Abstract
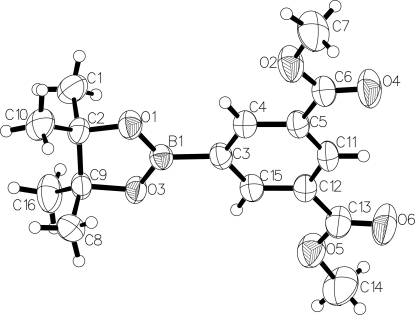
The title compound, C16H21BO6, has has approximate C 2 symmetry, but no crystallographically imposed molecular symmetry. In the crystal, molecules are packed into parallel columns along the a axis. Short intermolecular C—H⋯O contacts stabilize the crystal packing.
Related literature
For the synthesis of organoboronic esters, see: Kikuchi et al. (2008 ▶). For the synthesis of the title compound, see: Coventry et al. (2005 ▶).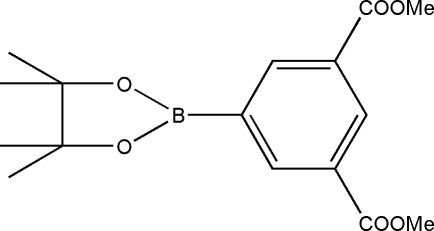
Experimental
Crystal data
C16H21BO6
M r = 320.14
Orthorhombic,
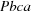
a = 7.2163 (2) Å
b = 20.9627 (4) Å
c = 22.4624 (4) Å
V = 3397.96 (13) Å3
Z = 8
Mo Kα radiation
μ = 0.09 mm−1
T = 296 K
1.00 × 0.40 × 0.20 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2005 ▶) T min = 0.766, T max = 0.982
22413 measured reflections
3864 independent reflections
2177 reflections with I > 2σ(I)
R int = 0.043
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.142
S = 1.01
3864 reflections
215 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.14 e Å−3
Data collection: APEX2 (Bruker, 2005 ▶); cell refinement: SAINT (Bruker, 2005 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811054559/bt5730sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811054559/bt5730Isup4.hkl
Supplementary material file. DOI: 10.1107/S1600536811054559/bt5730Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16C⋯O4i | 0.96 | 2.53 | 3.381 (3) | 148 |
Symmetry code: (i) 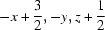 .
.
supplementary crystallographic information
Comment
Arylboronic acids and arylborates are the key reagents in Suzuki-Miyaura coupling reaction. Transition metal-catalyzed C—H borylation of arenes is a direct, economical and environmentally kind method to synthesize organoboronic esters and is widely studied (Kikuchi et al., 2008). Recently, we have synthesized the title compound by direct Ir-catalyzed borylation and determined its X-ray structure.
The two meta-position methoxycarbonyl are almost co-planar with the benzene ring. The C—B bond is 1.556 (2) Å. Because the borolane ring adopts a somewhat twist conformation and C2, C9 atoms displace to opposite sides of the BO2 plane, the title molecule has no crystallographic symmetry.
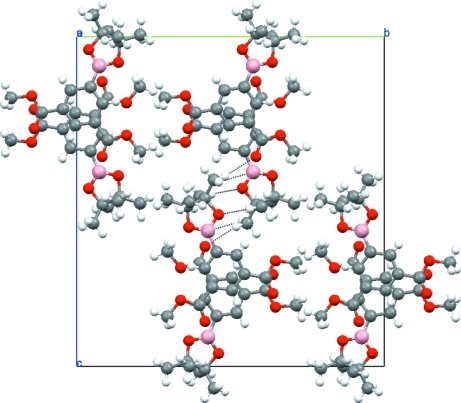
The molecules pack into columns along the a- axis uniformly and exhibit paralled patterns. Besides, there are some short intermolecular O···H (for example, O3···H8a, O4···H16c distances are 2.636 Å, 2.530 Å, respectively). We believe that these short intermolecular contacts are helpful for stabilization of the molecular packing in crystals.
