Abstract
Bacteria, the most abundant organisms on the planet, are outnumbered by a factor of 10 to 1 by phages that infect them. Faced with the rapid evolution and turnover of phage particles, bacteria have evolved various mechanisms to evade phage infection and killing, leading to an evolutionary arms-race. The extensive co-evolution of both phage and host has resulted in considerable diversity on the part of both bacterial and phage defensive and offensive strategies. Here, we discuss the unique and common features of phage resistance mechanisms and their role in global biodiversity. The commonalities between defense mechanisms suggest avenues for the discovery of novel such mechanisms based on their evolutionary traits.
Keywords: arms-race, phage, bacteria, evolution, resistance
Introduction
Phage-host relationships have been studied intensively since the early days of molecular biology. In the late 1970s, while viruses were found to be ubiquitous, it was assumed that they were present in relatively low numbers and that their effect on microbial communities was low [1]. With the increasing availability of new molecular techniques that allow studies of microbial communities without the need to culture them [2], it is now realized that viruses greatly outnumber bacteria in the ocean and other environments, with viral numbers (~107–108 ml−1) often tenfold larger than bacterial cell counts (~106 ml−1) [3–5]. Thus, bacteria are confronted with a constant threat of phage predation.
The Red Queen hypothesis (Box 1) posits that competitive environmental interactions, such as those displayed by hosts and parasites, will lead to continuous variation and selection towards adaptation of the host, and counter-adaptations on the side of the parasite. Arguably, nowhere is this evolutionary trend so pronounced as in phage-microbe interactions. This is due to the extremely rapid evolution and turnover of phage particles [6], causing acute pressure on microbial communities to evade infection and killing by phages. In fact, the arms-race between phage and bacteria is predicted to have had an impact on global nutrient cycling [7], on global climate [6, 7], on the evolution of the biosphere [8], and also on the evolution of virulence in human pathogens [9].
This review focuses on the evolution of three of the most well studied microbial defense mechanisms against phage: the restriction-modification system, the recently discovered CRISPR (clustered regularly interspersed palindromic repeats) loci together with their associated cas genes, and the abortive infection system (summarized in Table 1). We first describe these defense systems, as well as the counter-adaptations that evolved in the phage to allow escape from bacterial defense. Next, we discuss features that are common to many microbial defense systems, such as rapid evolution, tendency for lateral gene transfer (LGT), and the selfish nature of these systems. We elaborate on defense systems which have gained new functions in the host genome. Finally, the exciting hypothesis that many other prokaryotic defense systems are still yet to be discovered is discussed. Our review mainly focuses on the evolutionary angle of the phage-host arms-race; for deeper mechanistic descriptions of phage resistance systems we refer the readers to an excellent recent review published elsewhere [11]. In addition, the discussion here is restricted to active defense mechanisms; passive host adaptations, such as mutations at the phage receptors, are not discussed here.
Table 1.
A summary of the major defense mechanisms described in the manuscript.
| Name | Mechanism | Phylogenetic breadth |
|---|---|---|
| Restriction-modification | The restriction enzyme cleaves specific patterns in the incoming foreign DNA, while the modification enzyme protects host DNA from cleavage by unique biochemical modification. | Appear in ~90% of all sequenced prokaryotic genomes [13] |
| CRISPR/Cas | Fragments of phage DNA are integrated into CRISPR loci, which are then transcribed and processed into short non-coding RNAs. These RNAs, along with the associated Cas proteins, guide the way to interfere with the phage nucleic acids, in a yet unknown mechanism. | Appear in ~40% and ~90% of all sequenced bacterial and archaeal genomes, respectively [27]. The distribution of CRISPR/Cas-bearing species across the phylogenetic tree of life is highly patchy [38, 59]. |
| Abortive infection | Premature cellular death occurs upon phage entry, blocking the expansion of the phage to neighboring cells. Notably, abortive infection systems include a large collection of mechanisms with little or no known evolutionary relationship, apart from a very similar phenotype. | Currently known abortive infection systems display a sporadic phylogenetic distribution in Gamma-proteobacteria and in Firmicutes [44], yet recent reports suggest that some systems may have an even broader phylogenetic range [54, 111]. |
Restriction-modification systems: Degradation of foreign DNA
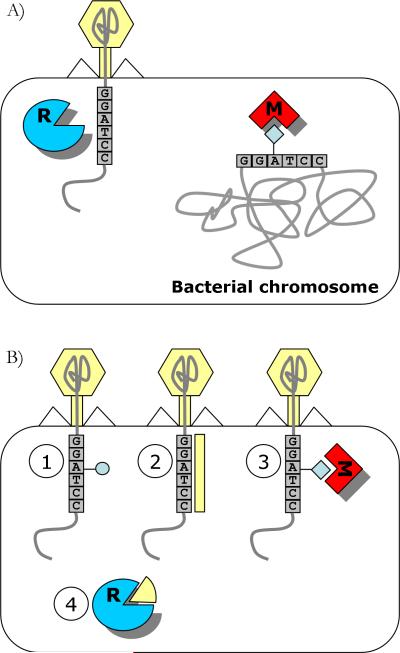
Arguably, the most well studied phage defense mechanism is the restriction-modification (RM) system [12], which is present in over 90% of sequenced bacterial and archaeal genomes [13]. This system is composed of two activities: one that restricts incoming foreign genetic material, and one that protects host genetic material from restriction (Fig. 1A). Both activities are mediated by recognition of a specific DNA sequence, on average 4–8 base-pairs (bp) long. Protection is normally conferred by modification (usually methylation) of specific bases in this recognition sequence in the host genome. Accordingly, all non-methylated DNA recognition sequences are recognized as foreign and are cleaved. Genetically, the minimal composition of RM systems consists of a methyltransferase (MTase) gene that perform the defense activity and a restriction endonuclease (REase) gene that performs the foreign restriction activity, and since the REases undergo rapid evolution, it is often the presence of a MTase that serves as the basis for identification of RM systems in newly sequenced genomes [14].
