3° |
We investigate how regions with a shorter unit height may contribute to a decrease in persistence length. Table 1 shows the rotating angles at different regions for both the heterotrimer and homotrimer. In the 3rd and 4th triplets (2–4 and 3–5), the angles of rotation for both heterotrimer and homotrimer collagen molecules are small. In regions where the unit height is short (12th residue in heterotrimer and 5th and 12th in the homotrimer), there is an increase in the angle of rotation. When a large rotation (larger than 15°) is present, we call this kink indicating a local bend in the molecule (Fig. 4). Both the heterotrimer and homotrimer have kinks at the microunfolding region but the kink angle of the homotrimer is about three times larger than the heterotrimer (63.9° vs. 22.4°). Moreover, one more kink region (the 5th triplet) is found in the homotrimer. Therefore, we conclude that the loss of the α-2 chain leads to the ability to kink with larger angles at specific regions.
The projections of the rotating vectors on the cross section of the collagen molecule at the 5th residue are shown in Fig. 5 c. The inner circle represents the line with a rotating angle of 20° and the outer circle represents the line with a rotating angle of 50°. It is seen that the projections of the rotating vectors of the heterotrimer collagen molecule lie within the inner circle and are closed to the origin, with an average of 10.1 ± 5.3° (Table 1). Projections of the rotating vectors of the homotrimer collagen molecule are much larger with an average value of 37.3 ± 7.6°. The numbers shown in the figure indicate the simulation time in ns unit, and indicate that the homotrimer rotates first counterclockwise (10 to 28 ns), and then clockwise (28 to 48 ns) at this location. From the results, we observe a very specific rotating angle for the homotrimer at this region, which is reflected in the overall shorter persistence length. The specific rotating behavior identified here provides the molecular origin of the result that the homotrimer is more flexible and has an overall smaller bending stiffness.
We also investigate the effects of the structural changes between the heterotrimer and heterotrimer on the mechanical responses of the collagen molecules. Under uniaxial pulling of the molecules, we observe significantly different stress-strain curves of the heterotrimer and the homotrimer (Fig. S10), which shows that the heterotrimer is stiffer and that the homotrimer is much more flexible. This is because the homotrimer has kinks, which results in a shorter end-to-end distance at equilibrium, and leading to softer stress-strain responses compared to the heterotrimer.
In our simulations we consider a segment of collagen that is 57 amino acids (≈160 Å) long, which is only a small portion of the 1014 amino acids (≈3000 Å) of the entire molecule, but computationally feasible to model at the atomic level. Despite modeling only a small portion (which can be limiting), we are able to clearly characterize differences in mechanical flexibility and local structural behavior. There is a possibility that throughout the molecule there are similar local regions of microunfolding in both homotrimer and heterotrimer cases. We anticipate that kinking also occurs at other regions, predominately in the homotrimer.
The behavior of the collagen molecule can affect the mechanical properties of many tissues including bone, tendon, and ligament. In these tissues, collagen molecules are aligned in quarter staggered arrangements to form fibrils (Fig. 6). The kinking and freely rotating behavior of homotrimer collagen is likely to affect the packing of the molecules into fibrils. The lateral space used by the homotrimer collagen molecule is larger because it has larger kink angles. Therefore, the larger kink angles are likely to lead to a larger distance between homotrimer molecules in the fibril. The larger distance between collagen molecules could lead to the reduction of cross-linking between collagen molecules and the loss of mechanical integrity of the tissue. Our simulation results provide evidence of the increase in possible lateral configurations for the homotrimer collagen molecule. As shown directly in Fig. 4 and Fig. 6, the explored lateral distance is increased in the kink regions. Indeed, from computations of the volume fraction of the water, Miles et al. (28) suggested an increase in the lateral packing of 1.4 Å between collagen molecules in homotrimer collagen compared to the heterotrimer collagen, which is consistent with our findings. Our studies on the structural and mechanical behavior of a single collagen molecule provide possible insights and relevance for the loss of mechanical integrity of the molecule at the atomistic level.
Figure 6.
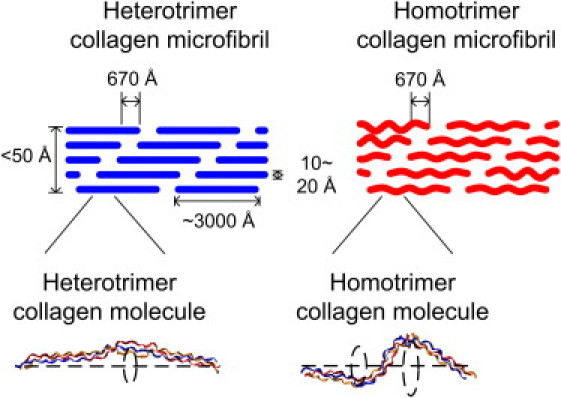
Role of the kink of collagen molecule in the microfibril. The kinking and freely rotating behavior of homotrimer collagen is likely to affect the packing of the molecules into fibrils. The lateral space used by the homotrimer collagen molecule is larger since it has larger kink angles, which are likely to lead to a larger lateral distance in the microfibril.