Experimental
The title compound was synthesized via Ir-catalyzed borylation (Coventry et al., 2005) of diethyl isophthalate. Catalyst precursor [Ir(COD)]Cl]2 (60 mg) and ligand dtbpy (4,4'-di-tert-bultyl-2,2'-dipyridyl) (120 mg) with a small amount of B2pin2 (pin=O2C2Me4) ((160 mg) were loaded in a Schlenk tube and dissolved in 2 ml THF under argon. The mixture was stirred vigorously till the bluish violet solution turned to amaranthine. Then diethyl isophthalate (1.94 g, 10.0 mmol) and B2pin2 (2.54 g, 10.0 mmol) in 15 ml THF was added into this tube. The mixture was heated in oil bath at 80°C for 24 h. After cooling to room temperature, the mixture then went through a short silica pad to remove the residual Ir catalyst and the final compound was purified by column chromatography using dichloromethane, giving 2.56 g (80% yield) white crystalline product. Crystals were grown by slow evaporation of a hexane solution.
Figures
Fig. 1.
Molecular structure of the title compound. Displacement elliposoid is drawn at 50% probability level.
Fig. 2.
The packing pattern of the title molecules in crystal viewed down the a- axis. O···H and B···H intermolecular short contacts are also showed.
Crystal data
| C16H21BO6 | F(000) = 1360 |
| Mr = 320.14 | Dx = 1.252 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 3703 reflections |
| a = 7.2163 (2) Å | θ = 2.7–20.6° |
| b = 20.9627 (4) Å | µ = 0.09 mm−1 |
| c = 22.4624 (4) Å | T = 296 K |
| V = 3397.96 (13) Å3 | Pod, colourless |
| Z = 8 | 1.00 × 0.40 × 0.20 mm |
Data collection
| Bruker APEXII CCD diffractometer | 3864 independent reflections |
| Radiation source: fine-focus sealed tube | 2177 reflections with I > 2σ(I) |
| graphite | Rint = 0.043 |
| φ and ω scans | θmax = 27.5°, θmin = 1.8° |
| Absorption correction: multi-scan (SADABS; Bruker, 2005) | h = −9→9 |
| Tmin = 0.766, Tmax = 0.982 | k = −27→26 |
| 22413 measured reflections | l = −27→29 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.046 | H-atom parameters constrained |
| wR(F2) = 0.142 | w = 1/[σ2(Fo2) + (0.0653P)2 + 0.3179P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 3864 reflections | Δρmax = 0.18 e Å−3 |
| 215 parameters | Δρmin = −0.14 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0031 (5) |
Special details
| Experimental. SADABS (Bruker, 2005) was used for absorption correction. R(int) was 0.0776 before and 0.0475 after correction. The Ratio of minimum to maximum transmission is 0.9190. The λ/2 correction factor is Not present. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.67152 (17) | 0.13558 (5) | 0.42187 (5) | 0.0525 (4) | |
| O2 | 0.6570 (2) | 0.16197 (6) | 0.20099 (6) | 0.0739 (5) | |
