Abstract
We show that the folding rates (kFs) of RNA are determined by N, the number of nucleotides. By assuming that the distribution of free-energy barriers separating the folded and the unfolded states is Gaussian, which follows from central limit theorem arguments and polymer physics concepts, we show that . Remarkably, the theory fits experimental rates spanning over 7 orders of magnitude with . Our finding suggests that the speed limit of RNA folding is ∼1 ms, just as it is in the folding of globular proteins.
RNA molecules are evolved biopolymers whose folding has attracted a great deal of attention (1–3) because of the crucial role they play in a number of cellular functions. The slightly branched polymeric nature of RNA implies that the shapes, relaxation dynamics, and even their folding rates must depend on N. In support of this assertion, it has been shown that the radius of gyration of the folded states, obtained with the use of data available in the Protein Data Bank, scales as Å, where the Flory exponent ν varies from 0.33 to 0.40 (4–6). Although this result is expected from the perspective of polymer physics, it is surprising from the viewpoint of structural biology because one might argue that the sequence and complexity of secondary and tertiary structure organization could lead to substantial deviations from the predictions based on Flory-like theory. Here, we show that the folding rates, kFs, of RNA are also primarily determined by N, thus adding to the growing evidence that it is possible to understand RNA folding by using polymer physics principles.
Theoretical Considerations
Theoretical arguments based on the dynamics of activated transitions in disordered systems suggest that
| (1) |
where β should be 0.5 (7). The rationale for this finding hinges on the observation that favorable basepairing interactions and the hydrophobic nature of the bases tend to collapse RNA, whereas the charged phosphate residues are better accommodated by extended structures. Thus, the distribution of activation free energy, , between the folded and unfolded states is a sum of favorable and unfavorable terms. We expect from central limit theorem that the distribution of should be roughly Gaussian with dispersion . Thus, with β = 1/2.
We analyzed the available experimental data (see Table 1 for a list of RNA molecules) on RNA folding rates by assuming that grows as Nβ with β as a free parameter. The theoretical value for β is 0.5. The folding rates of RNA spanning over 7 orders of magnitude is well fit using with a correlation coefficient of 0.98 (Fig. 1). The fit yields μs, α = 0.91, and β ≈ 0.46. In the inset we show the fit obtained by fixing β = 0.5. Apart from the moderate differences in the values, the theoretical prediction and the numerical fits are in agreement, which demonstrates that the major determining factor in determining RNA folding rates is N.
Table 1.
RNA length versus folding rate
| RNA | N | kf (sec-1) | |
|---|---|---|---|
| GCUUCGGC (16) | 8 | 6.7 × 104 | Tetraloop hairpin |
| GCUUCGGC (16) | 8 | 27.2 × 104 | Tetraloop hairpin |
| GGUUCGCC (16) | 8 | 1.3 × 104 | Tetraloop hairpin |
| GGUUCGCC (16) | 8 | 4.7 × 104 | Tetraloop hairpin |
| GGACUUUUGUCC (16) | 12 | 6.1 × 104 | Tetraloop hairpin |
| GGACUUCGGUCC (16) | 12 | 4.5 × 104 | Tetraloop hairpin |
| A6C6U6 (17) | 18 | 3.4 × 104 | Tetraloop hairpin |
| Extra arm of tRNAser (yeast) (18) | 21 | 1 × 105 | tRNA |
| pG half of tRNAPhe (yeast) (18) | 36 | 9 × 103 | tRNA |
| CCA half of tRNAPhe (yeast) (18) | 39 | 8.5 × 103 | tRNA |
| CCA half of tRNAPhe (wheat) (18) | 39 | 8 × 103 | tRNA |
| tRNAPhe (yeast) (19) | 76 | 5.3 × 102 | tRNA |
| tRNAAla (yeast) (18) | 77 | 9 × 102 | tRNA |
| Y4 hairpin (20) | 14 | 5.75 × 104 | Hairpin (5 × 2 + 4) |
| Y9 hairpin (20) | 19 | 2.29 × 104 | Hairpin (5 × 2 + 9) |
| Y19 hairpin (20) | 29 | 8.70 × 102 | Hairpin (5 × 2 + 19) |
| Y34 hairpin (20) | 44 | 6.03 × 102 | Hairpin (5 × 2 + 34) |
| VPK pseudoknot (21) | 34 | 9.09 × 102 | Pseudoknot |
| Hairpin ribozyme (four-way junction) (22,23) | 125 | 6 | Natural form of hairpin ribozyme |
| P5abc (24) | 72 | 50 | Group I intron T. ribozyme |
| P4-P6 domain(Tetrahymena ribozyme) (24) | 160 | 2 | Group I intron T. ribozyme |
| Azoarcus ribozyme (23,25) | 205 | 7 ∼ 14 | |
| B. subtilis RNase P RNA catalytic domain (26) | 225 | 6.5 ± 0.2 | |
| Ca.L-11 ribozyme (27) | 368 | 0.03 | |
| E. coli RNase P RNA (28) | 377 | 0.011 ± 0.001 | |
| B. subtilis RNase P RNA (28) | 409 | 0.008 ± 0.002 | |
| Tetrahymena ribozyme (23,29) | 414 | 0.013 | Group I intron T. ribozyme |
Figure 1.