Conclusion
In summary, we carried out fully atomistic simulations of mus type I heterotrimer and homotrimer in explicit water to explore the role of the loss of α-2 chain in collagen molecules. The simulations ranged over 50 ns to systematically explore the different structural and mechanical behaviors between heterotrimer and homotrimer collagen molecules. We found that the persistence length of the heterotrimer is twice that of the homotrimer, indicating the homotrimer collagen molecule is significantly more flexible and hence softer. This has also been confirmed in direct molecular pulling simulations (Fig. S10). Specifically, structural analyses of collagen molecules show that the greater flexibility of the homotrimer collagen molecule, smaller bending stiffness, and smaller persistence length, results from the emergence of kinking at two specific regions. The observation of these freely rotating behaviors at specific regions confirms the experimental and computational findings that the collagen molecules are not homogeneous along their length and that the behavior is dominated by the local variation of amino acid sequences. Local kinking is likely to disrupt the lateral distance between collagen molecules in a fibril. This provides a possible mechanism that accounts for the increase in the lateral intermolecular distance that has been suggested by experimental data. We believe that local kinking would not affect the axial spacing of the collagen molecules, which is why the D-period remains relatively unchanged in homotrimeric collagen.
Microunfolding regions are known to be important for protein digestion; in triple helical conformation, the protein is protected, but unfolded regions are susceptible to cleavage by proteolytic enzymes. Our atomistic model suggests that we can predict such cleavage sites from fundamental structural analyses of the molecule. Experimental studies have revealed that homotrimeric oim collagen is more resistant to digestion than heterotrimers (26), which is in accordance with the higher denaturation temperature of oim collagen (27,28,57). In the short section of collagen considered here, microunfolding occurs at only one site in both the homotrimeric and heterotrimeric collagen. Future work will sample multiple fragments of the collagen molecule to determine the location and relative number of microunfolding sites in heterotrimeric and homotrimeric collagen. Much research has investigated the oim model, not only to understand and develop treatments for osteogenesis imperfecta, but also to probe the function of the α-2 chain in collagen. Indeed, the oim model provides an interesting platform for investigating the ramifications of alterations in molecular structure on the mechanics and structural behavior of bone throughout its hierarchy. At the organ level, oim bones are extremely fragile, with smaller cross sections, and little or no plastic deformation resulting in brittle-type fractures (9,21–25). At the tissue level, oim has more disorganized woven bone than organized lamellar bone (26). At the matrix level, oim bones have higher mineral density, with smaller apatite crystals closely packed together (58,59). All of these changes are caused fundamentally by alterations at the molecular level. Previous experimental studies have found molecular differences in cleavage sites and denaturation temperatures (27,28), as well as cross-linking (60,61), however these studies by necessity are all carried out at the fibril level. Atomistic modeling allows us to examine the behavior of a single collagen molecule, helping to define the basic structural and mechanical differences responsible for the changes throughout the hierarchy.
Acknowledgments
We acknowledge support from a National Science Foundation-CAREER award, with additional support from the Department of Defense-Presidential Early Career Award for Scientists and Engineers, and the Leverhulme Foundation. The authors declare no conflict of interest of any kind.