| O3 | 0.74902 (17) | 0.03889 (5) | 0.46030 (5) | 0.0484 (3) | |
| O4 | 0.6855 (2) | 0.08265 (7) | 0.13639 (6) | 0.0817 (5) | |
| O5 | 0.8187 (2) | −0.14641 (6) | 0.32004 (7) | 0.0785 (5) | |
| O6 | 0.8121 (3) | −0.13786 (7) | 0.22123 (7) | 0.0903 (6) | |
| C1 | 0.4427 (3) | 0.13373 (9) | 0.49842 (10) | 0.0679 (6) | |
| H1A | 0.3750 | 0.1642 | 0.4751 | 0.102* | |
| H1B | 0.4192 | 0.1411 | 0.5399 | 0.102* | |
| H1C | 0.4037 | 0.0914 | 0.4880 | 0.102* | |
| C2 | 0.6479 (2) | 0.14086 (7) | 0.48620 (8) | 0.0439 (4) | |
| C3 | 0.7180 (2) | 0.04498 (8) | 0.34685 (7) | 0.0452 (4) | |
| C4 | 0.6938 (2) | 0.08372 (9) | 0.29731 (8) | 0.0474 (4) | |
| H4 | 0.6698 | 0.1269 | 0.3028 | 0.057* | |
| C5 | 0.7043 (2) | 0.05958 (8) | 0.23990 (8) | 0.0462 (4) | |
| C6 | 0.6823 (3) | 0.10082 (9) | 0.18671 (9) | 0.0546 (5) | |
| C7 | 0.6358 (4) | 0.20504 (11) | 0.15139 (9) | 0.0952 (9) | |
| H7A | 0.7436 | 0.2027 | 0.1264 | 0.143* | |
| H7B | 0.6216 | 0.2478 | 0.1659 | 0.143* | |
| H7C | 0.5282 | 0.1933 | 0.1288 | 0.143* | |
| C8 | 0.9702 (3) | 0.10025 (10) | 0.51525 (9) | 0.0702 (6) | |
| H8A | 1.0391 | 0.0619 | 0.5225 | 0.105* | |
| H8B | 0.9880 | 0.1293 | 0.5478 | 0.105* | |
| H8C | 1.0128 | 0.1197 | 0.4790 | 0.105* | |
| C9 | 0.7647 (3) | 0.08423 (8) | 0.50955 (7) | 0.0461 (4) | |
| C10 | 0.7157 (3) | 0.20608 (8) | 0.50516 (9) | 0.0651 (6) | |
| H10A | 0.8413 | 0.2118 | 0.4922 | 0.098* | |
| H10B | 0.7102 | 0.2094 | 0.5477 | 0.098* | |
| H10C | 0.6386 | 0.2383 | 0.4876 | 0.098* | |
| C11 | 0.7376 (3) | −0.00497 (9) | 0.23160 (8) | 0.0515 (5) | |
| H11 | 0.7437 | −0.0216 | 0.1933 | 0.062* | |
| C12 | 0.7619 (2) | −0.04497 (8) | 0.28033 (8) | 0.0481 (5) | |
| C13 | 0.7995 (3) | −0.11372 (10) | 0.26942 (10) | 0.0621 (6) | |
| C14 | 0.8615 (5) | −0.21352 (10) | 0.31398 (12) | 0.1040 (9) | |
| H14A | 0.7605 | −0.2348 | 0.2945 | 0.156* | |
| H14B | 0.8803 | −0.2318 | 0.3527 | 0.156* | |
| H14C | 0.9721 | −0.2184 | 0.2907 | 0.156* | |
| C15 | 0.7531 (2) | −0.01959 (8) | 0.33725 (8) | 0.0488 (5) | |
| H15 | 0.7709 | −0.0463 | 0.3698 | 0.059* | |
| C16 | 0.6931 (4) | 0.05342 (10) | 0.56561 (9) | 0.0764 (7) | |
| H16A | 0.5692 | 0.0383 | 0.5591 | 0.115* | |
| H16B | 0.6928 | 0.0841 | 0.5973 | 0.115* | |
| H16C | 0.7717 | 0.0182 | 0.5761 | 0.115* | |
| B1 | 0.7131 (3) | 0.07327 (9) | 0.41089 (9) | 0.0421 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0751 (9) | 0.0423 (7) | 0.0400 (7) | 0.0040 (6) | −0.0016 (6) | −0.0003 (5) |
| O2 | 0.1243 (13) | 0.0555 (9) | 0.0418 (8) | −0.0010 (8) | −0.0037 (8) | 0.0037 (6) |
| O3 | 0.0700 (9) | 0.0377 (6) | 0.0375 (7) | 0.0065 (5) | −0.0026 (6) | −0.0054 (5) |
| O4 | 0.1359 (14) | 0.0728 (10) | 0.0364 (8) | 0.0053 (9) | 0.0016 (8) | −0.0029 (7) |
