Abstract
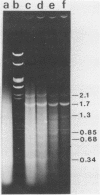
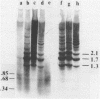
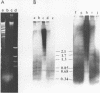
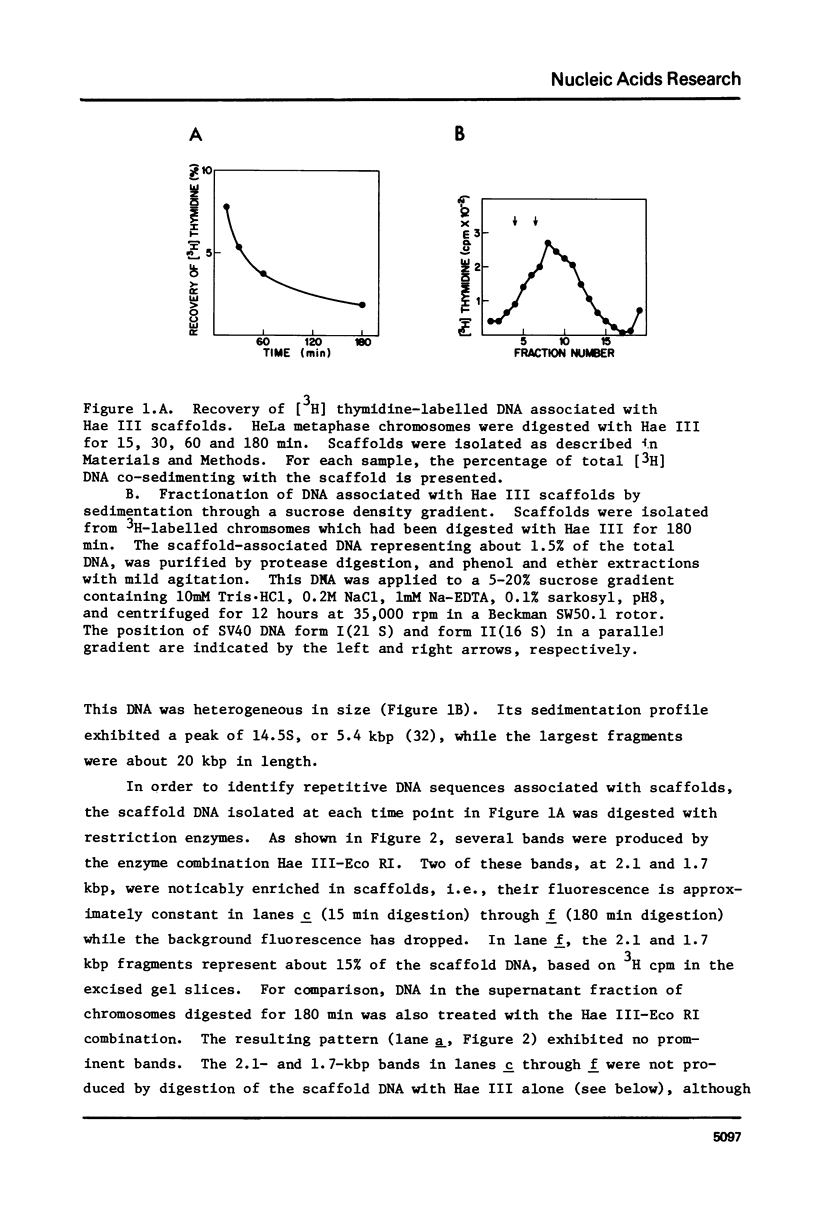
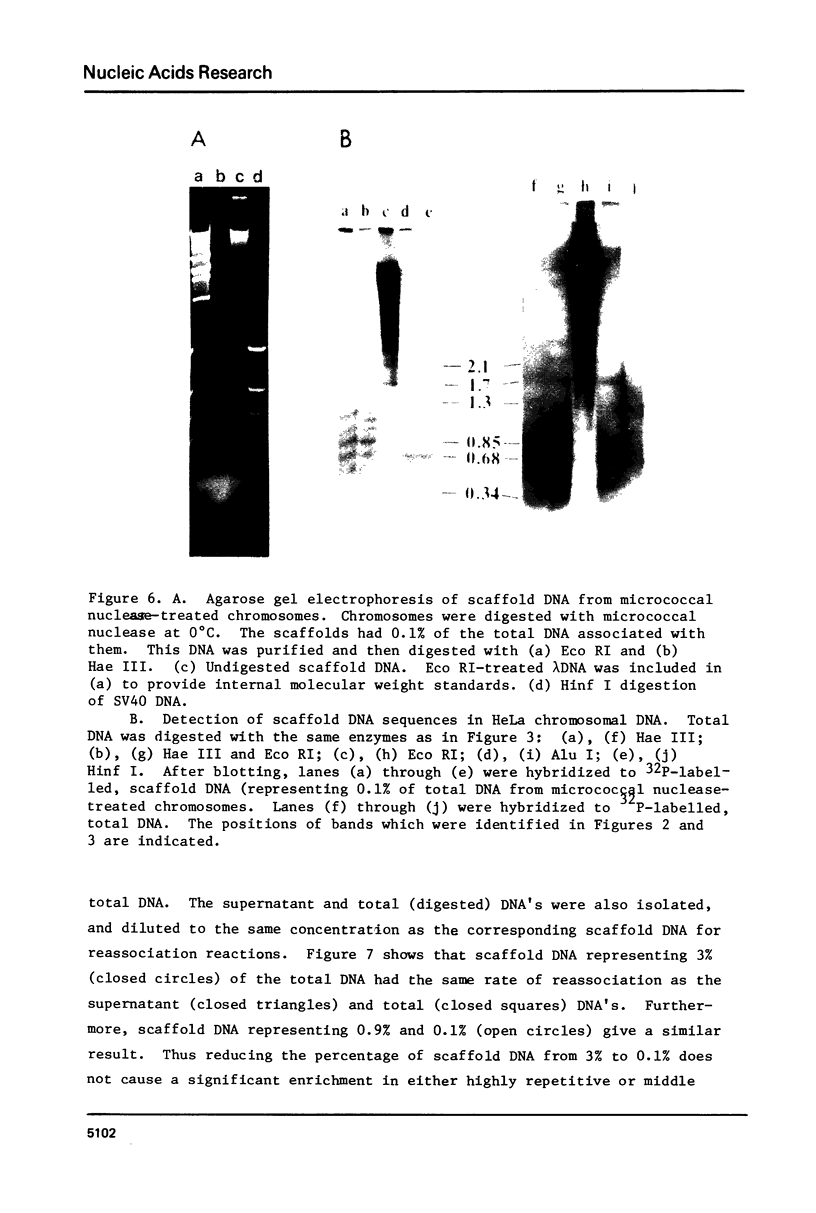
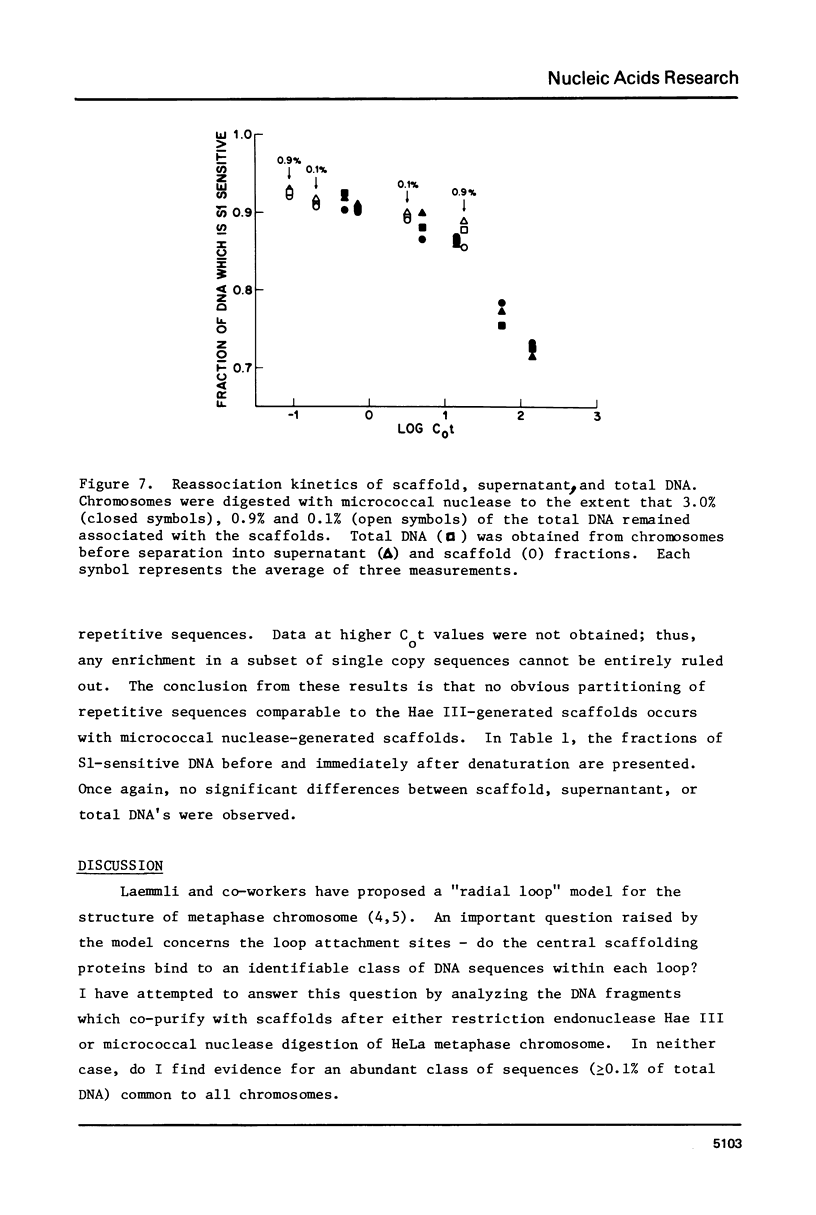
Following extensive digestion of HeLa metaphase chromosomes with either Hae III endonuclease or micrococcal nuclease, nonhistone protein scaffolds may be isolated. Scaffolds isolated after Hae III digestion have about 1.5% of the chromosomal DNA attached to them. This DNA is heterogeneous in size, ranging from about 0.2 to 20 kbp. It can be cleaved with either Eco RI or Hae III - Eco RI, producing a series of repeated fragments, of which the most abundant is 1.7 kbp in length. The 1.7-kdp fragment is tandemly repeated and is enriched (about 50-fold) in the scaffold-associated DNA. It is located primarily on human chromosome 1 and is probably a component of human satellites II and III. Scaffolds isolated after micrococcal nuclease digestion have about 0.1% of chromosomal DNA attached. This DNA is present in two size classes - fragments larger than 10 kbp and fragments approximately 0.2 kbp long. Restriction enzyme digestion of this DNA gives no prominent repeated fragments. Its reassociation kinetics are similar to those of total DNA, indicating that it is not enriched in either highly repetitive or middle repetitive sequences.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adolph K. W., Cheng S. M., Laemmli U. K. Role of nonhistone proteins in metaphase chromosome structure. Cell. 1977 Nov;12(3):805–816. doi: 10.1016/0092-8674(77)90279-3. [DOI] [PubMed] [Google Scholar]
