Abstract
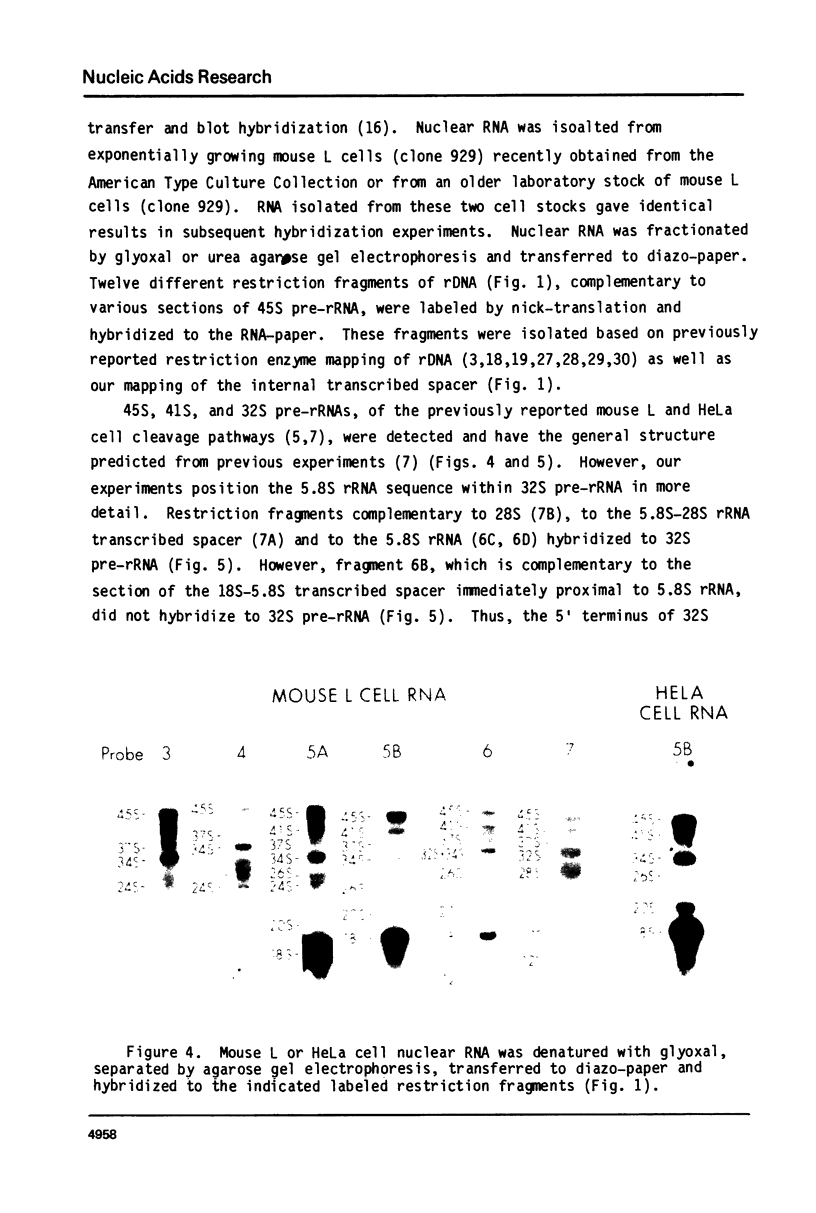
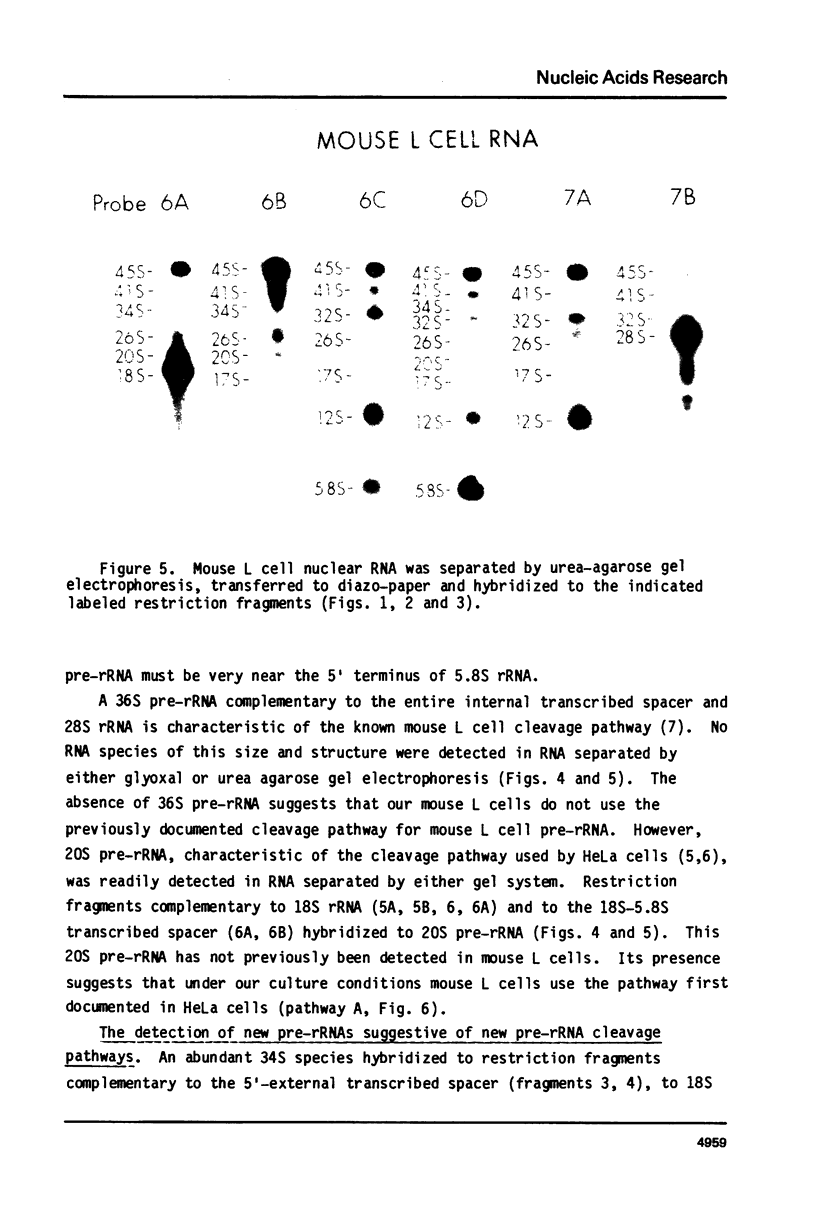
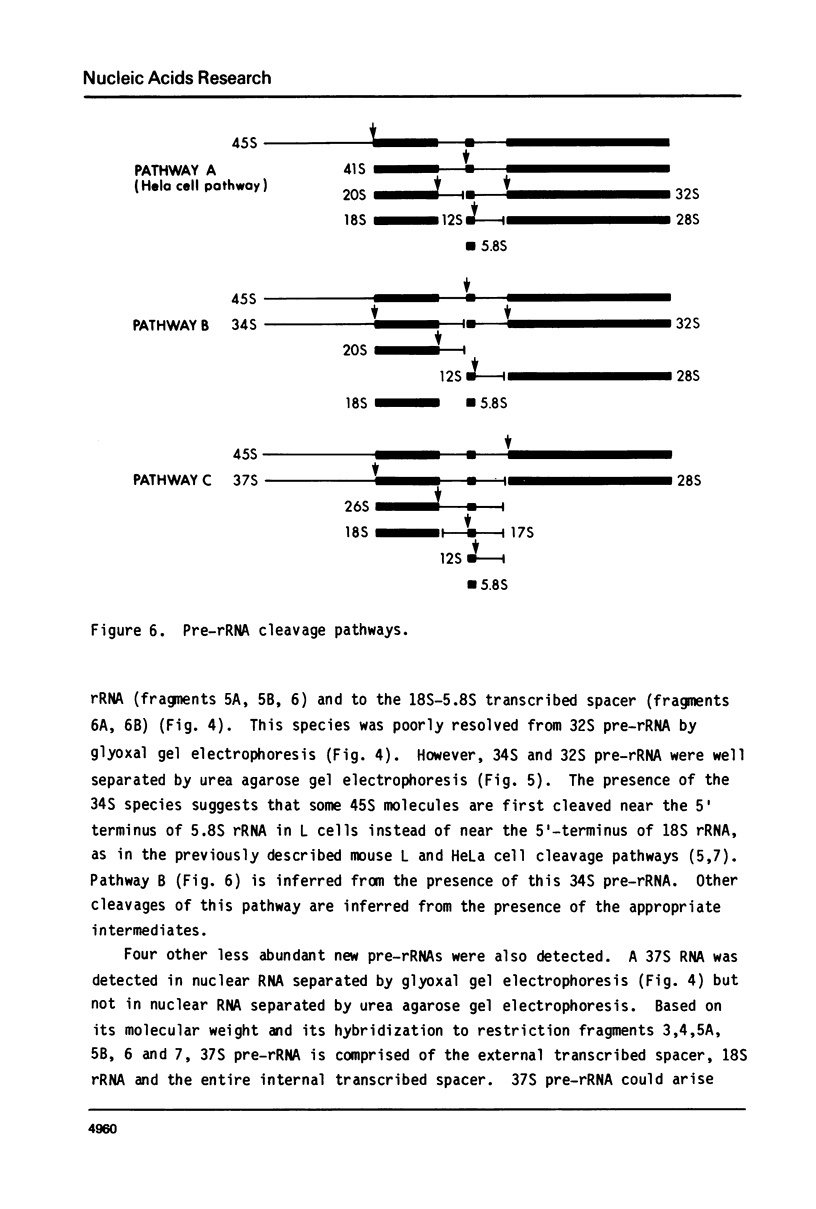
The sequence content of mouse L cell pre-rRNA was examined by RNA gel transfer and blot hybridization. Nuclear RNAs were separated by agarose gel electrophoresis, transferred to diazo-paper, and hybridized to twelve different restriction fragments that are complementary to various sections of 45S pre-rRNA. An abundant new 34S pre-rRNA and less abundant new 37S, 26S and 17S pre-rRNAs were detected. The presence of these new pre-rRNAs suggests the existence of at least two new pre-rRNA cleavage pathways. 34S and 26S pre-rRNAs were also detected in HeLa cells suggesting that these new cleavage pathways are characteristic of mammalian cells. Further, an abundant new 12S precursor to 5.8S rRNA was also detected and is common to all the proposed cleavage pathways. The previously identified 45S, 41S, 32S and 20S pre-rRNAs were readily detected and their general structure confirmed. The 20S pre-rRNA is characteristic of the known pathway used by HeLa and other cells, and its presence suggests that growing mouse L cells use this pre-rRNA cleavage pathway. The 36S pre-rRNA characteristic of the previously described mouse L cell cleavage pathway was not detected. In all these cleavage pathways pre-rRNA cleavage sites are apparently identical and occur at or near the termini of the mature 18S, 5.8S and 28S rRNA sequences. The pathways differ only in the temporal order of cleavage at these sites.