| O5 | 0.1204 (13) | 0.0542 (9) | 0.0610 (10) | 0.0147 (8) | 0.0004 (9) | −0.0028 (7) |
| O6 | 0.1523 (17) | 0.0612 (9) | 0.0573 (10) | 0.0077 (9) | 0.0085 (10) | −0.0174 (7) |
| C1 | 0.0513 (12) | 0.0673 (13) | 0.0853 (16) | 0.0041 (10) | 0.0048 (11) | −0.0188 (11) |
| C2 | 0.0520 (11) | 0.0382 (9) | 0.0417 (10) | 0.0004 (7) | 0.0012 (8) | −0.0070 (7) |
| C3 | 0.0483 (10) | 0.0526 (10) | 0.0349 (10) | −0.0010 (8) | 0.0002 (8) | −0.0016 (8) |
| C4 | 0.0514 (11) | 0.0490 (10) | 0.0419 (10) | −0.0024 (8) | 0.0019 (8) | −0.0032 (8) |
| C5 | 0.0492 (11) | 0.0538 (11) | 0.0357 (10) | −0.0045 (8) | 0.0019 (8) | −0.0023 (8) |
| C6 | 0.0643 (13) | 0.0580 (12) | 0.0415 (12) | −0.0046 (9) | 0.0020 (9) | −0.0024 (9) |
| C7 | 0.165 (3) | 0.0633 (14) | 0.0578 (15) | 0.0008 (15) | −0.0084 (15) | 0.0172 (12) |
| C8 | 0.0604 (14) | 0.0768 (14) | 0.0736 (15) | 0.0113 (10) | −0.0213 (11) | −0.0182 (11) |
| C9 | 0.0623 (12) | 0.0421 (9) | 0.0337 (10) | 0.0029 (8) | −0.0058 (8) | −0.0074 (7) |
| C10 | 0.0740 (14) | 0.0416 (10) | 0.0797 (16) | −0.0033 (9) | −0.0017 (11) | −0.0137 (10) |
| C11 | 0.0601 (12) | 0.0596 (11) | 0.0349 (10) | −0.0051 (9) | 0.0023 (8) | −0.0088 (9) |
| C12 | 0.0517 (11) | 0.0502 (10) | 0.0424 (11) | −0.0024 (8) | 0.0007 (8) | −0.0064 (8) |
| C13 | 0.0777 (15) | 0.0574 (12) | 0.0514 (13) | −0.0001 (10) | 0.0035 (11) | −0.0059 (10) |
| C14 | 0.163 (3) | 0.0530 (14) | 0.096 (2) | 0.0214 (15) | 0.0036 (18) | 0.0028 (13) |
| C15 | 0.0546 (11) | 0.0509 (11) | 0.0410 (10) | −0.0012 (8) | 0.0001 (8) | −0.0019 (8) |
| C16 | 0.128 (2) | 0.0618 (13) | 0.0393 (12) | 0.0006 (12) | 0.0063 (12) | 0.0024 (10) |
| B1 | 0.0470 (11) | 0.0404 (10) | 0.0390 (12) | −0.0019 (8) | −0.0001 (9) | 0.0004 (9) |
Geometric parameters (Å, °)
| O1—B1 | 1.363 (2) | C7—H7A | 0.9600 |
| O1—C2 | 1.459 (2) | C7—H7B | 0.9600 |
| O2—C6 | 1.334 (2) | C7—H7C | 0.9600 |
| O2—C7 | 1.442 (2) | C8—C9 | 1.526 (3) |
| O3—B1 | 1.349 (2) | C8—H8A | 0.9600 |
| O3—C9 | 1.4629 (19) | C8—H8B | 0.9600 |
| O4—C6 | 1.193 (2) | C8—H8C | 0.9600 |
| O5—C13 | 1.335 (2) | C9—C16 | 1.506 (3) |
| O5—C14 | 1.447 (2) | C10—H10A | 0.9600 |
| O6—C13 | 1.198 (2) | C10—H10B | 0.9600 |
| C1—C2 | 1.514 (3) | C10—H10C | 0.9600 |
| C1—H1A | 0.9600 | C11—C12 | 1.390 (2) |
| C1—H1B | 0.9600 | C11—H11 | 0.9300 |
| C1—H1C | 0.9600 | C12—C15 | 1.386 (2) |
| C2—C10 | 1.513 (2) | C12—C13 | 1.487 (3) |
| C2—C9 | 1.547 (2) | C14—H14A | 0.9600 |
| C3—C4 | 1.389 (2) | C14—H14B | 0.9600 |
| C3—C15 | 1.394 (2) | C14—H14C | 0.9600 |
| C3—B1 | 1.556 (2) | C15—H15 | 0.9300 |
| C4—C5 | 1.387 (2) | C16—H16A | 0.9600 |
| C4—H4 | 0.9300 | C16—H16B | 0.9600 |
| C5—C11 | 1.387 (3) | C16—H16C | 0.9600 |
| C5—C6 | 1.483 (3) | ||
| B1—O1—C2 | 106.12 (12) | O3—C9—C16 | 109.10 (14) |