- Adolphs K. W., Cheng S. M., Paulson J. R., Laemmli U. K. Isolation of a protein scaffold from mitotic HeLa cell chromosomes. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4937–4941. doi: 10.1073/pnas.74.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmain A., Birnie G. D. Nick translation of mammalian DNA. Biochim Biophys Acta. 1979 Jan 26;561(1):155–166. doi: 10.1016/0005-2787(79)90499-4. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Bonner J. J., Pardue M. L. Ecdysone-stimulated RNA synthesis in imaginal discs of Drosophila melanogaster. Assay by in situ hybridization. Chromosoma. 1976 Oct 12;58(1):87–99. doi: 10.1007/BF00293443. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Mapping sequences in loops of nuclear DNA by their progressive detachment from the nuclear cage. Nucleic Acids Res. 1980 Jul 11;8(13):2895–2906. doi: 10.1093/nar/8.13.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Cooke H. J., Hindley J. Cloning of human satellite III DNA: different components are on different chromosomes. Nucleic Acids Res. 1979 Jul 25;6(10):3177–3197. doi: 10.1093/nar/6.10.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Gates D. M., Bekhor I. DNA sequence selection by tightly-bound nonhistone chromosomal proteins. Nucleic Acids Res. 1979 Apr;6(4):1617–1630. doi: 10.1093/nar/6.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo-Kemenes T., Greil W., Zachau H. G. Prepartation of soluble chromatin and specific chromatin fractions with restriction nucleases. Nucleic Acids Res. 1977 Oct;4(10):3387–3400. doi: 10.1093/nar/4.10.