Figure 1. The restriction-modification defense system.
A: A general illustration of function, exemplified by type II R–M enzymes. B: Examples of strategies employed by phage to evade restriction. (1) Incorporation of unusual bases protects from restriction [15]; (2) Masking of the restriction sites by phage proteins[112]; (3) Stimulation of MTase activity causes the phage DNA to be protected; (4) Neutralization of REase by phage proteins that mimic DNA [113].
Phages have evolved to evade the ubiquitous RM systems in a variety of ways (Fig. 1B). Some phages have acquired an MTase, or stimulate the host MTase so that it confers protection to the phage genome [15]. Other phages code for proteins that target and shut down the REase. One interesting example is the Ocr protein of phage T7, which blocks the active site of some REases by mimicking 24 bp of bent B-form DNA [16]. Alternatively, some phages incorporate unusual bases in their genomes, thus throwing off the REase. For example, some Bacillus subtilis phages replace thymine with 5-hydroxymethyluracil [15]. These phages code a protein that then further inhibits the host protein uracil-DNA glycosalyse from cleaving uracil bases from the phage DNA [17, 18]. Interestingly, this inhibitory protein is also a DNA mimic (reviewed in [19]). The T-even phages T2, T4, and T6 also contain unusual bases in their genomes and may further post-synthetically glycosylate their DNA to avoid REase restriction [15]. Yet another evasion mechanism that has been reported is alteration of the restriction recognition sites. For example, some phages employ “palindrome avoidance”: since Type II REases often recognize symmetrical (palindromic) sequences, some phages tend to avoid containing such sites in their genome [20].
The well-studied model organisms Escherichia coli and T4 phage display a fascinating example of a co-evolutionary arms-race [11]. The battle purportedly begins with the T4 phage genome, which contains the modified base hydroxymethylcytosine instead of cytosine [15]. To counter-attack the phage, E. coli K-12 possesses a unique form of REase, the McrBC enzyme, which cleaves only modified DNA substrates (such as that of T4) [21]. The T4 phage rises to the challenge by glucosylating its genome, and is thus impervious to McrBC [15]. However, E. coli cT596 encodes the GmrS-GmrD system that can also restrict glucosylated DNA [22, 23]. In continuation of the battle, some T4 phages encode the IPI protein, which in its processed form (IPI*) disables the GmrS-GmrD system [24]. Evidently, the battle is far from an end, since bacterial strains have been found to overcome the IPI* protein of T4 [24]. In fact, it is likely that many bacterial defense systems, coupled with their cognate phage evasion strategies, have undergone similar attack and counter-attack cycles, where a change on one side selects for changes that can overcome the opponent.
CRISPR/Cas: Acquired adaptive immunity
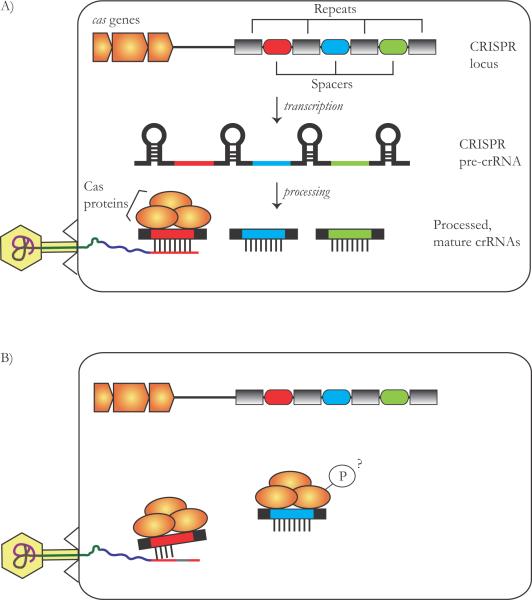
The existence of a unique nucleotide arrangement in E. coli, comprised of a cluster of direct repeats interspersed with variable sequences (termed spacers; both around 30 bp long), was first described in 1987 [25]. These clusters, together with several CRISPR-associated (cas) genes (Fig. 2A), were later found to exist in ~40% of sequenced bacterial genomes and ~ 90% of archaeal genomes [26–28]. The first inkling that this system comprises a phage-defense mechanism arose when spacer sequences were found to be highly similar to DNA from foreign origin, i.e. from phage, plasmid or transposon DNA [29–31]. In 2007, experimental evidence was presented that this system indeed confers resistance to phage infection [32]. This led to the striking insight that similar to higher eukaryotes, bacteria and archaea possess acquired (and inherited) immunity [27, 33–35]. Following phage infection, it has been found that a small portion of bacterial cells integrated new spacers identical to the phage genomic sequence (termed proto-spacer), resulting in CRISPR-mediated phage resistance [32, 36, 37]. Further experiments have shown that the CRISPR locus is transcribed into a single RNA transcript, which is then further cleaved by the Cas proteins to generate smaller CRISPR RNA (crRNA) units, each including one targeting spacer [37, 38]. These units then interfere with the incoming foreign genetic material by complementary base-pairing with either DNA [36, 37] or RNA [39] from the foreign element (Fig. 2A).