| C6—O2—C7 | 115.49 (16) | O3—C9—C8 | 106.34 (14) |
| B1—O3—C9 | 106.86 (13) | C16—C9—C8 | 110.94 (17) |
| C13—O5—C14 | 116.17 (17) | O3—C9—C2 | 101.54 (12) |
| C2—C1—H1A | 109.5 | C16—C9—C2 | 115.20 (16) |
| C2—C1—H1B | 109.5 | C8—C9—C2 | 112.88 (15) |
| H1A—C1—H1B | 109.5 | C2—C10—H10A | 109.5 |
| C2—C1—H1C | 109.5 | C2—C10—H10B | 109.5 |
| H1A—C1—H1C | 109.5 | H10A—C10—H10B | 109.5 |
| H1B—C1—H1C | 109.5 | C2—C10—H10C | 109.5 |
| O1—C2—C10 | 108.03 (14) | H10A—C10—H10C | 109.5 |
| O1—C2—C1 | 106.63 (14) | H10B—C10—H10C | 109.5 |
| C10—C2—C1 | 110.76 (15) | C5—C11—C12 | 120.31 (16) |
| O1—C2—C9 | 102.36 (12) | C5—C11—H11 | 119.8 |
| C10—C2—C9 | 114.95 (15) | C12—C11—H11 | 119.8 |
| C1—C2—C9 | 113.30 (15) | C15—C12—C11 | 119.27 (17) |
| C4—C3—C15 | 117.82 (16) | C15—C12—C13 | 122.17 (17) |
| C4—C3—B1 | 121.00 (16) | C11—C12—C13 | 118.56 (17) |
| C15—C3—B1 | 121.15 (16) | O6—C13—O5 | 123.02 (19) |
| C5—C4—C3 | 121.65 (17) | O6—C13—C12 | 124.89 (19) |
| C5—C4—H4 | 119.2 | O5—C13—C12 | 112.09 (17) |
| C3—C4—H4 | 119.2 | O5—C14—H14A | 109.5 |
| C11—C5—C4 | 119.35 (16) | O5—C14—H14B | 109.5 |
| C11—C5—C6 | 118.62 (16) | H14A—C14—H14B | 109.5 |
| C4—C5—C6 | 122.02 (16) | O5—C14—H14C | 109.5 |
| O4—C6—O2 | 122.50 (18) | H14A—C14—H14C | 109.5 |
| O4—C6—C5 | 125.10 (18) | H14B—C14—H14C | 109.5 |
| O2—C6—C5 | 112.40 (16) | C12—C15—C3 | 121.59 (17) |
| O2—C7—H7A | 109.5 | C12—C15—H15 | 119.2 |
| O2—C7—H7B | 109.5 | C3—C15—H15 | 119.2 |
| H7A—C7—H7B | 109.5 | C9—C16—H16A | 109.5 |
| O2—C7—H7C | 109.5 | C9—C16—H16B | 109.5 |
| H7A—C7—H7C | 109.5 | H16A—C16—H16B | 109.5 |
| H7B—C7—H7C | 109.5 | C9—C16—H16C | 109.5 |
| C9—C8—H8A | 109.5 | H16A—C16—H16C | 109.5 |
| C9—C8—H8B | 109.5 | H16B—C16—H16C | 109.5 |
| H8A—C8—H8B | 109.5 | O3—B1—O1 | 113.93 (15) |
| C9—C8—H8C | 109.5 | O3—B1—C3 | 123.55 (16) |
| H8A—C8—H8C | 109.5 | O1—B1—C3 | 122.52 (16) |
| H8B—C8—H8C | 109.5 |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H16C···O4i | 0.96 | 2.53 | 3.381 (3) | 148 |
Symmetry codes: (i) −x+3/2, −y, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5730).
References
- Bruker (2005). APEX2, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Coventry, D. N., Batsanov, A. S., Goeta, A. E. H., Judith, A. K., Marder, T. B. & Perutz, R. N. (2005). Chem. Commun. 16, 2172–2174. [DOI] [PubMed]
- Kikuchi, T., Takagi, J., Isou, H., Ishiyama, T. & Miyaura, N. (2008). Chem. Asian J. 3, 2082–2090. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811054559/bt5730sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811054559/bt5730Isup4.hkl
Supplementary material file. DOI: 10.1107/S1600536811054559/bt5730Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report