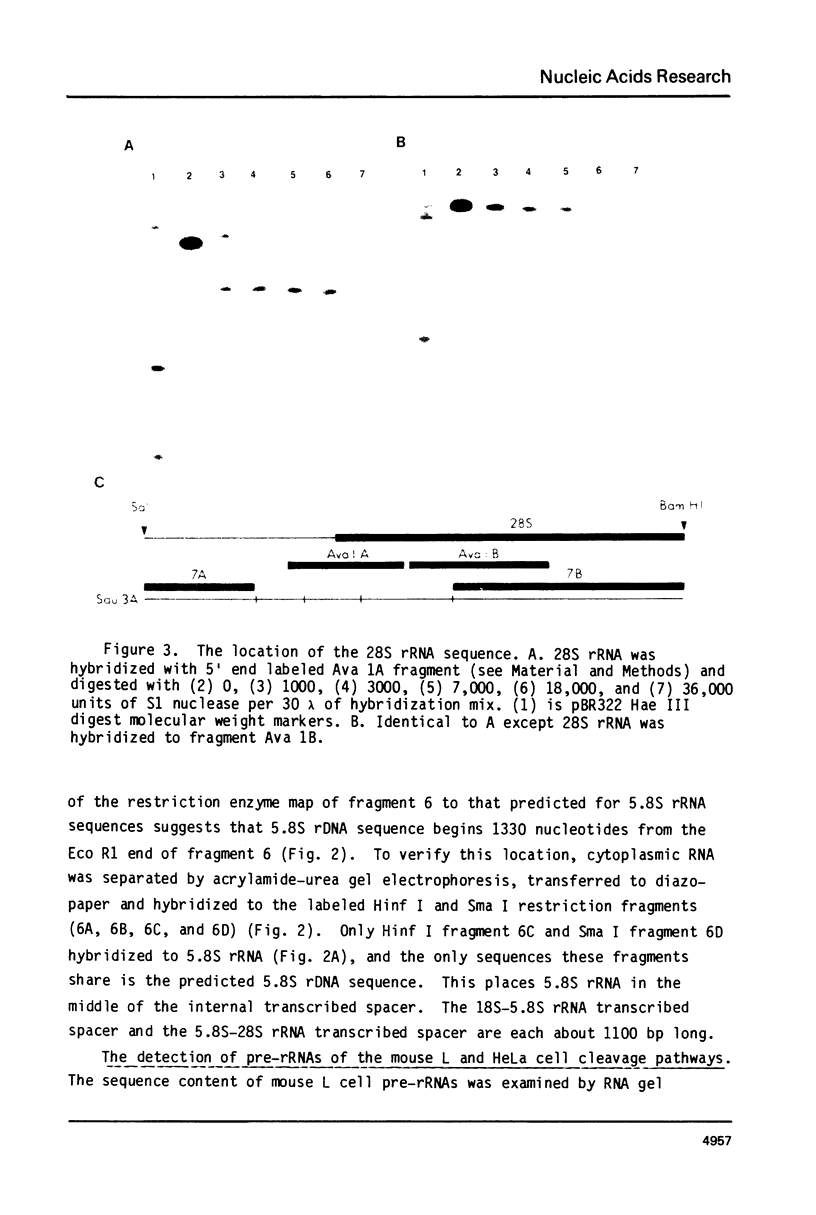
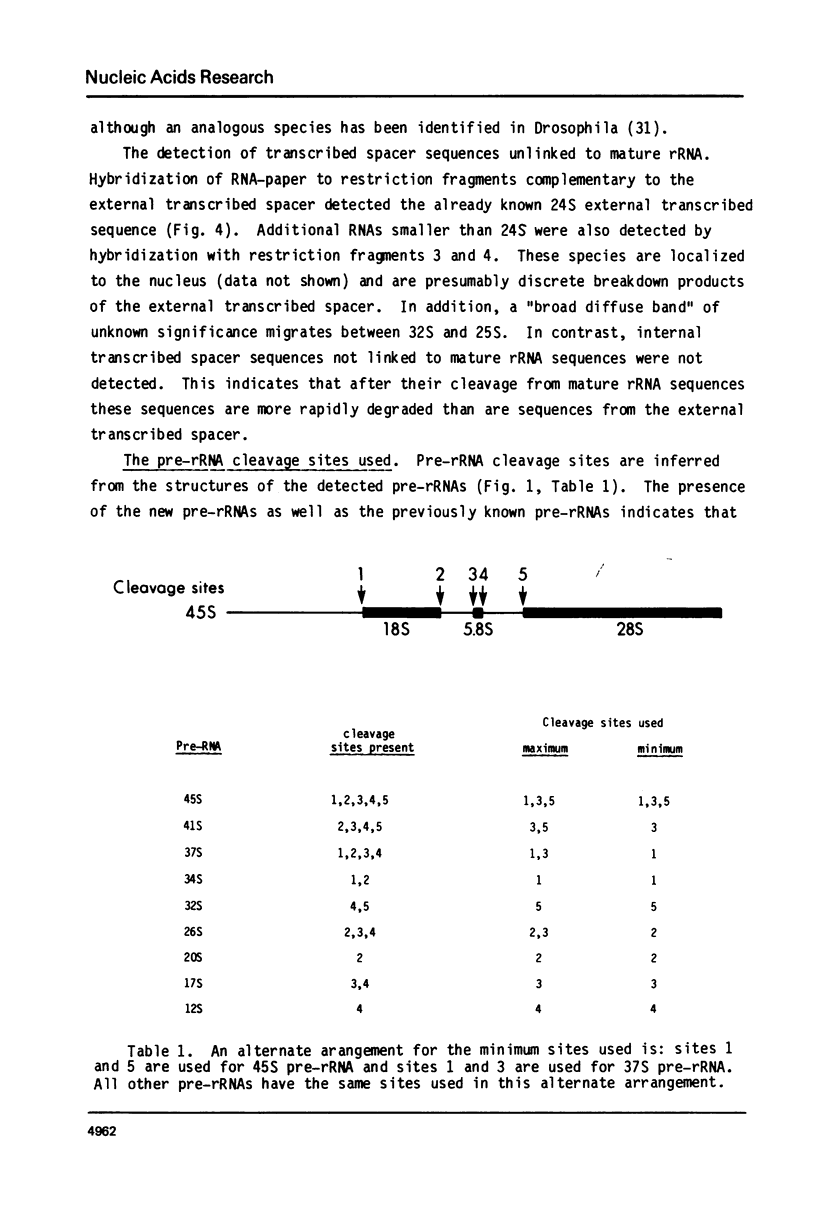
The position of the 5.8S rRNA sequence was located within the internal transcribed spacer. The known and conserved sequence of 5.8S rRNA from several organisms predicts a characteristic pattern of restriction enzyme sites for 5.8S rDNA. Internal transcribed spacer rDNA was mapped with restriction enzymes, and the characteristic pattern was found near the midpoint of the internal transcribed spacer. This places the 5.8S rRNA sequence at or near the 5′ terminus of 32S pre-rRNA.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Arnheim N., Southern E. M. Heterogeneity of the ribosomal genes in mice and men. Cell. 1977 Jun;11(2):363–370. doi: 10.1016/0092-8674(77)90053-8. [DOI] [PubMed] [Google Scholar]
- Azad A. A., Deacon N. J. The 3'-terminal primary structure of five eukaryotic 18S rRNAs determined by the direct chemical method of sequencing. The highly conserved sequences include an invariant region complementary to eukaryotic 5S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4365–4376. doi: 10.1093/nar/8.19.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Cory S., Adams J. M. A very large repeating unit of mouse DNA containing the 18S, 28S and 5.8S rRNA genes. Cell. 1977 Aug;11(4):795–805. doi: 10.1016/0092-8674(77)90292-6. [DOI] [PubMed] [Google Scholar]
- Dabeva M. D., Dudov K. P., Hadjiolov A. A., Emanuilov I., Todorov B. N. Intranuclear maturation pathways of rat liver ribosomal ribonucleic acids. Biochem J. 1976 Dec 15;160(3):495–503. doi: 10.1042/bj1600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz E., Nakashima K., Shatkin A. J. Base-pairing in conserved 3' end of 18 S rRNA as determined by psoralen photoreaction and RNase sensitivity. J Biol Chem. 1980 Jun 