Abstract
Upon blue-light irradiation, the bacterium Halorhodospira halophila is able to modulate the activity of its flagellar motor and thereby evade potentially harmful UV radiation. The 14 kDa soluble cytosolic photoactive yellow protein (PYP) is believed to be the primary mediator of this photophobic response, and yields a UV/Vis absorption spectrum that closely matches the bacterium's motility spectrum. In the electronic ground state, the para-coumaric acid (pCA) chromophore of PYP is negatively charged and forms two short hydrogen bonds to the side chains of Glu-46 and Tyr-42. The resulting acid triad is central to the marked pH dependence of the optical-absorption relaxation kinetics of PYP. Here, we describe an NMR approach to sequence-specifically follow all tyrosine side-chain protonation states in PYP from pH 3.41 to 11.24. The indirect observation of the nonprotonated 13Cγ resonances in sensitive and well-resolved two-dimensional 13C-1H spectra proved to be pivotal in this effort, as observation of other ring-system resonances was hampered by spectral congestion and line-broadening due to ring flips. We observe three classes of tyrosine residues in PYP that exhibit very different pKa values depending on whether the phenolic side chain is solvent-exposed, buried, or hydrogen-bonded. In particular, our data show that Tyr-42 remains fully protonated in the pH range of 3.41–11.24, and that pH-induced changes observed in the photocycle kinetics of PYP cannot be caused by changes in the charge state of Tyr-42. It is therefore very unlikely that the pCA chromophore undergoes changes in its electrostatic interactions in the electronic ground state.
Introduction
Photoactive yellow protein (PYP) is a soluble, cytosolic protein from the purple phototropic eubacterium Halorhodospira halophila. PYP shows a UV absorption spectrum that closely matches the wavelength dependence of the escape of the bacterium from potentially harmful blue-light radiation (1). Consequently, PYP has become an important example of a soluble bacterial light sensor, and a rich model system for the study of PER-ARNT-SIM (PAS) domain signaling (2).
PYP consists of 125 residues that form a six-stranded β-sheet flanked by five α-helices in an α/β fold (3). In the active site, the para-coumaric acid (pCA) chromophore forms a thioester bond to the side chain of C69. This chromophore is stabilized by the formation of two short hydrogen bonds (H-bonds) between pCA and E46, and between pCA and Y42 (4,5), as shown in Fig. 1.
Figure 1.
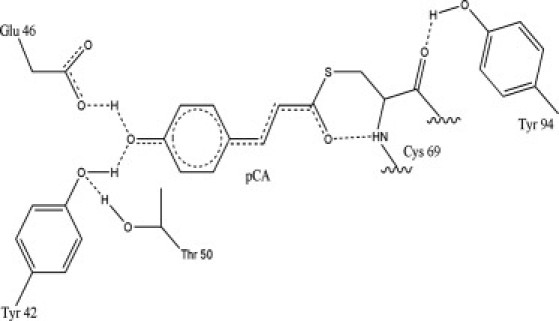
H-bond network of PYP in the active center.
Recent high-resolution neutron and x-ray crystallographic structures reveal a shared low-barrier H-bond between pCA and E46, whereas the H-bond between pCA and Y42 is qualified as a short ionic H-bond (6). Sigala et al. (7) detected the two H-bonds using one-dimensional (1D) 1H NMR. As depicted in Fig. 1, the phenolic oxygen of Y42 also participates as a proton acceptor in the H-bond formation with the hydroxyl proton of T50. Y94 is also involved in the H-bond network in the active center by donating its side-chain proton to the backbone oxygen of C69 (4,8).
Upon blue-light irradiation, PYP undergoes a number of changes in its optical properties that are associated with changes in its structure (9–11). Initially, the pCA chromophore undergoes a series of rapid bond isomerizations on the picosecond timescale to the pR state. This process is followed by proton transfer on the microsecond timescale, which involves the dissolution of the shared low-barrier H-bond (6) and protonation of pCA to form the pB′ state (12,13). Spectroscopic evidence has shown that the N-terminal domain of PYP dissociates from its PAS domain and becomes unfolded. This state is then recognized as pB, in which λmax of the chromophore absorption changes from 446 nm (in the ground state, pG) to 355 nm (in the pB state) (14,15). Finally, the intermediate (pB) relaxes slowly (subsecond) to the initial pR state, a process that is associated with formation of the central β-sheet and part of the helical structures, and is succeeded by a consolidation of structure around the pCA chromophore (14). The timescales indicated here are only approximate and display a strong pH dependence (8,16,17). Central to the mechanism of photoactivation is the presence of a low-barrier H-bond between pCA and E46 in the ground state. Photoactivation causes a breakage of this low-barrier H-bond, followed by significant conformational rearrangement of all helices except part of helix α5 (14). Thus, a distinct epitope is presented for interaction with downstream partners in the phototactic response, although the corresponding signaling pathway remains unknown (9).
To study the role of electrostatic interactions in protein function, it is important to assemble a set of NMR experiments that can accurately determine the protonation states of titratable amino acid side chains. One can determine individual pKa values by following chemical shifts of nuclei in the amino acid side chain. The first, obvious choice is 1H NMR (18), which because of its high NMR receptivity does not require any isotopic enrichment. However, there are several challenges to be overcome: the 1H NMR spectrum of larger proteins is highly crowded, the response of 1H chemical shifts to protonation state is often relatively small, and 1H chemical shifts are sensitive to ionization equilibria other than that of the amino acid to which the proton is attached (18). The chemical shifts of 13C or 15N nuclei at the site of (de)protonation tend to undergo larger (>1 ppm) chemical-shift changes, and are therefore ideal reporters of protonation states. Moreover, such large variations in nuclear shielding are caused mainly by changes in the electron distribution within the side chain, rather than by charges that develop in the environment, and can therefore be used as more-selective probes of protonation states and the pKa values of amino acid side chains.
