Abstract
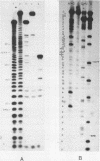
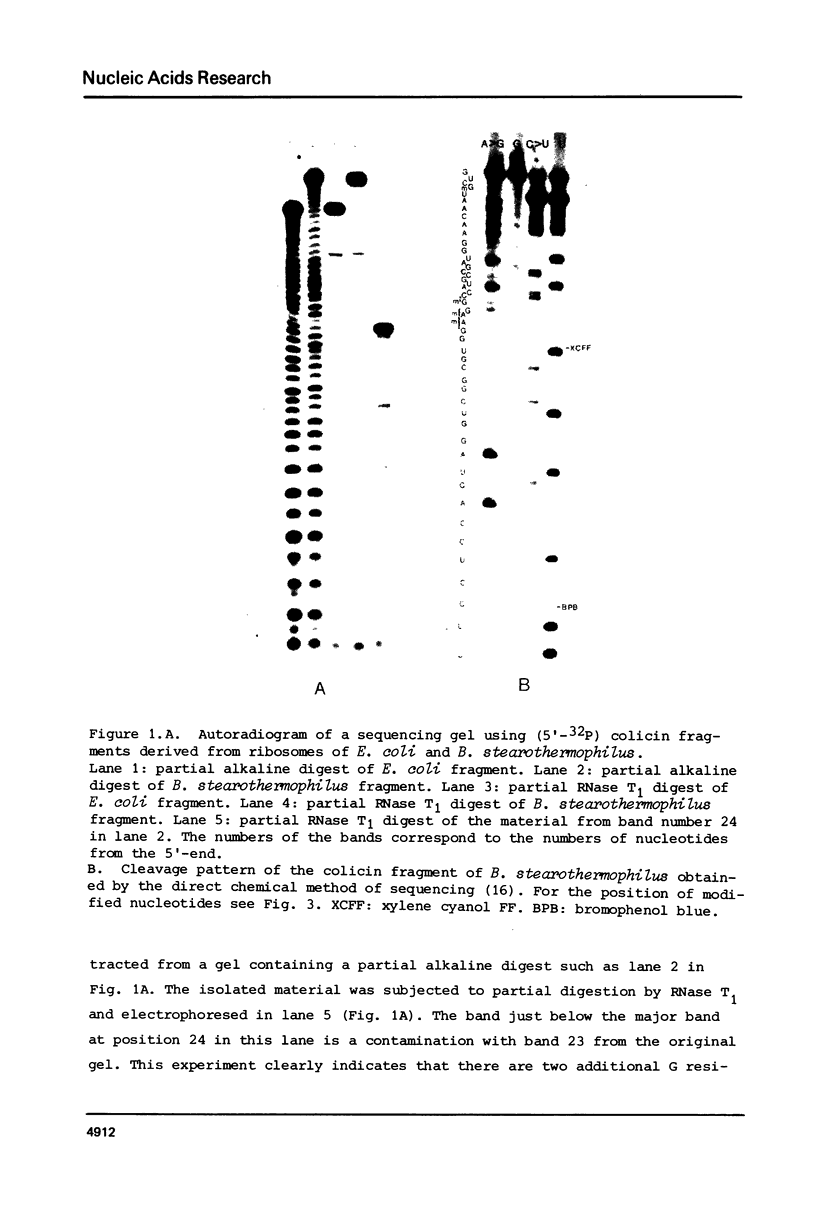
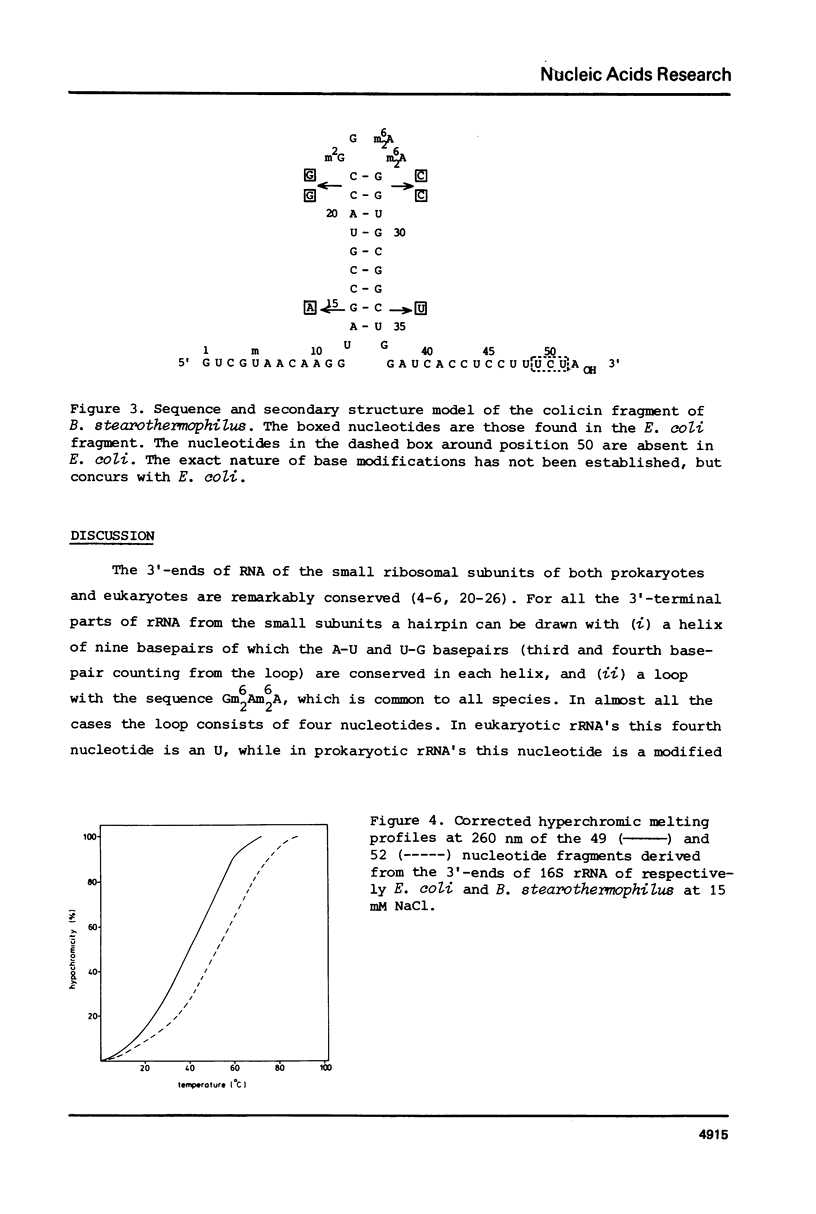
The sequence and the position of post-transcriptionally modified residues of the 3' -terminal end of Bacillus stearothermophilus 16S ribosomal RNA have been determined from the fragment that is cleaved off by bacteriocin treatment. The fragment contains 52 nucleotides, as compared to the 49 nucleotides of the corresponding fragment from E. coli ribosomes, The additional nucleotides are present in the sequence UCU very next to the 3' -terminus as was published earlier (1). The remainder of the sequence is identical to the one of E. coli except at six positions, due to the UV melting properties of the colicin fragment from B. stearothermophilus in comparison to the same fragment of E. coli show that the RNA from the thermophile has a more stable secondary structure.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberty H., Raba M., Gross H. J. Isolation from rat liver and sequence of a RNA fragment containing 32 nucleotides from position 5 to 36 from the 3' end of ribosomal 18S RNA. Nucleic Acids Res. 1978 Feb;5(2):425–434. doi: 10.1093/nar/5.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A. A., Deacon N. J. The 3'-terminal primary structure of five eukaryotic 18S rRNAs determined by the direct chemical method of sequencing. The highly conserved sequences include an invariant region complementary to eukaryotic 5S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4365–4376. doi: 10.1093/nar/8.19.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A. A. Intermolecular base-paired interaction between complementary sequences present near the 3' ends of 5S rRNA and 18S (16S) rRNA might be involved in the reversible association of ribosomal subunits. Nucleic Acids Res. 1979 Dec 11;7(7):1913–1929. doi: 10.1093/nar/7.7.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baan R. A., Hilbers C. W., Van Charldorp R., Van Leerdam E., Van Knippenberg P. H., Bosch L. High-resolution proton magnetic resonance study of the secondary structure of the 3'-terminal 49-nucleotide fragment of 16S rRNA from Escherichia coli. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1028–1031. doi: 10.1073/pnas.74.3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baan R. A., van Charldorp R., van Leerdam E., van Knippenberg P. H., Bosch L., de Rooij J. F., van Boom J. H. The 3'-terminus of 16 S ribosomal RNA of Escherichia coli. Isolation and purification of the terminal 49-nucleotide fragment at a milligram scale. FEBS Lett. 1976 Dec 1;71(2):351–355. doi: 10.1016/0014-5793(76)80968-4. [DOI] [PubMed] [Google Scholar]