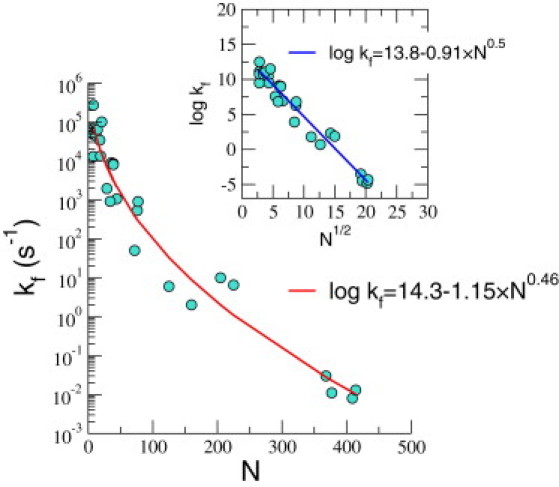
Dependence of the folding rates of RNA on N. The circles are experimental data and the line is the fit obtained using , with β used as an adjustable parameter. Inset shows the fit obtained by fixing β to the predicted theoretical value of 0.5.
It is known that RNAs, such as Tetrahymena ribozyme, fold by multiple pathways that are succinctly described by the kinetic partitioning mechanism (8). According to this mechanism, a fraction, Φ, of molecules reaches the native states rapidly and the remaining fraction is trapped in an ensemble of misfolded intermediates. For Tetrahymena ribozyme Φ ∼ 0.1 (9). The N dependence given by Eq. 1 holds for the majority of molecules that fold to the native state from the compact intermediates, which form rapidly under folding conditions (10).
Conclusions
Our findings indicate that the inverse of the prefactor, μs, is almost 6 orders of magnitude larger than the transition-state theory estimate of h / kBT ≈ 0.16 ps. The value of τ0, which coincides with the typical basepairing time (11), is the speed limit for RNA folding. Of interest, general arguments based on the kinetics of loop formation have been used to predict that the speed limit for protein folding is also ∼1 ms (12–14). It remains to be ascertained whether the common folding speed limit for proteins and RNA is due to evolutionary pressure on the folding of evolved sequences. It is worth pointing out that Dill et al. (15) recently showed that the rates and stabilities of protein folding depend only on the number of amino acids, which in turn places strict constraints on their functions in the cellular context. Taken together, these studies show that despite the complexity of protein and RNA folding, it is possible that only a few variables determine their global properties. This suggests that certain simple principles may determine biological functions.
Acknowledgments
This work was supported by grants from the Korea Research Foundation (2010-0000602) (C.H.) and the National Science Foundation (CHE 09-14033) (D.T.).
Contributor Information
Changbong Hyeon, Email: hyeoncb@kias.re.kr.