To accurately determine the protonation states of Tyr residues, one can use several 13C chemical shifts as reporters for the charge state of the Tyr ring, as shown in Table 1 and Fig. 2 (19).
Table 1.
13C chemical shift reporters in the Tyr side chain and their corresponding chemical shift changes upon deprotonation
| Carbon chemical shift reporter | Δδ (ppm) |
|---|---|
| Cγ | −6.2 |
| Cδ | 0 |
| Cε | +3.3 |
| Cζ | +10.4 |
Values from Norton and Bradbury (19).
Figure 2.
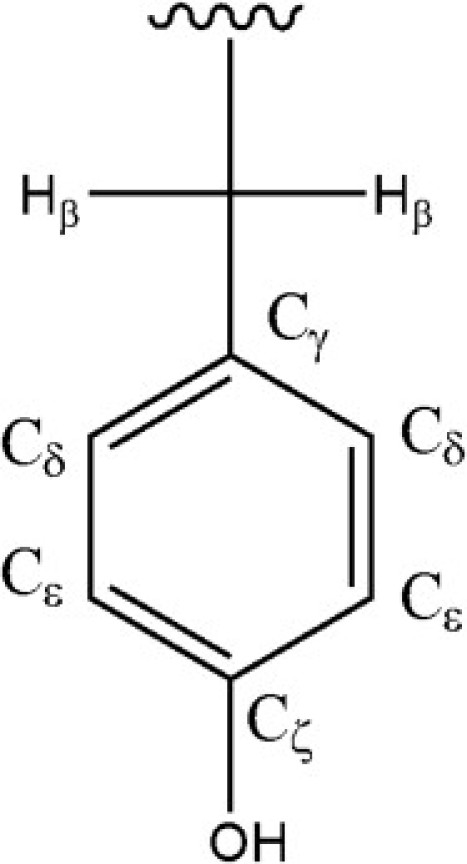
Tyrosine side chain.
1D 13C NMR on uniformly 13C-enriched protein samples is hindered by the presence of 13C-13C splittings in the spectrum and additional overlap with the 13Cζ signals of arginines. Therefore, 1D 13C NMR has been combined with selective incorporation of 13C-Tyr (20–22). In this approach, one obtains the assignments by removing individual Tyr residues through mutagenesis, which requires the parallel production and purification of multiple protein samples. Furthermore, one must ensure that the removal of each Tyr does not significantly perturb the structure, and thus the spectral change can be unambiguously attributed to the mutation site.
Alternatively, two-dimensional (2D) 1H-13C or 1H-15N NMR experiments applied to uniformly 13C/15N enriched proteins are ideally suited for recording the heteronuclear chemical shifts as a function of pH because they offer excellent sensitivity and resolution, and permit comprehensive and residue-specific assignments of individual amino acid resonances. So far, experiments of this type have been described for Asp, Glu, Lys, Arg, and His side chains, as well as the C- and N-termini (23–25), but an analogous method for addressing Tyr charge states was lacking until recently (26).
In this study, we present a strategy for comprehensively determining the pKa values of the Tyr residues of PYP using indirect-detection 2D NMR spectroscopy of a uniformly 13C-enriched protein. We investigate the pH-dependent protonation state of Y42, which forms a short H-bond to the pCA chromophore, stabilizing its delocalized negative charge. The presence of a short H-bond under physiological conditions has been firmly established on the basis of x-ray and neutron crystallographic structures (6), NMR spectroscopic observation (14,7), and other spectroscopic data and theoretical calculations (27). However, the basis for the pH dependence of the photocycle kinetics is not fully understood at atomic resolution. Here, we show that Y42 remains fully protonated in the pH range of 3.41–11.24, indicating that the pH dependence of the PYP photocycle kinetics cannot be caused by changes in the charge state of Y42, and therefore it is very unlikely that the pCA chromophore undergoes changes in its electrostatic interactions in the electronic ground state.
Material and Methods
Sample preparation
Uniformly 13C, 15N-labeled wild-type PYP was produced in M9 minimal medium containing uniformly 13C-enriched glucose and 15NH4Cl and purified as described previously (28). NMR samples contained ∼1.0 mM of doubly labeled [13C, 15N] PYP, 0.15 mM DSS, 10% D2O, and either 15 mM sodium acetate –d3, 5 mM potassium phosphate, or 15 mM sodium bicarbonate as buffer for the pH ranges of 3.4–5.8, 5.9–8.6, and 8.7–11.24, respectively. The pH was changed in steps of 0.2 pH units by addition of a few microliters of concentrated HCl or NaOH solution.
To calibrate the pH meter (PB-11-P11; Sartorius Mechatronics, Nieuwegein, The Netherlands), we used standard calibration buffers of pH 4.0, 7.0, and 10.0. For measurements below pH 4 and above pH 10, we used a 0.1 M HCl solution (pH 1.0) and a 10 mM NaOH solution (pH 12) for calibration.
NMR spectroscopy
All NMR experiments were carried out on a Varian Unity INOVA 600 MHz spectrometer equipped with a pulsed field-gradient probe at 293 K. All aromatic proton-carbon correlations were established from 2D constant-time 1H-13C heteronuclear single quantum coherence (HSQC) spectra. This experiment was performed with the carrier position at 125 ppm in the 13C domain. We recorded 512 (Haro) × 128 (Caro) complex points with maximum acquisition times of 64 and 16 ms for 1H and 13C, respectively. An interscan delay of 1 s was used with two scans per free induction decay (FID), giving rise to a measurement time of 9 min.