Figure 2. The CRISPR/Cas system.
A: Mechanism of action: transcription from the repeat-spacer CRISPR locus generates a long non-coding RNA, with repeats that may sometimes assume a secondary structure. Cleavage of the repeat sequences by the Cas proteins generates crRNAs that target the phage DNA or RNA, and interfere with phage infection. B: Phages can evade CRISPR interference by mutation or recombination of the targeted proto-spacer sequence. Another putative evasion mechanism is phosphorylation of the Cas proteins. This remains, however, to be verified.
While the study of CRISPR/Cas is still in its infancy, examples of phages that are resistant to CRISPR/Cas interference have nevertheless been noted (Fig. 2B). Following the first round of infection and acquisition of novel spacers by the bacterial CRISPR, phages which have mutated, recombined or lost their proto-spacer target sequence in the second round of infection are now resistant to CRISPR [32, 40–42]. It also seems likely that phages have evolved mechanisms that directly target the CRISPR/Cas machinery. To date, only vague hints of such mechanisms exist. For example, it has been shown that one of the proteins encoded by the T7 phage phosporylates the CasB protein [43]. It remains to be shown whether this feat of the phage affects CRISPR/Cas functioning.
Abortive infection: Cellular suicide
If a phage has successfully entered the host cell and avoided restriction by the host RM systems and by CRISPR, it proceeds to develop, replicate, and release its progeny. Abortive infection (Abi) is a collective term describing host mechanisms that interrupt with phage development at different stages of phage transcription, genome replication, and phage packaging [44]. Abi-mediated resistance leads to death (“suicide”) of the cell, and is thought to occur since corruption of host functions has already been initiated by the phage. However, this death confers an advantage to surrounding bacterial cells since it confines the infection to the sacrificed cell and prevents the spread of infectious particles.
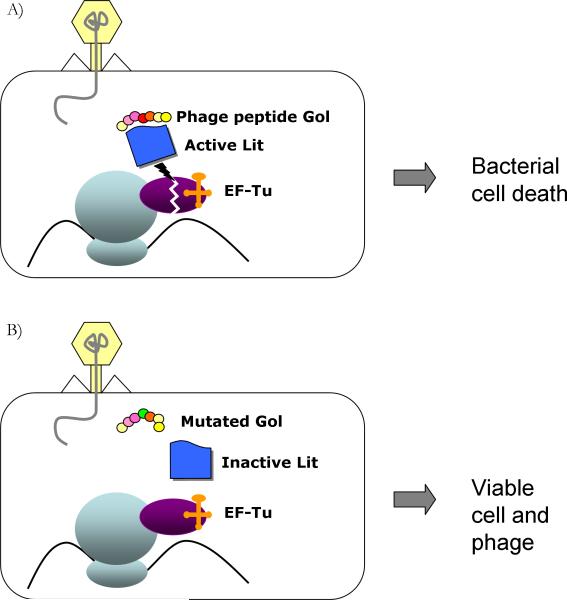
Although several Abi systems have been discovered, the majority of them encoded on plasmids, the mechanism by which they operate remains largely unknown. Frequently, abortive infection is mediated by a single gene encoded on a plasmid or on a prophage, which displays little or no homology to any known proteins. Some Abi genes have been shown to target phage genes involved in DNA replication [45, 46], and others have been shown to target the host translation apparatus [47, 48] . For instance, in E. coli K-12 the Lit protease, encoded by the defective e14 prophage, is only activated in the presence of a short polypeptide called Gol, which is produced by the T4 phage. Once active, this protease cleaves the translation elongation factor Ef-Tu, thus leading to translational arrest and cell death (Fig. 3A). Similarly, the Prr protein (also part of a defective prophage in E. coli cT196) cleaves tRNALys in response to the presence of the T4 peptide Stp [49]. Mutations in these phage peptides suppress activation of Lit or Prr and rescue the infecting phage [47, 49] (Fig. 3B).
Figure 3. The abortive infection system.