D. Thirumalai, Email: thirum@umd.edu.
References and Footnotes
- 1.Thirumalai D., Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 2.Treiber D.K., Williamson J.R. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 3.Woodson S.A. Compact intermediates in RNA folding. Annu. Rev. Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyeon C., Dima R.I., Thirumalai D. Size, shape, and flexibility of RNA structures. J. Chem. Phys. 2006;125:194905. doi: 10.1063/1.2364190. [DOI] [PubMed] [Google Scholar]
- 5.Yoffe A.M., Prinsen P., Ben-Shaul A. Predicting the sizes of large RNA molecules. Proc. Natl. Acad. Sci. USA. 2008;105:16153–16158. doi: 10.1073/pnas.0808089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajdin C.E., Ding F., Weeks K.M. On the significance of an RNA tertiary structure prediction. RNA. 2010;16:1340–1349. doi: 10.1261/rna.1837410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thirumalai D. From minimal models to real proteins: time scales for protein folding kinetics. J. Phys. 1995;5:1457–1467. [Google Scholar]
- 8.Guo Z., Thirumalai D. Kinetics of protein folding: nucleation mechanism, time scales, and pathways. Biopolymers. 1995;36:83–102. [Google Scholar]
- 9.Pan J., Thirumalai D., Woodson S.A. Folding of RNA involves parallel pathways. J. Mol. Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 10.Thirumalai D., Lee N., Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 11.Pörschke D., Uhlenbeck O., Martin F. Thermodynamics and kinetics of the helix-coil transition of oligomers containing GC base pairs. Biopolymers. 1973;12:1313–1335. [Google Scholar]
- 12.Hagen S.J., Hofrichter J., Eaton W.A. Diffusion-limited contact formation in unfolded cytochrome c: estimating the maximum rate of protein folding. Proc. Natl. Acad. Sci. USA. 1996;93:11615–11617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W.Y., Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 14.Kubelka J., Hofrichter J., Eaton W.A. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Dill K.A., Ghosh K., Schmit J.D. Physical limits of cells and proteomes. Proc. Natl. Acad. Sci. USA. 2011;108:17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proctor D.J., Ma H., Bevilacqua P.C. Folding thermodynamics and kinetics of YNMG RNA hairpins: specific incorporation of 8-bromoguanosine leads to stabilization by enhancement of the folding rate. Biochemistry. 2004;43:14004–14014. doi: 10.1021/bi048213e. [DOI] [PubMed] [Google Scholar]
- 17.Porschke D. Thermodynamic and kinetic parameters of an oligonucleotide hairpin helix. Biophys. Chem. 1974;1:381–386. doi: 10.1016/0301-4622(74)85008-8. [DOI] [PubMed] [Google Scholar]
- 18.Riesner D., Römer R. Thermodynamics and kinetics of conformational transitions in oligonucleotides and tRNA. In: Duchesne J., editor. Vol. 2. Academic Press; London: 1973. pp. 237–318. (Physico-Chemical Properties of Nucleic Acids). [Google Scholar]
- 19.Serebrov V., Clarke R.J., Kisselev L. Mg2+-induced tRNA folding. Biochemistry. 2001;40:6688–6698. doi: 10.1021/bi002241p. [DOI] [PubMed] [Google Scholar]
- 20.Kuznetsov S.V., Ren C.C., Ansari A. Loop dependence of the stability and dynamics of nucleic acid hairpins. Nucleic Acids Res. 2008;36:1098–1112. doi: 10.1093/nar/gkm1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan R., Velmurugu Y., Ansari A. Fast folding of RNA pseudoknots initiated by laser temperature-jump. J. Am. Chem. Soc. 2011;133:18767–18774. doi: 10.1021/ja205737v. [DOI] [PubMed] [Google Scholar]
- 22.Tan E., Wilson T.J., Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. USA. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sosnick T.R., Pan T. Reduced contact order and RNA folding rates. J. Mol. Biol. 2004;342:1359–1365. doi: 10.1016/j.jmb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Deras M.L., Brenowitz M., Woodson S.A. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 25.Rangan P., Masquida B., Woodson S.A. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc. Natl. Acad. Sci. USA. 2003;100:1574–1579. doi: 10.1073/pnas.0337743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang X., Pan T., Sosnick T.R. Mg-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol. 1999;12:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- 27.Xiao M., Leibowitz M.J., Zhang Y. Concerted folding of a Candida ribozyme into the catalytically active structure posterior to a rapid RNA compaction. Nucleic Acids Res. 2003;31:3901–3908. doi: 10.1093/nar/gkg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarrinkar P.P., Wang J., Williamson J.R. Slow folding kinetics of RNase P RNA. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 29.Sclavi B., Sullivan M., Woodson S.A. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]