To obtain the signal from two-bond correlations between Cζ and Hε, we performed a constant-time 1H-13C HSQC experiment with the INEPT delay set to 24 ms (29). We recorded 512 (Haro) × 128 (Caro) complex points with maximum acquisition times of 64 and 16 ms for 1H and 13C, respectively. The number of scans per FID was 96, and the interscan delay was 1 s. The total experiment time was 7.5 h.
We recorded Cγ-Hβ correlations in Tyr side chains using a 2D CG(CB)HB experiment with constant-time Cγ evolution (30). We acquired the spectra using 90 (Cγ) × 512 (Hβ) complex points with maximum evolution times equal to 15 and 64 ms, respectively. The carrier position was placed at 130 ppm in the 13C domain to excite the Cγ region. An interscan delay of 1 s was used, and 20 scans per FID were recorded, giving a total experiment time of ∼1 h for every 2D spectrum.
We processed all of the spectra using NMRPipe (31) and analyzed them using SPARKY (32). Mirror-image linear prediction was applied during processing to extend the 13C time-domain signal and improve the spectral resolution. All chemical shifts were referenced to DSS based on the IUPAC recommendation (33).
All Tyr peaks were assigned based on the strategy described by Oktaviani et al. (34). The complete assignment of PYP will be presented in a forthcoming publication.
Data analysis
The titration data for all Tyr chemical shifts were fitted to the Henderson-Hasselbalch equation, which is appropriate for rapid exchange of the nuclei between the environments associated with the neutral and charged states of the side chain:
| (1) |
where δAH denotes the chemical shift for the protonated form, Δδ = δA– – δAH is the change in chemical shift upon deprotonation, and nH is the Hill coefficient (nH > 1 indicates apparent positive cooperativity). All calculations were performed with the use of Mathematica (Wolfram).
Results and Discussion
Because the chemical-shift response to (de)protonation is sufficiently large for 13Cε, in principal one could unambiguously determine the charge state of individual Tyr side chains by recording 2D HSQC or heteronuclear multiple-quantum correlation (HMQC) spectra, which exhibit high sensitivity. The 2D 1H-13C HSQC spectrum for PYP is shown in Fig. 3.
Figure 3.
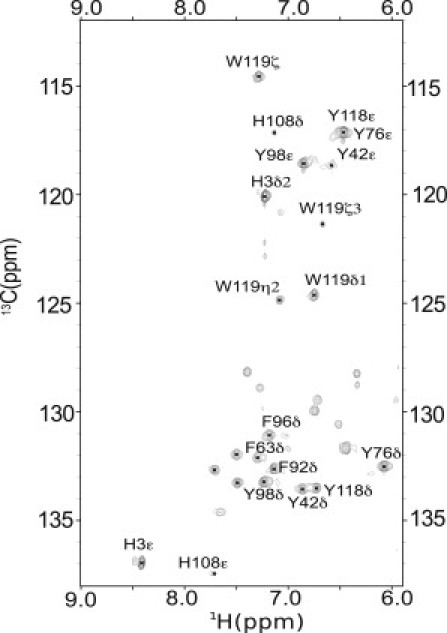
Region of the 2D aromatic 13C-1H HSQC spectrum for PYP containing the Tyr C-H correlations. The peak of Y118 Cε overlaps with Y76 Cε. Signals for Trp, and His side chains also fall in this spectral region. Assignments of the correlations are indicated.
Signals for the surface-exposed residue Y98 are easily identified. Y76 overlaps with Y118, but resonances for Y42, Y94 are weak or invisible. This is probably due to the slow rotation of several tyrosines in the protein interior leading to exchange broadening. This may be a serious limitation for experiments that aim to use the Hδ or Hε ring-proton resonances for other proteins as well.
Alternatively, 2D experiments that correlate 13C with a 1H nucleus two bonds removed via successive homonuclear and heteronuclear transfer through large one-bond couplings have been designed for the determination of acidic side-chain groups of Asp and Glu and the C-terminus (25). These experiments offer excellent sensitivity and resolution. In a recent study (26), a 2D HE(CE)CZ pulse sequence that correlates Hε and Cζ was successfully applied to determine the Tyr pKa in Bacillus circulans xylanase. An approach to assign Tyr 13Cζ in the context of SAIL isotope labeling (29) employs the two-bond Hε-Cζ coupling. This experiment is also applicable to uniformly enriched samples, where three-bond Hδ-Cγ correlations may also be observed. Unfortunately, these experiments suffer from sensitivity losses in the case of ring flips, when the Hδ and Hε protons move between different magnetic environments. In the case of PYP, only signals from Y76 and Y98 are observed in the latter experiment, as shown in Fig. 4.
Figure 4.
2D 13C-1H HSQC spectrum that correlates Hδ and Hε with Cζ for Tyr residues of PYP. Assignments of the correlations are indicated. Signals for Y42, Y94, and Y118 were not observed.