Supporting Material
References
- 1.Fratzl P., editor. Collagen: Structure and Mechanics. Springer; New York: 2008. [Google Scholar]
- 2.Buehler M.J., Yung Y.C. Deformation and failure of protein materials in physiologically extreme conditions and disease. Nat. Mater. 2009;8:175–188. doi: 10.1038/nmat2387. [DOI] [PubMed] [Google Scholar]
- 3.Rainey J.K., Wen C.K., Goh M.C. Hierarchical assembly and the onset of banding in fibrous long spacing collagen revealed by atomic force microscopy. Matrix Biol. 2002;21:647–660. doi: 10.1016/s0945-053x(02)00101-4. [DOI] [PubMed] [Google Scholar]
- 4.Tang Y., Ballarini R., Eppell S.J. Deformation micromechanisms of collagen fibrils under uniaxial tension. J. R. Soc. Interface. 2010;7:839–850. doi: 10.1098/rsif.2009.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Z.L., Dodge M.R., Eppell S.J. Stress-strain experiments on individual collagen fibrils. Biophys. J. 2008;95:3956–3963. doi: 10.1529/biophysj.107.124602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eppell S.J., Smith B.N., Ballarini R. Nano measurements with micro-devices: mechanical properties of hydrated collagen fibrils. J. R. Soc. Interface. 2006;3:117–121. doi: 10.1098/rsif.2005.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuznetsova N.V., Forlino A., Leikin S. Structure, stability and interactions of type I collagen with GLY349-CYS substitution in alpha 1(I) chain in a murine Osteogenesis Imperfecta model. Matrix Biol. 2004;23:101–112. doi: 10.1016/j.matbio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Kuznetsova N., McBride D.J., Jr., Leikin S. Osteogenesis imperfecta murine: interaction between type I collagen homotrimers. J. Mol. Biol. 2001;309:807–815. doi: 10.1006/jmbi.2001.4682. [DOI] [PubMed] [Google Scholar]
- 9.McBride D.J., Choe V., Brodsky B. Altered collagen structure in mouse tail tendon lacking the alpha2(I) chain. J. Mol. Biol. 1997;270:275–284. doi: 10.1006/jmbi.1997.1106. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez S.A., Bashey R.I., Yankowski R. Identification of collagen alpha1(I) trimer in embryonic chick tendons and calvaria. Biochem. Biophys. Res. Commun. 1977;78:1354–1361. doi: 10.1016/0006-291x(77)91441-3. [DOI] [PubMed] [Google Scholar]
- 11.Rojkind M., Giambrone M.A., Biempica L. Collagen types in normal and cirrhotic liver. Gastroenterology. 1979;76:710–719. [PubMed] [Google Scholar]
- 12.Narayanan A.S., Page R.C., Meyers D.F. Characterization of collagens of diseased human gingiva. Biochemistry. 1980;19:5037–5043. doi: 10.1021/bi00563a016. [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich H.P., Brown H., White B.S. Evidence for type V and I trimer collagens in Dupuytren's Contracture palmar fascia. Biochem. Med. 1982;28:273–284. doi: 10.1016/0006-2944(82)90080-1. [DOI] [PubMed] [Google Scholar]
- 14.Makareeva E., Han S., Leikin S. Carcinomas contain a matrix metalloproteinase-resistant isoform of type I collagen exerting selective support to invasion. Cancer Res. 2010;70:4366–4374. doi: 10.1158/0008-5472.CAN-09-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeClerck Y.A., Bomann E.T., Biedler J.L. Differential collagen biosynthesis by human neuroblastoma cell variants. Cancer Res. 1987;47:6505–6510. [PubMed] [Google Scholar]
- 16.Lesot H., Karcher-Djuricic V., Ruch J.V. Synthesis of collagen type I, type I trimer and type III by embryonic mouse dental epithelial and mesenchymal cells in vitro. Biochim. Biophys. Acta. 1981;656:206–212. doi: 10.1016/0005-2787(81)90088-5. [DOI] [PubMed] [Google Scholar]
- 17.Little C.D., Church R.L., Ruddle F.H. Procollagen and collagen produced by a teratocarcinoma-derived cell line, TSD4: evidence for a new molecular form of collagen. Cell. 1977;10:287–295. doi: 10.1016/0092-8674(77)90222-7. [DOI] [PubMed] [Google Scholar]
- 18.Minafra S., Luparello C., Pucci-Minafra I. Collagen biosynthesis by a breast carcinoma cell strain and biopsy fragments of the primary tumour. Cell Biol. Int. Rep. 1988;12:895–905. doi: 10.1016/0309-1651(88)90053-7. [DOI] [PubMed] [Google Scholar]
- 19.Rupard J.H., Dimari S.J., Haralson M.A. Synthesis of type I homotrimer collagen molecules by cultured human lung adenocarcinoma cells. Am. J. Pathol. 1988;133:316–326. [PMC free article] [PubMed] [Google Scholar]
- 20.Han S.J., Makareeva E., Leikin S. Molecular mechanism of type I collagen homotrimer resistance to mammalian collagenases. J. Biol. Chem. 2010;285:22276–22281. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saban J., Zussman M., King D. Heterozygous oim mice exhibit a mild form of osteogenesis imperfecta. Bone. 1996;19:575–579. doi: 10.1016/s8756-3282(96)00305-5. [DOI] [PubMed] [Google Scholar]