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. G., Bankier A. T. A partial characterization of DNA fragments protected from nuclease degradation in histone depleted metaphase chromosomes of the Chinese hamster. Nucleic Acids Res. 1979 Sep 11;7(1):49–67. doi: 10.1093/nar/7.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. G., Bankier A. T., Sanders L. Non-histone proteins and the structure of metaphase chromosomes. Exp Cell Res. 1978 Sep;115(2):293–302. doi: 10.1016/0014-4827(78)90284-7. [DOI] [PubMed] [Google Scholar]
- Jones K. W., Corneo G. Location of satellite and homogeneous DNA sequences on human chromosomes. Nat New Biol. 1971 Oct 27;233(43):268–271. doi: 10.1038/newbio233268a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Cheng S. M., Adolph K. W., Paulson J. R., Brown J. A., Baumbach W. R. Metaphase chromosome structure: the role of nonhistone proteins. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):351–360. doi: 10.1101/sqb.1978.042.01.036. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Chromosomal localization of complex and simple repeated human DNAs. Chromosoma. 1978 Mar 22;66(1):23–32. doi: 10.1007/BF00285813. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Repeating restriction fragments of human DNA. Nucleic Acids Res. 1976 Nov;3(11):3063–3076. doi: 10.1093/nar/3.11.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden M. P., Laemmli U. K. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979 Aug;17(4):849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Maxwell I. H., Van Ness J., Hahn W. E. Assay of DNA-RNA hybrids by S1 nuclease digestion and adsorption to DEAE-cellulose filters. Nucleic Acids Res. 1978 Jun;5(6):2033–2038. doi: 10.1093/nar/5.