An alternative approach would be to indirectly detect 13Cγ chemical shifts, which can be correlated to Hβ chemical shifts via successive magnetization transfers through the large CγCβ (1JCC ∼45 Hz) and CβHβ (1JCH ∼130 Hz) coupling constants. This could be done via an out-and-back HB(CB)CG experiment, but for larger proteins, one could obtain increased sensitivity by avoiding one of the long and lossy homonuclear 13C-magnetization transfer steps, by starting on the nonprotonated 13Cγ (30). CG(CB)HB spectra are generally well resolved, even for larger proteins (>20 kDa), because they exhibit significant variation in the Cγ chemical shift as a function of amino acid type and secondary structure, and because there are two nonequivalent protons available to read out the Cγ shift in the resulting 2D spectrum. Partial deuteration will improve the sensitivity of the technique for proteins with higher molecular weights (35,36). Improved sensitivity of HB(CB)CG-type measurements has been achieved with a ∼50% level of deuteration (37). For PYP, we observed all Tyr side-chain signals with good sensitivity, and there was not a single instance of overlap in the spectrum (Fig. 5). In addition, we were able to detect the Cγ resonance of H3 (which showed a pKa value of 6.5) but not that of H118. We note that His Cγ detection could be improved by 15N decoupling during Cγ evolution, which was not done here.
Figure 5.
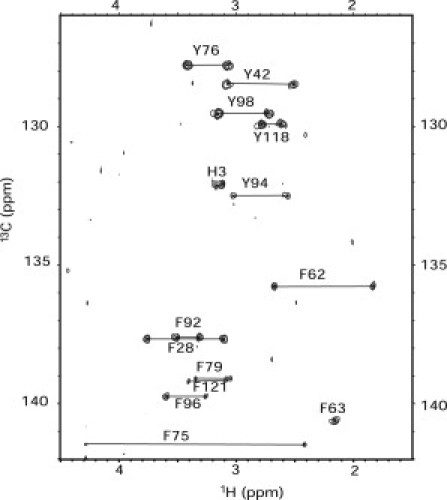
2D CG(CB)HB spectrum that correlates Hβ2/Hβ3 with Cγ for Tyr residues of PYP. His correlations were also detected. Assignments are indicated.
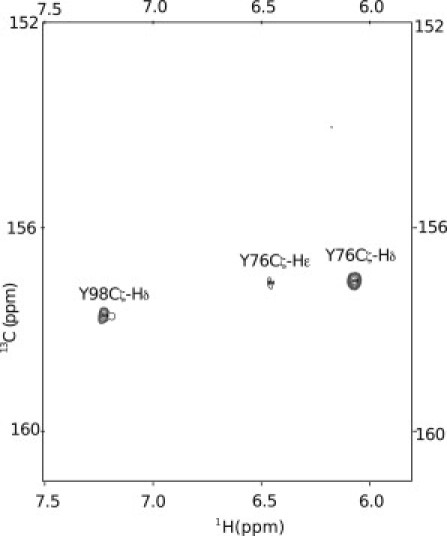
We followed the various chemical shifts that can be used as reporters of the side-chain charge state for the two Tyr residues that titrate below pH 11 (see Table 2). For Y76 and Y98, we can confirm that the chemical shifts of Cγ and Cε stand out as responsive reporters for the side-chain protonation state, and that the Cδ resonance positions cannot be employed for this purpose. The small pH-dependent changes measured for the ring-proton chemical shifts yield pKa values that are within 0.1 from those determined by 13C chemical shifts. However, small changes in 1H shifts can also result from changes in the charge states of nearby side chains. In particular, the pKa values of Lys amino groups are very similar to those of Tyr side chains, and this could lead to misinterpretation. The fact that this does not seem to occur for PYP is due to the spatial separation of the titrating groups. The fact that 1H chemical shifts are particularly reactive to changes in the local distribution of charges is documented in the literature (38).
Table 2.
Best-fit values for the pH-dependent 13C chemical-shift changes and pKa values of Y76 and Y98 of PYP
| Residue | Nucleus∗ | δ0 (ppm) | Δδ (ppm) | pKa | nH |
|---|---|---|---|---|---|
| Y76 | Cδ | 132.47 | – | – | – |
| Cε | 117.1 ± 0.1 | 2.23 ± 0.04 | 10.14 ± 0.02 | 1.24 ± 0.06 | |
| Cγ (at Hβ2/3) | 127.8 ± 0.1 | −3.77 ± 0.08 | 10.21 ± 0.02 | 1.16 ± 0.04 | |
| Hβ | 3.003 | – | – | – | |
| Hδ | 6.05 | – | – | – | |
| Hε | 6.46 ± 0.04 | 0.162 ± 0.004 | 10.1 ±0.03 | 1.3 ± 0.1 | |
| Global fit† | Cδ | 132.47 ± 0.02 | −0.08 ± 0.02 | ||
| Cε | 117.1 ± 0.1 | 2.30 ± 0.04 | 10.20 ± 0.03 | 1.18 ± 0.06 | |
| Cγ | 127.7 ± 0.1 | −3.73 ± 0.06 | |||
| Y98 | Cδ | 133.00 | – | – | – |
| Cε | 118.6 ± 0.1 | 2.8 ± 0.1 | 10.32 ± 0.03 | 1.12 ± 0.08 | |
| Cγ (at Hβ2/3) | 129.5 ± 0.1 | −4.8 ± 0.1 | 10.20 ± 0.01 | 1.54 ± 0.06 | |
| Hβ | 3.15 | – | – | – | |
| Hδ | 7.231 ± 0.004 | 0.157 ± 0.04 | 10.43 ± 0.03 | 1.05 ± 0.05 | |
| Hε | 6.85 ± 0.01 | 0.28 ± 0.01 | 10.37 ± 0.04 | 1.14 ± 0.1 | |
| Global fit† | Cδ | 133.00 ± 0.02 | −0.23 ± 0.02 | ||
| Cε | 118.6 ± 0.1 | 2.5 ± 0.1 | 10.21 ± 0.03 | 1.48 ± 0.08 | |
| Cγ | 129.5 ± 0.1 | −4.8 ± 0.1 |
Cδ, Cε, Hδ, and Hε resonances were monitored by 13C-1H HSQC spectroscopy; Cγ and Hβ resonances were recorded by CG(CB)HB spectroscopy.
