Abstract
Embryonic stem cells derived from nuclear transfer embryos (ntESCs) are particularly valuable for regenerative medicine, as they are a patient-specific and histocompatible cell source for the treatment of varying diseases. However, currently, little is known about their cellular and molecular profile. In the present study, in a mouse model different donor cell-derived ntESCs from various genetic backgrounds were compared with reference ESCs and analyzed comprehensively at the cellular level. A number of pluripotency marker genes were compared by flow cytometry and immunocytochemistry analysis. Significant differences at the protein level were observed for POU5F1, SOX2, FGF4, NANOG, and SSEA-1. However, such differences had no effect on in vitro cell differentiation and cell fate: derivatives of the three germ layers were detected in all ntESC lines. The neural and cardiac in vitro differentiation revealed minor differences between the cell lines, both at the mRNA and protein level. Karyotype analyses and cell growth studies did not reveal any significant variations. Despite some differences observed, the present study revealed that ntESC lines had similar differentiation competences compared to other ESCs. The results indicate that the observed differences may be related to the genotype rather than to the nuclear transfer technology.
Introduction
Derivation of embryonic stem cell (ESC) lines from nuclear transfer (NT) embryos is an advantageous method for production of histocompatible cells/tissue, which could be used for the treatment of numerous human diseases. Currently, human somatic cell nuclear transfer (hSCNT) as a step in the derivation of autologous ESCs for research and clinical treatment remains subject to ethical debate. Incredibly, the milestone in producing hSCNT blastocysts has already been achieved (Wun and Dittman, 2008), and very recently NT-derived ESCs established (Noggle et al., 2011). There are great expectations for new and promising methods that avoid the process of NT, such as induced pluripotent stem cells (iPS) derived from somatic cell cultures (Okita et al., 2007; Park et al., 2008; Takahashi et al., 2007; Wernig et al., 2007). A number of advantages of these cells include easy and simple isolation, a wider donor cell range, and the ablation of ethical concerns over embryo sacrifice (in case of human iPS lines) (Hipp and Atala, 2008; Kim et al., 2009; Nishikawa et al. 2008). However, recent publications have pointed out some current limitations of the iPS technology. Lanza and colleagues described that although the capacity of human iPS cells to differentiate into a variety of cell types was almost the same as that of human ESCs, cells differentiated from iPS cells exhibited significantly increased apoptosis, severely limited growth and expansion capability compared to their human ESC derivatives (Feng et al., 2010). In addition, reactivation of transgenes such as c-Myc has led to early death and tumor formation in chimeric mice, which raises further safety concerns over lines generated from this oncogene (Nakagawa et al., 2008; Okita et al., 2008; Wernig et al., 2007). However, using modified reprogramming approaches, for example, the exclusion of c-Myc, and viral vector-free or genome integration-free induction of reprogramming, these may reduce the tumor formation in iPS-derived chimeric mice (Kaji et al., 2009; Nakagawa et al., 2008; Okita et al., 2008). However, a recent report shown immunogenicity of iPSCs compared to ESCs (Zhao et al., 2011). These results indicate a need for further in-depth studies before safe clinical use of iPS-derived cells can be achieved and further investigations using nuclear transfer embryonic stem cells (ntESCs) are still very relevant.