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B., Coffey D. S. A fixed site of DNA replication in eucaryotic cells. Cell. 1980 Feb;19(2):527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Paulson J. R., Laemmli U. K. The structure of histone-depleted metaphase chromosomes. Cell. 1977 Nov;12(3):817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Goldsmith M., Hamkalo B. A. Chromatin organization during meiotic prophase of Bombyx mori. Chromosoma. 1980;79(2):215–224. doi: 10.1007/BF01175187. [DOI] [PubMed] [Google Scholar]
- Razin S. V., Mantieva V. L., Georgiev G. P. DNA adjacent to attachment points of deoxyribonucleoprotein fibril to chromosomal axial structure is enriched in reiterated base sequences. Nucleic Acids Res. 1978 Dec;5(12):4737–4751. doi: 10.1093/nar/5.12.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S. V., Mantieva V. L., Georgiev G. P. The similarity of DNA sequences remaining bound to scaffold upon nuclease treatment of interphase nuclei and metaphase chromosomes. Nucleic Acids Res. 1979 Nov 24;7(6):1713–1735. doi: 10.1093/nar/7.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wray W., Stubblefield E. A new method for the rapid isolation of chromosomes, mitotic apparatus, or nuclei from mammalian fibroblasts at near neutral pH. Exp Cell Res. 1970 Mar;59(3):469–478. doi: 10.1016/0014-4827(70)90656-7. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Gall J. G. A single integrated gene for ribosomal RNA in a eucaryote, Tetrahymena pyriformis. Cell. 1977 Sep;12(1):121–132. doi: 10.1016/0092-8674(77)90190-8. [DOI] [PubMed] [Google Scholar]