For global fitting, the four titrations pertaining to the same Tyr ring (probed at CγHβ2, CγHβ3, CδHδ, and CεHε) were combined and fitted simultaneously to a model with one pKa and one nH for that Tyr. Standard deviations (SDs) for the best-fit parameters resulting from the individual fits were estimated by analysis of the χ2 function (43). For the global-fit parameters, the SDs were estimated via a Monte Carlo protocol in which estimated SDs per data point (0.05 in pH and 0.03 ppm in chemical shift) were used. See text for further discussion about the accuracy of these results.
Fig. 6 A shows the location of the two solvent-exposed Tyr residues in PYP, together with titration curves of their 13Cγ resonances. The pKa values for the solvent-exposed residues Y76 and Y98 are 10.20 and 10.21, respectively, which is 0.5 units above the intrinsic pKa reported for Tyr in aqueous solution (16). Because the net protein charge is approximately −6 at pH 9, this upshift of 0.5 units (which corresponds to an energy difference of ∼2.8 kJ/mol) is probably due to coulombic forces that favor the neutral form of Tyr.
Figure 6.
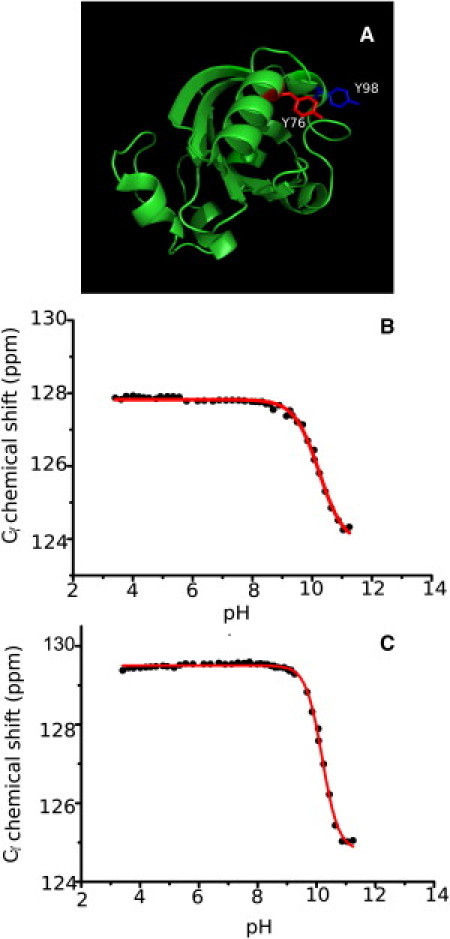
(A) PYP structure with the solvent-exposed residues Y76 and Y98 indicated. pH titration profile of Y76 (B) and Y98 (C) using the 13Cγ chemical shifts as reporters.
Fits of Eq. 1 to the experimental data improved significantly when the Hill parameters were allowed to become >1, especially for Y98. This is unusual for pH titrations in compactly folded proteins, where electrostatic interactions between nearby groups that titrate in the same pH range are expected to lead to an apparent negative cooperativity (nH < 1). Even more puzzling is the observation that two titrations pertaining to the same residue (e.g,. Y98; see Table 2) yield different values for the Hill parameter. We conclude that small systematic errors (e.g., in the measurements of the highest pH values) must be part of the explanation. However, we also note that structural rearrangements in the direct environment of Tyr residues (in response to the increasing density of negative charges) can in principle explain the apparent positive cooperativity in their titration curves. The fact that the 13Cδ resonance shifts 0.23 ppm downfield in the pH range of ∼10.5 (Table 2) is another indication that structural rearrangements must occur, because the 13Cδ chemical shift is normally unresponsive to ionization of the side chain (19).
As shown in Fig. 7 A, Y118 is partly (9%) buried within the protein interior, but its hydroxyl group is not involved in H-bonding. The titration curve for the 13Cγ of Y118 is shown in Fig. 7 B. At the highest pH value, the 13Cγ resonance has moved only −0.7 ppm. Assuming that Δδ = −4 ppm (similar to the value measured for Y76), its estimated pKa based on Eq. 1 is 11.6, which is higher than the values of the solvent-exposed residues Y76 and Y98. Substitution of Δδ values within the range of −3 to −6 ppm yielded estimated pKa values between 11.4 and 12.0. The difference in the pKa value of 1.4 pH units between solvent-exposed Tyr and buried Tyr can be explained by the different dielectric properties of water as a solvent and the protein interior, which is more hydrophobic (39). The more polar the environment, the more easily an acid/base can be ionized. Thus, in the hydrophobic environment of Y118, its pKa has shifted to a higher value.
Figure 7.