10;255(11):4973–4975. [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dudov K. P., Dabeva M. D., Hadjiolov A. A., Todorov B. N. Processing and migration of ribosomal ribonculeic acids in the nucleolus and nucleoplasm of rat liver nuclei. Biochem J. 1978 May 1;171(2):375–383. doi: 10.1042/bj1710375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1979 Jan;6(1):r29–r44. doi: 10.1093/nar/6.1.419-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Soellner C., Scholz I. Characterization of a cloned ribosomal fragment from mouse which contains the 18S coding region and adjacent spacer sequences. Nucleic Acids Res. 1979 Apr;6(4):1351–1369. doi: 10.1093/nar/6.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov A. A., Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol. 1976;31(2):95–144. doi: 10.1016/0079-6107(78)90006-8. [DOI] [PubMed] [Google Scholar]
- Hamada H., Kominami R., Muramatsu M. 3'-terminal processing of ribosomal RNA precursors in mammalian cells. Nucleic Acids Res. 1980 Feb 25;8(4):889–903. [PMC free article] [PubMed] [Google Scholar]
- Kominami R., Hamada H., Fujii-Kuriyama Y., Muramatsu M. 5'-Terminal processing of ribosomal 28S RNA. Biochemistry. 1978 Sep 19;17(19):3965–3970. doi: 10.1021/bi00612a014. [DOI] [PubMed] [Google Scholar]
- Locker J. Analytical and preparative electrophoresis of RNA in agarose-urea. Anal Biochem. 1979 Oct 1;98(2):358–367. doi: 10.1016/0003-2697(79)90154-4. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J Mol Biol. 1980 Apr 25;138(4):873–878. doi: 10.1016/0022-2836(80)90070-4. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Robertson J. S. Demonstration of the "5-8 S" ribosomal sequence in HeLa cell ribosomal precursor RNA. J Mol Biol. 1974 Aug 5;87(2):227–235. doi: 10.1016/0022-2836(74)90145-4. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Salim M., Summers D. F. Maturation pathway for ribosomal RNA in the Hela cell nucleolus. Nat New Biol. 1972 May 3;237(70):5–9. doi: 10.1038/newbio237005a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y., Kominami R., Honjo T., Muramatsu M. Cloning and determination of a putative promoter region of a mouse ribosomal deoxyribonucleic acid fragment. Biochemistry. 1980 Aug 5;19(16):3780–3786. doi: 10.1021/bi00557a020. [DOI] [PubMed] [Google Scholar]
- Nazar R. N. Studies on the 5' termini of Novikoff ascites hepatoma ribosomal precursor RNA. Biochemistry. 1977 Jul 12;16(14):3215–3219. doi: 10.1021/bi00633a027. [DOI] [PubMed] [Google Scholar]
- Perry R. P. Processing of RNA. Annu Rev Biochem. 1976;45:605–629. doi: 10.1146/annurev.bi.45.070176.003133. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeier D. C., Tilghman S. M., Leder P. Purification and cloning of a mouse ribosomal gene fragment in coliphage lambda. Gene. 1977;2(3-4):173–191. doi: 10.1016/0378-1119(77)90016-6. [DOI] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Processing of 45 s nucleolar RNA. J Mol Biol. 1970 Jan 28;47(2):169–178. doi: 10.1016/0022-2836(70)90337-2. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of RNA: processing of HeLa ribosomal RNA. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2827–2831. doi: 10.1073/pnas.70.10.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winicov I. Alternate temporal order in ribosomal RNA maturation. J Mol Biol. 1976 Jan 15;100(2):141–155. doi: 10.1016/s0022-2836(76)80145-3. [DOI] [PubMed] [Google Scholar]
- Wolf S. F., Schlessinger D. Nuclear metabolism of ribosomal RNA in growing, methionine-limited, and ethionine-treated HeLa cells. Biochemistry. 1977 Jun 14;16(12):2783–2791. doi: 10.1021/bi00631a031. [DOI] [PubMed] [Google Scholar]