Several studies have proven that it is feasible to establish ESCs from NT embryos of mice (Kawase et al., 2000; Munsie et al., 2000), primates (Byrne et al., 2007), bovine (Cibelli et al., 1998; Wang et al., 2005), rabbit (Fang et al., 2006), and recently from porcine (Vassiliev et al., 2011) and human (Noggle et al., 2011), by using different nuclear donor cells. However, the number of nuclear donor cell types used for derivation of ntESCs is lower than the number of cell types used in NT for production of live offspring (Wakayama et al., 2008a). So far, mouse ntESCs have been established from embryos cloned from freshly isolated cells, for example, cumulus cells (Munsie et al., 2000), tail tip fibroblasts (Wakayama et al., 2001), fetal neuronal cells (Kawase et al., 2000), or tooth pulp cells (Gurer et al., 2009), as well as from cultured cells like ESCs (Wakayama et al., 2001, 2005a, 2006), testicular Sertoli cells (Wakayama et al., 2005b) or mouse embryonal carcinoma (EC) cell lines (Blelloch et al., 2006). Furthermore, ntESCs have been used as nuclei donor cells in a second round of NT, that is, serial NT, although, this did not significantly improve the efficiency of live offspring production, compared to somatic cells from the same individual (Wakayama et al., 2005b). Surprisingly, generation of ntESCs has been achieved by using mouse tissues frozen without any cryoprotectant for different time periods as donor cell source. These ntESCs were able to rescue the nuclear genome of the tissue donor through ntESC chimeras (Li and Mombaerts, 2008), or serial NT to produce healthy cloned mice (Wakayama et al., 2008b). Recently, iPS cells were used as nuclear donors to produce cloned offspring, where the efficiency was similar to that of using ESCs derived via normal fertilization (Kou et al., 2010). Although, the production of cloned embryos, ntESCs, or cloned offspring by the aforementioned experiments were successful, most studies revealed that this procedure is highly variable according to both the epigenetic and genetic status of the original genomes (Inoue et al., 2007; Oback and Wells, 2007; Wakayama, 2007). The success rate for producing live offspring by cloning is highly affected by the mouse genotype: in particular the hybrid stains (e.g., B6D2F1) and 129SV have been the most amenable to reprogramming. However, new protocols (such as using histone deacetylase inhibitor, trichostatin A (TSA) treatment) have shown that inbreed (e.g., ICR) strains and other “nonpermissive” strains (C57Bl/6 or C3H/He) can be used for cloning with comparable success to hybrid strains (Kishigami et al., 2006; Wakayama, 2007).
Murine ntESC lines derived from different donor cells have been shown to express pluripotent stem cell markers and are capable of forming simple embryoid bodies (EBs) in suspension culture (Zhao et al., 2007). These ntESC are able to differentiated into neural or myogenic cells (Munsie et al., 2000); moreover, insulin-producing cells were also generated in vitro (Jiang et al., 2008). Previous studies have reported that ntESCs possess the same characteristics for self-renewal and differentiation as ESCs derived from natural (i.e., fertilized) blastocysts. These cells have the ability to differentiate into embryonic tissues in vivo and contribute to the germ line (Kawase et al., 2000; Wakayama et al., 2001; Zhao et al., 2007). They have been shown to rescue degenerative phenotypes, for example, the differentiated dopaminergic neurons from ntESCs have improved symptoms in Parkinsonian mice (Barberi et al., 2003).
Although several articles described the possibility of ntESC establishment and examined thoroughly the potential of these cell lines (for review, see Yang et al., 2007), only a few articles compared them comprehensively with fertilized embryo-derived ESCs, both at the biological and molecular level (Brambrink et al., 2006; Fan et al., 2008; Wakayama et al., 2006). Furthermore, no studies are available that compares different nuclear transfer method derived ntESCs.
Herein, we focused on the comprehensive evaluation of ntESCs derived from different donor cell types. We evaluated if any critical factors or differences could be detected between ntESCs and their ESC counterparts, and whether any differences could be detected between cell lines of the same nuclear donor origin. In the first part of the present study, we performed flow cytometry and immunocytochemistry analysis of pluripotency marker proteins to compare ESC lines derived from NT and control embryos. Cell lines were differentiated in vitro through EBs both with and without induction. Neural and cardiac lineages (as induced in vitro differentiation) were also analysed. Cell growth and karyotype of cell lines were also compared.