- Baer R., Dubin D. T. The 3'-terminal sequence of the small subunit ribosomal RNA from hamster mitochondria. Nucleic Acids Res. 1980 Nov 11;8(21):4927–4941. doi: 10.1093/nar/8.21.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Dubin D. T. The 3'-terminal sequence of the small subunit ribosomal RNA from hamster mitochondria. Nucleic Acids Res. 1980 Nov 11;8(21):4927–4941. doi: 10.1093/nar/8.21.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ebel J. P., Ehresmann C. The sequence of the ribosomal 16S RNA from Proteus vulgaris. Sequence comparison with E. coli 16S RNA and its use in secondary model building. Nucleic Acids Res. 1981 May 25;9(10):2325–2333. doi: 10.1093/nar/9.10.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Eperon I. C., Anderson S., Nierlich D. P. Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature. 1980 Jul 31;286(5772):460–467. doi: 10.1038/286460a0. [DOI] [PubMed] [Google Scholar]
- Graf L., Kössel H., Stutz E. Sequencing of 16S--23S spacer in a ribosomal RNA operon of Euglena gracilis chloroplast DNA reveals two tRNA genes. Nature. 1980 Aug 28;286(5776):908–910. doi: 10.1038/286908a0. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Sequence of the 3'-terminal portion of Drosophila melanogaster 18 S rRNA and of the adjoining spacer: comparison with corresponding prokaryotic and eukaryotic sequences. FEBS Lett. 1980 Aug 11;117(1):227–231. doi: 10.1016/0014-5793(80)80951-3. [DOI] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Rushlow K. E., Dodd J. R., Hallick R. B. Euglena gracilis chloroplast ribosomal RNA transcription units. II. Nucleotide sequence homology between the 16 S--23 S ribosomal RNA spacer and the 16 S ribosomal RNA leader regions. J Biol Chem. 1980 Nov 25;255(22):10997–11003. [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldermans B., Bakker H., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16S ribosomal RNA of Escherichia coli. IV. The effect of the methylgroups on ribosomal subunit interaction. Nucleic Acids Res. 1980 Jan 11;8(1):143–151. doi: 10.1093/nar/8.