A: Illustration of the mechanism of the E. coli K-12 abortive infection Lit system. When the T4 phage peptide Gol is synthesized, it binds and activates the bacterial (prophage-encoded) Lit protein, which then cleaves the elongation factor EF-Tu. This leads to the arrest of protein synthesis and to bacterial cell death, with the phage trapped inside. B: A mutation at the Gol polypeptide reduces the activation of Lit, and rescues the phage.
Since Abi genes have a toxic effect on their host, they are under tight regulation [50]. In fact, many similarities can be drawn between Abi systems (and also RM systems), and toxin-antitoxin (TA) systems. TA systems are composed of a stable toxin and an unstable antitoxin [51]. Normally, the antitoxin binds and inhibits the toxin. However, a decrease in the levels of the unstable antitoxin activates the toxin and leads to growth arrest or cell death. It has been shown that this situation occurs in response to phage infection in two known TA modules , mazEF and hok-sok, which can thus cause abortive infection [52, 53]. Moreover, the AbiQ system was recently found to function as a protein-RNA TA pair [54], albeit how exactly this system interferes with phage replication remains unknown. Thus, it appears that some TA systems may be a subtype of Abi systems. Interestingly, RM systems may also be viewed as TA systems, since loss of the MTase (which is often unstable, similar to the antitoxin) results in a toxic effect of the REase [55, 56].
Commonalities among phage defense systems
One of the most striking features common to all phage defense systems is their high genetic variability, which occurs as a consequence of the co-evolutionary arms-race with phages [57]. This is manifested in the enormous numbers and types of RM systems [12, 58], of CRISPR subtypes [59, 60], and in the variety of abortive infection systems [44]. Moreover, the gene sequences of these systems often display a high evolutionary rate ([61–63], Stern and Sorek, unpublished data). This variability is most easily exemplified in the diversity of RM systems: Four inherently different groups of RM systems exist, classified as Type I–IV based on their subunit composition, mode of action, and cofactor requirement ([reviewed in 12]). Each type of RM system includes an assortment of restriction enzymes that recognize different recognition sequences. For instance, nearly 4000 Type II enzymes are known of today, which are further divided into 11 overlapping sub-classes [13, 64]. Interestingly, Type II REase sequences have frequently been found to be ORFan sequences, i.e. have no significant similarity to any other protein [65, 66]. Initially, this was seen as evidence for convergent evolution [67]. However, crystallographic studies have shown structural similarity among Type II REases, most likely representing a rapid evolutionary rate since their divergence from a common ancestor [68].
Another trait common to all defense systems presented here is their propensity to undergo LGT, sometimes between distantly related prokaryotes [69–72]. Often these systems reside on plasmids, on prophages, on genomic islands of foreign origin, or are linked to transposase genes. This mobility allows rapid acquisition and dissemination of new systems to counteract the invading phage, thus contributing to the extensive co-evolution with the prokaryotic parasites. However, there is an interesting alternative view, not necessarily mutually exclusive, which may explain the high variability and mobility of phage defense systems. Accordingly, these systems are selfish elements, as will be elaborated in the next section.
Defense systems: A burden or a benefit?
In this section, the advantages and disadvantages of microbial immune systems are reviewed. Such a discussion is incomplete without examining the pros and cons of the phage-host relationship. Evidently, for a single cell, phage predation and killing of the host are a strong disadvantage. However, phages may also contribute directly to host fitness by supplying a pool of new and possibly beneficial genes [73]. As such, phages serve as vessels of LGT, which is one of the most important forces in microbial evolution (Box 2).
What is the associated cost of encoding phage defense mechanisms? An obvious primary cost is the energy cost linked to carrying additional genetic cargo. The fact that often, only a small portion of a given bacterial population bears a plasmid or a prophage with a given defense mechanism may be seen as evidence for this cost. A second intriguing cost of encoding a phage defense mechanism is the risk of autoimmunity. Autoimmunity, classically defined in mammalian immune systems, is the failure of the immune system to recognize what is self and what is foreign, resulting in an immune response against self. In RM systems, this is a clear danger since the restriction enzyme is often more stable than the protecting methylase [74]. Abortive infection mechanisms are equally lethal, since errant function of the Abi gene will lead to cell death. Finally, the CRISPR system is also not “immune” to errors, and the frequent acquisition of self genetic material also leads to spacers that target the self-genome, potentially incurring autoimmunity [75] (see also [76]).
Paradoxically phages themselves often bear anti-phage defense systems, enabling their acquisition by host cells. To cite a few examples, the HindIII RM gene complex was found on a cryptic prophage in the Haeomphilus influenzae genome [77], the abiN abortive infection gene is encoded on a prophage in Lactococcus lactis subsp. cremoris S114, and even a CRISPR array was found within a Clostridium difficile prophage [78]. These results initially seem counterintuitive, since what benefit is there for the phage to carry such systems? One possible explanation is that this allows superinfection exclusion, thus preventing other phages from infecting an already infected cell [11, 78, 79]. However, this phenomenon has also been viewed as evidence for the selfish nature of phage defense mechanisms.