- 22.Misof K., Landis W.J., Fratzl P. Collagen from the osteogenesis imperfecta mouse model (oim) shows reduced resistance against tensile stress. J. Clin. Invest. 1997;100:40–45. doi: 10.1172/JCI119519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camacho N.P., Hou L., Boskey A.L. The material basis for reduced mechanical properties in oim mice bones. J. Bone Miner. Res. 1999;14:264–272. doi: 10.1359/jbmr.1999.14.2.264. [DOI] [PubMed] [Google Scholar]
- 24.Miller E., Delos D., Pleshko Camacho N. Abnormal mineral-matrix interactions are a significant contributor to fragility in oim/oim bone. Calcif. Tissue Int. 2007;81:206–214. doi: 10.1007/s00223-007-9045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanleene M., Saldanha Z., Shefelbine S.J. Transplantation of human fetal blood stem cells in the osteogenesis imperfecta mouse leads to improvement in multiscale tissue properties. Blood. 2011;117:1053–1060. doi: 10.1182/blood-2010-05-287565. [DOI] [PubMed] [Google Scholar]
- 26.Chipman S.D., Sweet H.O., Shapiro J.R. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proc. Natl. Acad. Sci. USA. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznetsova N.V., McBride D.J., Leikin S. Changes in thermal stability and microunfolding pattern of collagen helix resulting from the loss of alpha2(I) chain in osteogenesis imperfecta murine. J. Mol. Biol. 2003;331:191–200. doi: 10.1016/s0022-2836(03)00715-0. [DOI] [PubMed] [Google Scholar]
- 28.Miles C.A., Sims T.J., Bailey A.J. The role of the alpha2 chain in the stabilization of the collagen type I heterotrimer: a study of the type I homotrimer in oim mouse tissues. J. Mol. Biol. 2002;321:797–805. doi: 10.1016/s0022-2836(02)00703-9. [DOI] [PubMed] [Google Scholar]
- 29.Kühn K. Relationship between amino acid sequence and higher structures of collagen. Connect. Tissue Res. 1982;10:5–10. doi: 10.3109/03008208209034401. [DOI] [PubMed] [Google Scholar]
- 30.Buehler M.J. Nanomechanics of collagen fibrils under varying cross-link densities: atomistic and continuum studies. J. Mech. Behav. Biomed. Mater. 2008;1:59–67. doi: 10.1016/j.jmbbm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Uzel S.G.M., Buehler M.J. Molecular structure, mechanical behavior and failure mechanism of the C-terminal cross-link domain in type I collagen. J. Mech. Behav. Biomed. Mater. 2011;4:153–161. doi: 10.1016/j.jmbbm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Arnold W.V., Fertala A., Prockop D.J. Recombinant procollagen II: deletion of D period segments identifies sequences that are required for helix stabilization and generates a temperature-sensitive N-proteinase cleavage site. J. Biol. Chem. 1998;273:31822–31828. doi: 10.1074/jbc.273.48.31822. [DOI] [PubMed] [Google Scholar]
- 33.Makareeva E., Cabral W.A., Leikin S. Molecular mechanism of alpha 1(I)-osteogenesis imperfecta/Ehlers-Danlos syndrome: unfolding of an N-anchor domain at the N-terminal end of the type I collagen triple helix. J. Biol. Chem. 2006;281:6463–6470. doi: 10.1074/jbc.M511830200. [DOI] [PubMed] [Google Scholar]
- 34.Makareeva E., Mertz E.L., Leikin S. Structural heterogeneity of type I collagen triple helix and its role in osteogenesis imperfecta. J. Biol. Chem. 2008;283:4787–4798. doi: 10.1074/jbc.M705773200. [DOI] [PubMed] [Google Scholar]
- 35.Steplewski A., Majsterek I., Fertala A. Thermostability gradient in the collagen triple helix reveals its multi-domain structure. J. Mol. Biol. 2004;338:989–998. doi: 10.1016/j.jmb.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 36.Bodian D.L., Radmer R.J., Klein T.E. Molecular dynamics simulations of the full triple helical region of collagen type I provide an atomic scale view of the protein's regional heterogeneity. Pac. Symp. Biocomput. 2011:193–204. doi: 10.1142/9789814335058_0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodian D.L., Madhan B., Klein T.E. Predicting the clinical lethality of osteogenesis imperfecta from collagen glycine mutations. Biochemistry. 2008;47:5424–5432. doi: 10.1021/bi800026k. [DOI] [PubMed] [Google Scholar]