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Samols D. R., Hagenbuchle O., Gage L. P. Homology of the 3' terminal sequences of the 18S rRNA of Bombyx mori and the 16S rRNA of Escherchia coli. Nucleic Acids Res. 1979 Nov 10;7(5):1109–1119. doi: 10.1093/nar/7.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidikaro J., Nomura M. Colicin E3-induced in vitro inactivation of ribosomes from colicin-insensitive bacterial species. FEBS Lett. 1973 Jan 1;29(1):15–19. doi: 10.1016/0014-5793(73)80005-5. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Steitz J. A., Grenley R. M., Stocking C. E. 3' terminal sequences of 16S rRNA do not explain translational specificity differences between E. coli and B. stearothermophilus ribosomes. Nature. 1977 Jun 2;267(5610):462–465. doi: 10.1038/267462a0. [DOI] [PubMed] [Google Scholar]
- Stanley J., Van Kammen A. Nucleotide sequences adjacent to the proteins covalently linked to the cowpea mosaic virus genome. Sequence determination after labelling in vitro using RNA ligase. Eur J Biochem. 1979 Nov 1;101(1):45–49. doi: 10.1111/j.1432-1033.1979.tb04214.x. [DOI] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H. 16S ribosomal RNA of Escherichia coli contains a N2-methylguanosine at 27 nucleotides from the 3' end. Nucleic Acids Res. 1981 Jun 25;9(12):2717–2725. doi: 10.1093/nar/9.12.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Charldorp R., Heus H. A., Van Knippenberg P. H. Adenosine dimethylation of 16S ribosomal RNA: effect of the methylgroups on local conformational stability as deduced from electrophoretic mobility of RNA fragments in denaturing polyacrylamide gels. Nucleic Acids Res. 1981 Jan 24;9(2):267–275. doi: 10.1093/nar/9.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duin J., Kurland C. G., Dondon J., Grunberg-Mangago M., Branlant C., Ebel J. P. New aspects of the IF3-ribosome interaction. FEBS Lett. 1976 Feb 15;62(2):111–114. doi: 10.1016/0014-5793(76)80030-0. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Yuan R. C., Steitz J. A., Moore P. B., Crothers D. M. The 3' terminus of 16S rRNA: secondary structure and interaction with ribosomal protein S1. Nucleic Acids Res. 1979 Dec 20;7(8):2399–2418. doi: 10.1093/nar/7.8.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf F. K., Niekus H. G., Klootwijk J. Inactivation of bacterial ribosomes in vivo and in vitro by cloacin DF13. FEBS Lett. 1973 Sep 1;35(1):161–165. doi: 10.1016/0014-5793(73)80601-5. [DOI] [PubMed] [Google Scholar]