The behavior of defense mechanisms as selfish mobile elements has been extensively discussed for the case of RM systems [55, 80], but is also applicable for many Abi systems that operate as TA systems, which also have “selfish” properties [51, 81]. The main lines of evidence in favor of this view is that (a) RM systems destroy any other invading RM system, (b) any attempt to lose the RM system will result in the death of the host, and (c) RM systems are prone to extensive mobility, and are often associated with plasmids, phages, transposons, and integrons [55]. These characteristics of the RM systems lead to an increase of their relative frequency in the bacterial population. Thus, according to this hypothesis the defense incurred by RM systems on host cells is a mere by-product of the fact that RM systems defend themselves.
Exaptations: Alternative functions of defense systems
Intriguingly, a number of anti-phage defense systems have evolved to gain a distinct function in cellular regulation that is independent of phage restriction. Such evolutionary events were coined “exaptations” [92], a term used to describe the use of a biological structure or function for a purpose other than that for which it initially evolved. For instance, several RM systems have lost their REase activity, leaving an orphan MTase that can now take part in epigenetic modifications. The Dam methylase in E. coli and the CcrM methylase in Caulobacter crescentus, both of which have originated from RM systems [93], are two such examples. Methylation by Dam has been linked to several important regulatory processes such as mismatch repair by the MutHLS complex, binding of the replication initiation complex to methylated OriC, and regulation of bacterial pathogenicity [93]. On the other hand, the CcrM methylase has been shown to affect the cell cycle in alpha proteobacteria that encode this gene [93].
Another interesting example of exaptation of RM systems is evident in phase-variable Type III RM systems [94], whose genes can be reversibly inactivated due to tandem repeat tracts in their sequences. These repeats initiate a mechanism called slipped-strand mispairing, leading to a change in the number of repeats after DNA replication and possible frame-shift mutations [94]. Several regulatory roles have been suggested for phase-variable RM systems: (a) to allow regulated removal of the barrier against foreign DNA, thus allowing potentially beneficial uptake of DNA [95, 96], (b) autolytic self-DNA degradation, or “bacterial suicide” [97–99], further discussed below, and (c) epigenetic gene regulation via differential methylation of the genome [100]. The latter phenomenon, which allows switching different genes on and off, has been linked to pathogenicity of bacterial species by allowing colonization, immune evasion and adaptation to novel environments [94].
TA modules, which include some abortive infection systems, are also known to participate in a variety of other cellular processes. For example, one of the most studied TA loci mazEF, aborts translation by cleaving mRNA molecules in response to different stress signals [101, 102], one of which is phage infection [52]. An ongoing debate exists whether this action is reversible or not: reversible effects have been attributed to bacteriostatic effects, which allow reduced growth rate of each cell during nutritional stress [101, 102]. On the other hand, irreversible effects of mazEF have been attributed to programmed cell death that occurs in a subpopulation of cells, permitting the survival of the population of a whole [103]. Interestingly, in Myxococcus xanthus it was shown that the toxin MazF exists without the antitoxin MazE, and has adopted a key transcriptional regulator as an alternative antitoxin [104]. MazF mediates programmed cell death during multicellular development of this organism. To summarize, these accounts exemplify the broad evolutionary diversification of different microbial defense mechanisms, and their potential to cross boundaries from phage-encoded mechanisms, to anti-phage systems, to regulatory host mechanisms.
Conclusions
Despite our growing understanding of microbial immunity, much still remains obscure. Do defense systems work separately or in unison? What is the cost of each system? Which phages are targeted by which systems? For instance, while most characterized defense systems work against double-stranded DNA phages, RNA viruses might also be abundant [105]. Defense systems that target such viruses are yet to be discovered.
The recently discovered CRISPR system epitomizes our incomplete understanding of the complexity of bacterial defense systems. The discovery that almost half of all prokaryotes possess acquired inherited immunity came as a surprise to the scientific community, given our initial tendency to view prokaryotes as less complex organisms. However, since it is now realized that prokaryotes are faced with a constant threat of predation, it is becoming clearer that the microbial immune system must be highly complex. Nevertheless, the CRISPR system and the Abi systems have a highly sporadic distribution across the prokaryotic phylogeny (Table 1). Furthermore, while RM systems are present in ~90% of prokaryotic species [13], this system is far from tight-proof, since it has been shown that phages have a non-negligible probability of escaping the REase and being methylated by the MTase. [106]. Thus, all three mechanisms reviewed here are either somewhat leaky, or span only part of the phylogenetic breadth of bacteria and archaea. Given the intensive arms-race between bacteria and phage, it seems probable that there are yet other unknown defense mechanisms waiting to be revealed.
So how does one set about the search for novel prokaryotic defense systems, and how does one learn more about existing ones? It is likely that many solutions to these questions will be made possible thanks to advances in sequencings technologies, such as whole-transcriptome studies and metagenomics. For instance, a seminal work by Andersson and Banfield [42] reconstructed viral and host population genomes from community genomic data, and showed how CRISPR spacers correlate with location of coexisting viruses and hosts. In addition, controlled experimental evolution studies, such as those performed in Pseudomonas fluorescens and its phage, enable a direct survey of the changes that occur in the phage and host population with and without infection [107–109]. These studies are also likely to shed light on current defense systems and possibly discover novel ones.