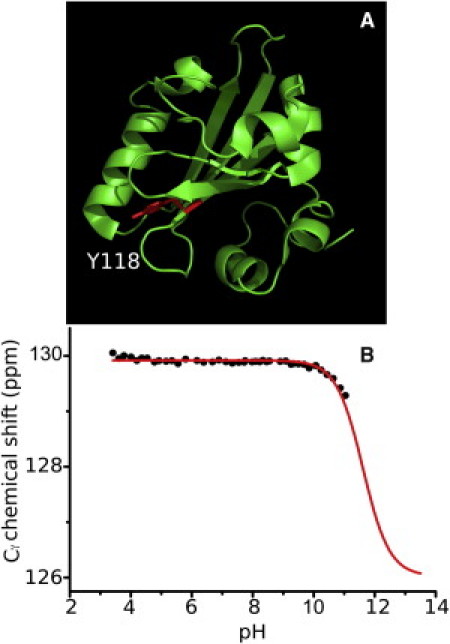
Buried Y118 in PYP structure (A) and its corresponding pH titration profile using the 13Cγ chemical shift as reporter (B).
The two remaining Tyr residues in PYP, Y42 and Y94, are 82% and 27% buried, respectively, and involved in H-bonding. Their location in the protein is indicated in Fig. 8 A. Because the changes in their 13Cγ chemical shifts are <0.1 ppm up to pH 11.24, the pKa values of those side chains are significantly higher than 13 and cannot be determined reliably. The pKa value of Y42 is of special mechanistic interest in the case of PYP, because this side chain shares a proton with the pCA chromophore, and forms a short H-bond. Our data demonstrate that this H-bond is extremely stable and remains intact over the entire pH range, from 3.41 to 11.24 (see Fig. 8 B). In this pH range, the protein is folded and the active center is intact. However, the disruption of the H-bonding network involving the chromophore at very low pH (<3) results in partial protein unfolding (40), whereas at very high pH (>11) it leads to hydrolysis of the thioester bond that connects the pCA chromophore to C69 (41). Y42 is also known to play an important role in stabilizing the native conformation of the pCA chromophore through H-bonding. The mutation of Y42 into Phe (Y42F) disrupts the H-bond network between Y42 and the pCA chromophore, as well as that between Y42 and the hydroxyl group of T50. This causes the distance between F42 and T50 to increase due to van der Waals repulsion, and movement of the pCA chromophore toward T50. As a consequence, a second conformation of the pCA chromophore can be observed in the protein population as a shoulder in the UV/Vis spectrum at 391 nm (8). However, the Y42F mutation does not have a significant effect on the pH dependence of the photocycle kinetics. The maximum rate constant for the transition of pR to pB in the Y42F mutant occurs at a similar pH compared with wild-type PYP (8). This finding agrees with our result that Y42 remains fully protonated in the pH range of 3.41–11.24, which makes it unlikely that pH-induced changes observed in the photocycle kinetics of PYP are caused by changes in the electrostatic interactions involving the chromophore in the ground state.
Figure 8.
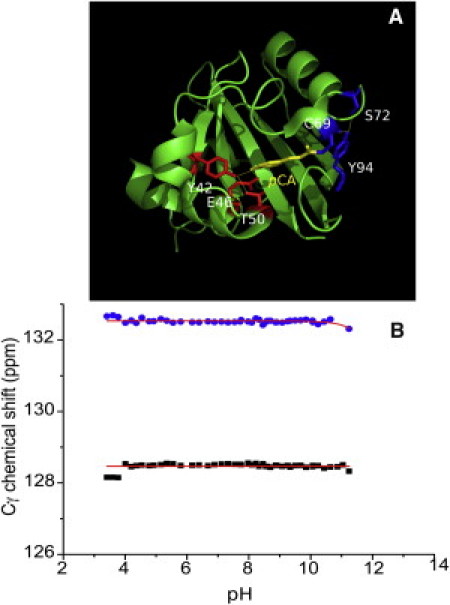
H-bonded Tyr side chains in PYP. (A) Y42 forms an H-bond to pCA and the hydroxyl group of T50, whereas Y94 forms H-bonds to the backbone of C69 and the hydroxyl group of S72. (B) pH titration profile of Y42 (squares) and Y94 (circles) using the 13Cγ chemical shift as reporter.
Y94 is also buried and donates an H-bond to the backbone of C69 and the hydroxyl side chain of S72. Fig. 8 B shows that the protonation state of Y94 is pH-independent, indicating that Y94 is protonated over the entire pH range of 3.41–11.24. Although there is no direct H-bond between Y94 and the pCA chromophore, the stability of the H-bond network over a wide pH range may also be important for the stability of the thioester bond between C69 and pCA. This result is supported by the fact that the mutation Y94A shifts the absorption maximum by 4 nm toward the blue (442 nm) (42). In agreement with these findings, spectroscopic studies also imply that the H-bond between the side chain of Y94 and the hydroxyl side chain of S72 is important for maintaining the helical secondary structure (42).
Conclusions
We have presented an NMR approach based on 2D Cγ-Hβ correlation spectroscopy to determine the residue-specific pKa values of individual Tyr side chains in native proteins with high sensitivity and resolution. This approach offers a number of advantages over existing practices. Our approach does not require mutagenesis to assign the NMR resonances, and facilitates complete and comprehensive analyses of electrostatic interactions involving Tyr side chains in proteins.
For PYP from H. halophila, we were able to determine the pH dependence of the protonation states of all individual Tyr side chains. In PYP we observe three classes of tyrosines based on their titration behavior: solvent-exposed (Y76 and Y98), buried in the hydrophobic environment (Y118), and H-bonded (Y42 and Y94), with pKa values of ∼10, 12, and above 13, respectively. Our study also shows that the short H-bonds to the pCA chromophore persist over the entire pH range in which the protein is chemically and thermodynamically stable. Our data indicate that previous observations of pH-dependent changes in PYP photocycle kinetics cannot be caused by changes in the charge state of Y42, and it is therefore very unlikely that the pCA chromophore undergoes changes in its electrostatic interactions in the electronic ground state.