- 38.Di Lullo G.A., Sweeney S.M., San Antonio J.D. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 39.Gautieri A., Uzel S., Buehler M.J. Molecular and mesoscale mechanisms of osteogenesis imperfecta disease in collagen fibrils. Biophys. J. 2009;97:857–865. doi: 10.1016/j.bpj.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uzel S.G.M., Buehler M.J. Nanomechanical sequencing of collagen: tropocollagen features heterogeneous elastic properties at the nanoscale. Integr Biol (Camb) 2009;1:452–459. doi: 10.1039/b906864c. [DOI] [PubMed] [Google Scholar]
- 41.Rainey J.K., Goh M.C. An interactive triple-helical collagen builder. Bioinformatics. 2004;20:2458–2459. doi: 10.1093/bioinformatics/bth247. [DOI] [PubMed] [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 43.Nelson M.T., Humphrey W., Schulten K. NAMD: a parallel, object oriented molecular dynamics program. Int. J. Supercomput. Appl. High Perform. Comput. 1996;10:251–268. [Google Scholar]
- 44.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.Anderson D. University of Toronto; Toronto, Canada: 2005. Collagen Self-Assembly: A Complementary Experimental and Theoretical Perspective. [Google Scholar]
- 46.Gautieri A., Vesentini S., Redaelli A. Mechanical properties of physiological and pathological models of collagen peptides investigated via steered molecular dynamics simulations. J. Biomech. 2008;41:3073–3077. doi: 10.1016/j.jbiomech.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 47.Gautieri A., Buehler M.J., Redaelli A. Deformation rate controls elasticity and unfolding pathway of single tropocollagen molecules. J. Mech. Behav. Biomed. Mater. 2009;2:130–137. doi: 10.1016/j.jmbbm.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Gautieri A., Vesentini S., Buehler M.J. Single molecule effects of osteogenesis imperfecta mutations in tropocollagen protein domains. Protein Sci. 2009;18:161–168. doi: 10.1002/pro.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srinivasan M., Uzel S.G.M., Buehler M.J. Alport syndrome mutations in type IV tropocollagen alter molecular structure and nanomechanical properties. J. Struct. Biol. 2009;168:503–510. doi: 10.1016/j.jsb.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Flory P.J. Interscience Publishers; New York: 1969. Statistical Mechanics of Chain Molecules. [Google Scholar]
- 51.Schäfer L., Elsner K. Calculation of the persistence length of a flexible polymer chain with short-range self-repulsion. Eur. Phys. J. E. Soft Matter. 2004;13:225–237. doi: 10.1140/epje/i2003-10071-1. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y.L., Luo Z.P., An K.N. Direct quantification of the flexibility of type I collagen monomer. Biochem. Biophys. Res. Commun. 2002;295:382–386. doi: 10.1016/s0006-291x(02)00685-x. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y.L., Luo Z.P., An K.N. Stretching type II collagen with optical tweezers. J. Biomech. 2004;37:1665–1669. doi: 10.1016/j.jbiomech.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 54.Buehler M.J., Wong S.Y. Entropic elasticity controls nanomechanics of single tropocollagen molecules. Biophys. J. 2007;93:37–43. doi: 10.1529/biophysj.106.102616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gautieri A., Russo A., Buehler M.J. Coarse-grained model of collagen molecules using an extended MARTINI force field. J. Chem. Theory Comput. 2010;6:1210–1218. [Google Scholar]
- 56.Rainey J.K., Goh M.C. A statistically derived parameterization for the collagen triple-helix. Protein Sci. 2002;11:2748–2754. doi: 10.1110/ps.0218502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leikina E., Mertts M.V., Leikin S. Type I collagen is thermally unstable at body temperature. Proc. Natl. Acad. Sci. USA. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fratzl P., Paris O., Landis W.J. Bone mineralization in an osteogenesis imperfecta mouse model studied by small-angle x-ray scattering. J. Clin. Invest. 1996;97:396–402. doi: 10.1172/JCI118428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camacho N.P., Landis W.J., Boskey A.L. Mineral changes in a mouse model of osteogenesis imperfecta detected by Fourier transform infrared microscopy. Connect. Tissue Res. 1996;35:259–265. doi: 10.3109/03008209609029199. [DOI] [PubMed] [Google Scholar]
- 60.Sims T.J., Miles C.A., Camacho N.P. Properties of collagen in OIM mouse tissues. Connect. Tissue Res. 2003;44(Suppl 1):202–205. [PubMed] [Google Scholar]
- 61.Pfeiffer B.J., Franklin C.L., Phillips C.L. Alpha 2(I) collagen deficient oim mice have altered biomechanical integrity, collagen content, and collagen cross-linking of their thoracic aorta. Matrix Biol. 2005;24:451–458. doi: 10.1016/j.matbio.2005.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