More specifically, the search for novel defense mechanisms may be based on the well established common properties of both eukaryotic and prokaryotic immune systems: their high rate of evolution, and their tendency to undergo horizontal gene transfer. Thus, it may be that a reservoir of novel defense mechanisms lies in the most variable regions of bacterial and archaeal genomes, known as genomic islands [110]. A strategy that focuses on such islands to search for novel phage resistance mechanisms might lead, in the future, to surprising discoveries.
Box 1. The Red Queen Hypothesis.
The Red Queen hypothesis was originally proposed by Leigh Van Valen (1973) [10], and is also termed the evolutionary arms-race hypothesis. As the Red Queen tells Alice in Lewis Carroll's “Through the Looking-Glass”: “Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else, you must run at least twice as fast as that!” The original theory proposed that in tight co-evolutionary interactions such as those in a prey-predator relationship, changes (e.g. running faster) on the one side may lead to near extinction of the other side. The only way the second side can maintain its fitness is by counter-adaptation (running even faster). This will lead to an uneasy balance between prey and predator, where species have to constantly evolve in order to stay at the same fitness level. The metaphor of an evolutionary arms-race has been found to be relevant for many biological processes, but nowhere is this metaphor as apt as in host-parasite relationships.
Box 2. Impact of defense systems on LGT.
LGT is the process whereby genetic material is incorporated from a non-parental organism. There are three major mechanisms of LGT: (a) transduction by phages; (b) transformation of naked DNA; and (c) bacterial conjugation, which involves transfer of DNA via direct connections generated between a pair of LGT is recognized as one of the most important mechanisms of genetic innovation in both bacteria and archaea [82–84]. In fact, it is now realized that much of the genomic diversity in prokaryotes is a consequence of LGT, rather than allelic differences at the same loci [85, 86]. However, when viewed from the point of view of a single cell, LGT has a negligible chance of contributing to the fitness of the organism. Evidently, transduction by lytic phages may severely compromise the survival of a bacterial colony. Plasmids and transposons may also decrease the fitness of an organism by integrating into crucial regions of the genomes, by adding an energetic cost involved with replication of excess DNA, and by expressing harmful genes [e.g. 87, 88, 89]. Finally, even if a new beneficial gene sequence is obtained, there is only a small probability that it will integrate into the current cellular network of an organism without causing deleterious effects [90, 91]. Thus, defense mechanisms such as those described here are crucial for maintaining the genetic identity of organisms, and protecting it against the constant bombardment of potentially detrimental foreign DNA.
Acknowledgements
R.S. is an EMBO Young Investigator. He was supported, in part, by the ISF-FIRST program (grant 1615/09), NIH R01AI082376-01, ERC-StG, the Wolfson Family Trust miRNA research program, the Minerva foundation, and the Yeda-Sela Center for basic research. A.S. was supported by the Clore postdoctoral fellowship.
Abbreviations
- Abi
Abortive infection
- cas
CRISPR associated sequence
- CRISPR
clustered regularly interspersed palindromic repeats
- crRNA
CRISPR RNA
- LGT
lateral gene transfer
- MTase
methyltransferase
- REase
restriction endonuclease
- RM
restriction modification
- TA
toxin-antitoxin
References
- 1.Torrella F, Morita RY. Evidence by electron micrographs for a high incidence of bacteriophage particles in the waters of Yaquina Bay, oregon: ecological and taxonomical implications. Appl Environ Microbiol. 1979;37:774–8. doi: 10.1128/aem.37.4.774-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–52. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 3.Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–8. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 4.Chibani-Chennoufi S, Bruttin A, Dillmann ML, Brussow H. Phage-host interaction: an ecological perspective. J Bacteriol. 2004;186:3677. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 7.Suttle CA. Marine viruses - major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 8.Comeau AM, Krisch HM. War is peace - dispatches from the bacterial and phage killing fields. Curr Opin Microbiol. 2005;8:488–94. doi: 10.1016/j.mib.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Valen L. A new evolutionary law. Evol Theor. 1973;1:1–30. [Google Scholar]
- 11.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 12.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–72. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE - a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–6. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE - restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruger DH, Bickle TA. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983;47:345–60. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay PK, Studier FW, Hamilton DL, Yuan R. Inhibition of the type I restriction-modification enzymes EcoB and EcoK by the gene 0.3 protein of bacteriophage T7. J Mol Biol. 1985;182:567–78. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor gene of bacteriophage PBS2 encodes a binding protein specific for uracil-DNA glycosylase. J Biol Chem. 1989;264:1163–71. [PubMed] [Google Scholar]
- 18.Wang Z, Mosbaugh DW. Uracil-DNA glycosylase inhibitor of bacteriophage PBS2: cloning and effects of expression of the inhibitor gene in Escherichia coli. J Bacteriol. 1988;170:1082–91. doi: 10.1128/jb.170.3.1082-1091.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair (Amst) 2005;4:1410–20. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Rocha EP, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–58. doi: 10.1101/gr.gr-1531rr. [DOI] [PubMed] [Google Scholar]