Acknowledgments
This work was supported by a VIDI career development award to F.A.A.M. from the Netherlands Organization for Scientific Research.
Footnotes
Frans A.A. Mulder's present address is Interdisciplinary Nanoscience Center (iNANO) and Department of Chemistry, University of Aarhus, Aarhus, Denmark.
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Sprenger W.W., Hoff W.D., Hellingwerf K.J. The eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption spectrum of the photoactive yellow protein. J. Bacteriol. 1993;175:3096–3104. doi: 10.1128/jb.175.10.3096-3104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellequer J.L., Wager-Smith K.A., Getzoff E.D. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc. Natl. Acad. Sci. USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borgstahl G.E., Williams D.R., Getzoff E.D. 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 4.Anderson S., Crosson S., Moffat K. Short hydrogen bonds in photoactive yellow protein. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1008–1016. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 5.Fisher S.Z., Anderson S., Schultz A.J. Neutron and X-ray structural studies of short hydrogen bonds in photoactive yellow protein (PYP) Acta Crystallogr. D Biol. Crystallogr. 2007;63:1178–1184. doi: 10.1107/S0907444907047646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamaguchi S., Kamikubo H., Kataoka M. Low-barrier hydrogen bond in photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 2009;106:440–444. doi: 10.1073/pnas.0811882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigala P.A., Tsuchida M.A., Herschlag D. Hydrogen bond dynamics in the active site of photoactive yellow protein. Proc. Natl. Acad. Sci. USA. 2009;106:9232–9237. doi: 10.1073/pnas.0900168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brudler R., Meyer T.E., Getzoff E.D. Coupling of hydrogen bonding to chromophore conformation and function in photoactive yellow protein. Biochemistry. 2000;39:13478–13486. doi: 10.1021/bi0009946. [DOI] [PubMed] [Google Scholar]
- 9.Imamoto Y., Kataoka M. Structure and photoreaction of photoactive yellow protein, a structural prototype of the PAS domain superfamily. Photochem. Photobiol. 2007;83:40–49. doi: 10.1562/2006-02-28-IR-827. [DOI] [PubMed] [Google Scholar]
- 10.Imamoto Y., Mihara K., Yoshihara K. Evidence for proton transfer from Glu-46 to the chromophore during the photocycle of photoactive yellow protein. J. Biol. Chem. 1997;272:12905–12908. doi: 10.1074/jbc.272.20.12905. [DOI] [PubMed] [Google Scholar]
- 11.Xie A., Hoff W.D., Hellingwerf K.J. Glu46 donates a proton to the 4-hydroxycinnamate anion chromophore during the photocycle of photoactive yellow protein. Biochemistry. 1996;35:14671–14678. doi: 10.1021/bi9623035. [DOI] [PubMed] [Google Scholar]
- 12.Carroll E.C., Song S.H., Larsen D.S. Subpicosecond excited-state proton transfer preceding isomerization during the photorecovery of photoactive yellow protein. J Phys Chem Lett. 2010;1:2793–2799. doi: 10.1021/jz101049v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ujj L., Devanathan S., Atkinson G.H. New photocycle intermediates in the photoactive yellow protein from Ectothiorhodospira halophila: picosecond transient absorption spectroscopy. Biophys. J. 1998;75:406–412. doi: 10.1016/S0006-3495(98)77525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Düx P., Rubinstenn G., Kaptein R. Solution structure and backbone dynamics of the photoactive yellow protein. Biochemistry. 1998;37:12689–12699. doi: 10.1021/bi9806652. [DOI] [PubMed] [Google Scholar]
- 15.van der Horst M.A., van Stokkum I.H., Hellingwerf K.J. The role of the N-terminal domain of photoactive yellow protein in the transient partial unfolding during signalling state formation. FEBS Lett. 2001;497:26–30. doi: 10.1016/s0014-5793(01)02427-9. [DOI] [PubMed] [Google Scholar]
- 16.Hendriks J., Hellingwerf K.J. pH Dependence of the photoactive yellow protein photocycle recovery reaction reveals a new late photocycle intermediate with a deprotonated chromophore. J. Biol. Chem. 2009;284:5277–5288. doi: 10.1074/jbc.M805904200. [DOI] [PubMed] [Google Scholar]
- 17.Imamoto Y., Harigai M., Kataoka M. Direct observation of the pH-dependent equilibrium between L-like and M intermediates of photoactive yellow protein. FEBS Lett. 2004;577:75–80. doi: 10.1016/j.febslet.2004.09.065. [DOI] [PubMed] [Google Scholar]
- 18.Karplus S., Snyder G.H., Sykes B.D. A nuclear magnetic resonance study of bovine pancreatic trypsin inhibitor. Tyrosine titrations and backbone NH groups. Biochemistry. 1973;12:1323–1329. doi: 10.1021/bi00731a012. [DOI] [PubMed] [Google Scholar]
- 19.Norton R., Bradbury J. Carbon-13 nuclear magnetic resonance study of tyrosine titrations. J. Chem. Soc. Chem. Commun. 1974;21:870–871. [Google Scholar]