- 21.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci USA. 1986;83:9070–4. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bair CL, Rifat D, Black LW. Exclusion of glucosyl-hydroxymethylcytosine DNA containing bacteriophages is overcome by the injected protein inhibitor IPI*. J Mol Biol. 2007;366:779–89. doi: 10.1016/j.jmb.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J Mol Biol. 2007;366:768–78. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rifat D, Wright NT, Varney KM, Weber DJ, et al. Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J Mol Biol. 2008;375:720–34. doi: 10.1016/j.jmb.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishino Y, Shinagawa H, Makino K, Amemura M, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 27.Sorek R, Kunin V, Hugenholtz P. CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 28.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–61. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 30.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 31.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 32.Barrangou R, Fremaux C, Deveau H, Richards M, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 33.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 34.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Oost J, Jore MM, Westra ER, Lundgren M, et al. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–7. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 36.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouns SJ, Jore MM, Lundgren M, Westra ER, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hale CR, Zhao P, Olson S, Duff MO, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deveau H, Barrangou R, Garneau JE, Labonte J, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–50. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- 43.Qimron U, Tabor S, Richardson CC. New Details about Bacteriophage T7-Host Interactions. Microbe. 2010;5:117–20. [Google Scholar]
- 44.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–9. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Durmaz E, Klaenhammer TR. Abortive phage resistance mechanism AbiZ speeds the lysis clock to cause premature lysis of phage-infected Lactococcus lactis. J Bacteriol. 2007;189:1417. doi: 10.1128/JB.00904-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bidnenko E, Ehrlich D, Chopin MC. Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J Bacteriol. 1995;177:3824. doi: 10.1128/jb.177.13.3824-3829.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georgiou T, Yu YTN, Ekunwe S, Buttner MJ, et al. Specific peptide-activated proteolytic cleavage of Escherichia coli elongation factor Tu. Proc Natl Acad Sci USA. 1998;95:2891. doi: 10.1073/pnas.95.6.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morad I, Chapman-Shimshoni D, Amitsur M, Kaufmann G. Functional expression and properties of the tRNA (Lys)-specific core anticodon nuclease encoded by Escherichia coli prrC. J Biol Chem. 1993;268:26842. [PubMed] [Google Scholar]
- 49.Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol Microbiol. 1995;15:415–20. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 50.Blower TR, Fineran PC, Johnson MJ, Toth IK, et al. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol. 2009;191:6029–39. doi: 10.1128/JB.00720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–82. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 52.Hazan R, Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol Genet Genomics. 2004;272:227–34. doi: 10.1007/s00438-004-1048-y. [DOI] [PubMed] [Google Scholar]
- 53.Pecota DC, Wood TK. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA. 2009;106:894–9. doi: 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–56. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–9. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- 57.Hoskisson PA, Smith MC. Hypervariation and phase variation in the bacteriophage 'resistome'. Curr Opin Microbiol. 2007;10:396–400. doi: 10.1016/j.mib.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:3552. doi: 10.1093/nar/gkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharp PM, Kelleher JE, Daniel AS, Cowan GM, et al. Roles of selection and recombination in the evolution of type I restriction-modification systems in enterobacteria. Proc Natl Acad Sci USA. 1992;89:9836–40. doi: 10.1073/pnas.89.20.9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray NE, Daniel AS, Cowan GM, Sharp PM. Conservation of motifs within the unusually variable polypeptide sequences of type I restriction and modification enzymes. Mol Microbiol. 1993;9:133–43. doi: 10.1111/j.1365-2958.1993.tb01675.x. [DOI] [PubMed] [Google Scholar]
- 63.Zheng Y, Roberts RJ, Kasif S. Identification of genes with fast-evolving regions in microbial genomes. Nucleic Acids Res. 2004;32:6347–57. doi: 10.1093/nar/gkh935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kroger M, Hobom G, Schutte H, Mayer H. Eight new restriction endonucleases from Herpetosiphon giganteus-divergent evolution in a family of enzymes. Nucleic Acids Res. 1984;12:3127. doi: 10.1093/nar/12.7.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mullings R, Bennett SP, Brown NL. Investigation of sequence homology in a group of type-II restriction/modification isoschizomers. Gene. 1988;74:245–51. doi: 10.1016/0378-1119(88)90297-1. [DOI] [PubMed] [Google Scholar]
- 67.Wilson G, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 68.Venclovas C, Timinskas A, Siksnys V. Five-stranded beta-sheet sandwiched with two alpha-helices: a structural link between restriction endonucleases EcoRI and EcoRV. Proteins. 1994;20:279–82. doi: 10.1002/prot.340200308. [DOI] [PubMed] [Google Scholar]
- 69.Nelson KE, Clayton RA, Gill SR, Gwinn ML, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–9. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 70.Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J Mol Evol. 1996;42:91–6. doi: 10.1007/BF02198833. [DOI] [PubMed] [Google Scholar]