- 20.Kato-Toma Y., Iwashita T., Ishiguro M. pKa measurements from nuclear magnetic resonance of tyrosine-150 in class C β-lactamase. Biochem. J. 2003;371:175–181. doi: 10.1042/BJ20021447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ugurbil K., Bersohn R. Tyrosine emission in the tryptophanless azurin from Pseudomonas fluorescens. Biochemistry. 1977;16:895–901. doi: 10.1021/bi00624a013. [DOI] [PubMed] [Google Scholar]
- 22.Wilbur D.J., Allerhand A. Titration behavior of individual tyrosine residues of myoglobins from sperm whale, horse, and red kangaroo. J. Biol. Chem. 1976;251:5187–5194. [PubMed] [Google Scholar]
- 23.André I., Linse S., Mulder F.A.A. Residue-specific pKa determination of lysine and arginine side chains by indirect 15N and 13C NMR spectroscopy: application to apo calmodulin. J. Am. Chem. Soc. 2007;129:15805–15813. doi: 10.1021/ja0721824. [DOI] [PubMed] [Google Scholar]
- 24.Hass M.A., Yilmaz A., Led J.J. Histidine side-chain dynamics and protonation monitored by 13C CPMG NMR relaxation dispersion. J. Biomol. NMR. 2009;44:225–233. doi: 10.1007/s10858-009-9332-0. [DOI] [PubMed] [Google Scholar]
- 25.Oda Y., Yamazaki T., Nakamura H. Individual ionization constants of all the carboxyl groups in ribonuclease HI from Escherichia coli determined by NMR. Biochemistry. 1994;33:5275–5284. doi: 10.1021/bi00183a034. [DOI] [PubMed] [Google Scholar]
- 26.Baturin S.J., Okon M., McIntosh L.P. Structure, dynamics, and ionization equilibria of the tyrosine residues in Bacillus circulans xylanase. J. Biomol. NMR. 2011;51:379–394. doi: 10.1007/s10858-011-9564-7. [DOI] [PubMed] [Google Scholar]
- 27.Boggio-Pasqua M., Robb M.A., Groenhof G. Hydrogen bonding controls excited-state decay of the photoactive yellow protein chromophore. J. Am. Chem. Soc. 2009;131:13580–13581. doi: 10.1021/ja904932x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mihara K., Hisatomi O., Tokunaga F. Functional expression and site-directed mutagenesis of photoactive yellow protein. J. Biochem. 1997;121:876–880. doi: 10.1093/oxfordjournals.jbchem.a021668. [DOI] [PubMed] [Google Scholar]
- 29.Takeda M., Jee J., Kainosho M. Hydrogen exchange rate of tyrosine hydroxyl groups in proteins as studied by the deuterium isotope effect on C(ζ) chemical shifts. J. Am. Chem. Soc. 2009;131:18556–18562. doi: 10.1021/ja907911y. [DOI] [PubMed] [Google Scholar]
- 30.Prompers J.J., Groenewegen A., Pepermans H.A.M. Two-dimensional NMR experiments for the assignment of aromatic side chains in 13C-labeled proteins. J. Magn. Reson. 1998;130:68–75. doi: 10.1006/jmre.1997.1277. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Goddard T., Kneller D. University of California; San Francisco: 2004. SPARKY 3. [Google Scholar]
- 33.Markley J.L., Bax A., Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids—IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. Eur. J. Biochem. 1998;256:1–15. doi: 10.1046/j.1432-1327.1998.2560001.x. [DOI] [PubMed] [Google Scholar]
- 34.Oktaviani N.A., Otten R., Mulder F.A.A. 100% complete assignment of non-labile (1)H, (13)C, and (15)N signals for calcium-loaded Calbindin D(9k) P43G. Biomol. NMR Assign. 2011;5:79–84. doi: 10.1007/s12104-010-9272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Constantine K.L., Mueller L., Farmer B.T., 2nd Characterization of NADP+ binding to perdeuterated MurB: backbone atom NMR assignments and chemical-shift changes. J. Mol. Biol. 1997;267:1223–1246. doi: 10.1006/jmbi.1997.0915. [DOI] [PubMed] [Google Scholar]
- 36.Gardner K.H., Kay L.E. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 37.Paquin R., Ferrage F., Bodenhausen G. Multiple-timescale dynamics of side-chain carboxyl and carbonyl groups in proteins by 13C nuclear spin relaxation. J. Am. Chem. Soc. 2008;130:15805–15807. doi: 10.1021/ja803794g. [DOI] [PubMed] [Google Scholar]
- 38.Webb H., Tynan-Connolly B.M., Nielsen J.E. Remeasuring HEWL pK(a) values by NMR spectroscopy: methods, analysis, accuracy, and implications for theoretical pK(a) calculations. Proteins. 2011;79:685–702. doi: 10.1002/prot.22886. [DOI] [PubMed] [Google Scholar]
- 39.Harris T.K., Turner G.J. Structural basis of perturbed pKa values of catalytic groups in enzyme active sites. IUBMB Life. 2002;53:85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 40.Craven C.J., Derix N.M., Kaptein R. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry. 2000;39:14392–14399. doi: 10.1021/bi001628p. [DOI] [PubMed] [Google Scholar]
- 41.Hoff W.D., Devreese B., Hellingwerf K.J. Chemical reactivity and spectroscopy of the thiol ester-linked p-coumaric acid chromophore in the photoactive yellow protein from Ectothiorhodospira halophila. Biochemistry. 1996;35:1274–1281. doi: 10.1021/bi951755z. [DOI] [PubMed] [Google Scholar]
- 42.Morishita T., Harigai M., Imamoto Y. Array of aromatic amino acid side chains located near the chromophore of photoactive yellow protein. Photochem. Photobiol. 2007;83:280–285. doi: 10.1562/2006-06-15-RA-929. [DOI] [PubMed] [Google Scholar]
- 43.Berendsen H.J.C. Cambridge University Press; Cambridge, UK: 2011. A student's guide to data and error analysis. [Google Scholar]