- 71.Haaber J, Moineau S, Hammer K. Activation and transfer of the chromosomal phage resistance mechanism AbiV in Lactococcus lactis. Appl Environ Microbiol. 2009;75:3358–61. doi: 10.1128/AEM.02538-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 73.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–87. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 74.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–9. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 75.Stern A, Keren L, Wurtzel O, Amitai G, et al. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–40. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–71. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hendrix RW, Smith MC, Burns RN, Ford ME, et al. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–7. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sebaihia M, Wren BW, Mullany P, Fairweather NF, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–86. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 79.Lu MJ, Henning U. Superinfection exclusion by T-even-type coliphages. Trends Microbiol. 1994;2:137–9. doi: 10.1016/0966-842x(94)90601-7. [DOI] [PubMed] [Google Scholar]
- 80.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci USA. 1998;95:6442–7. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–38. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 83.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–42. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA. 2008;105:10039–44. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitaker RJ, Banfield JF. Population genomics in natural microbial communities. Trends Ecol Evol. 2006;21:508–16. doi: 10.1016/j.tree.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 86.Medini D, Serruto D, Parkhill J, Relman DA, et al. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–30. doi: 10.1038/nrmicro1901. [DOI] [PubMed] [Google Scholar]
- 87.Modi RI, Adams J. Coevolution in bacterial-plasmid populations. Evolution. 1991;45:656–67. doi: 10.1111/j.1558-5646.1991.tb04336.x. [DOI] [PubMed] [Google Scholar]
- 88.Zund P, Lebek G. Generation time-prolonging R plasmids: correlation between increases in the generation time of Escherichia coli caused by R plasmids and their molecular size. Plasmid. 1980;3:65–9. doi: 10.1016/s0147-619x(80)90034-7. [DOI] [PubMed] [Google Scholar]
- 89.Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–2. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 90.Kedzierska B, Glinkowska M, Iwanicki A, Obuchowski M, et al. Toxicity of the bacteriophage lambda cII gene product to Escherichia coli arises from inhibition of host cell DNA replication. Virology. 2003;313:622–8. doi: 10.1016/s0042-6822(03)00376-3. [DOI] [PubMed] [Google Scholar]
- 91.Lercher MJ, Pal C. Integration of horizontally transferred genes into regulatory interaction networks takes many million years. Mol Biol Evol. 2008;25:559–67. doi: 10.1093/molbev/msm283. [DOI] [PubMed] [Google Scholar]
- 92.Brosius J, Gould SJ. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc Natl Acad Sci USA. 1992;89:10706–10. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marinus MG, Casadesus J. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev. 2009;33:488–503. doi: 10.1111/j.1574-6976.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, et al. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci USA. 2005;102:5547–51. doi: 10.1073/pnas.0501169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ando T, Xu Q, Torres M, Kusugami K, et al. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol Microbiol. 2000;37:1052–65. doi: 10.1046/j.1365-2958.2000.02049.x. [DOI] [PubMed] [Google Scholar]
- 96.Donahue JP, Israel DA, Peek RM, Blaser MJ, et al. Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol Microbiol. 2000;37:1066–74. doi: 10.1046/j.1365-2958.2000.02036.x. [DOI] [PubMed] [Google Scholar]
- 97.Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci USA. 1998;95:13923–8. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27:1091–8. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 99.Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol. 2006;59:376–85. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- 100.Seib KL, Peak IR, Jennings MP. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol Med Microbiol. 2002;32:159–65. doi: 10.1111/j.1574-695X.2002.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 101.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–10. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 102.Christensen SK, Pedersen K, Hansen FG, Gerdes K. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–19. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 103.Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 105.Lang AS, Rise ML, Culley AI, Steward GF. RNA viruses in the sea. FEMS Microbiol Rev. 2009;33:295–323. doi: 10.1111/j.1574-6976.2008.00132.x. [DOI] [PubMed] [Google Scholar]
- 106.Korona R, Levin BR. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution. 1993;47:556–75. doi: 10.1111/j.1558-5646.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- 107.Paterson S, Vogwill T, Buckling A, Benmayor R, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–8. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buckling A, Wei Y, Massey RC, Brockhurst MA, et al. Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc Biol Sci. 2006;273:45–9. doi: 10.1098/rspb.2005.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wichman HA, Badgett MR, Scott LA, Boulianne CM, et al. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–4. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 110.Juhas M, van der Meer JR, Gaillard M, Harding RM, et al. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev. 2009;33:376–93. doi: 10.1111/j.1574-6976.2008.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–76. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iida S, Streiff MB, Bickle TA, Arber W. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA- phages. Virology. 1987;157:156–66. doi: 10.1016/0042-6822(87)90324-2. [DOI] [PubMed] [Google Scholar]
- 113.Dryden DT, Tock MR. DNA mimicry by proteins. Biochem Soc Trans. 2006;34:317–9. doi: 10.1042/BST20060317. [DOI] [PubMed] [Google Scholar]