Materials and Methods
Materials for embryo culture and manipulation, unless specified otherwise, were purchased from Sigma-Aldrich Chem. Inc. (St. Louis, MO, USA; http://www.sigmaaldrich.com). All other materials, unless specified otherwise, were purchased from Invitrogen (Carlsbad, CA, USA; http://www.invitrogen.com).
Nuclear transfer and ESC establishment
The animal experiments were established in full compliance with European and Hungarian laws and regulations, and were approved by the Animal Experimentation Committee of the Agricultural Biotechnology Center.
Nuclear transfer was done by following the protocol of Ribas et al. (2005). The NT and control ESCs were established and cultured using the standard protocols published by Nagy et al. (2003). Further details of ntESCs used in this study are published previously (Kobolak et al., 2010). The mouse HM1 ESC (Selfridge et al., 1992) at passage 19 was kindly provided by Dr. Jim McWhir (Roslin Institute, Roslin, UK). The attributes of cell lines used in this study are summarized in Table 1.
Table 1.
Attributes of ESCs Used in the Study
| Name of the ESC line | Type | Nucleus donor cell | Genotype | Heterogeneity |
|---|---|---|---|---|
| HM1 | ESC | − | 129/Ola | homozygote |
| HM1 NT | ntESC | HM1 ESC | 129/Ola | homozygote |
| B6D2 | ESC | − | B6D2 F1 | heterozygote |
| B6D2 MEF NT | ntESC | B6D2 MEF | B6D2 F1 | heterozygote |
| B6D2 CUM NT | ntESC | B6D2 cumulus | B6D2 F1 | heterozygote |
| B6D2 CUM NT (PEM) | ntESC | B6D2 cumulus | B6D2 F1 | heterozygote |
| B6D2 PGA | pESC | B6D2 oocytea | B6D2 F1 | homozygote |
ESC, embryonic stem cell; NT/nt, nuclear transfer; MEF, mouse embryonic fibroblast; CUM, cumulus; PEM, piezoelectric micromanipulation; p, parthenogenetic.
Parthenogenetically activated.
For further details about ESC establishment see Kobolak et al., 2010.
Immunocytochemistry, flow cytometry (FACS), and karyotyping
Samples were fixed in 4% paraformaldehyde (PFA) fixative for 15 min, followed by three-times washing steps in phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.1% Triton X-100 for 10 min. For blocking, washing solution containing 10% fetal bovin serum (FBS) was used for 1 h. Primary antibodies were incubated overnight at 4°C in the indicated dilution listed in Supplementary Table S1 (see online supplementary data at www/liebertonline.com/cell). Samples were incubated at room temperature (RT) with the secondary antibody (see details in Supplementary Table S1) for 1 h on the following day. In the case of double or triple staining, the second or third primary antibody was incubated after the first one, whereas secondary antibodies were applied at the same time in a mixture. Samples were mounted with Vectashield-DAPI mounting media (Vector Laboratories, Burlingame, CA, USA; http://www.vectorlabs.com). The immunostainings were visualized with an AxioObserver Z.1 inverse fluorescent microscope and ApoTome slider system and AxioCam MRm camera system (Carl Zeiss GmbH, Germany; http://www.zeiss.de). Images were processed with AxioCam MRm own software, AxioVision 4.8, by using the multidimensional acquisition option.
For flow cytometry, the cells were pelleted and resuspended after the secondary labeling in 1 mL ice-cold PBS, and analyzed within 12 h, with FACS-CALIBUR (Becton Dickinson, Franklin Lakes, NJ, USA; http://www.bd.com). Three biological replicates for each sample (50,000 cells per replicate) were analyzed and the percentage (mean±SEM) of positive cells was calculated.
Alkaline phosphatase (ALP) staining and chromosome preparation was made as described by Nagy (2003). For analyzing euploidy, 200 metaphase nuclei were counted after DAPI staining for each cell line. Karyotype analyses were performed with FISH fluorescent-labeled StarFISH Mouse Chromosome-Specific Probes (mX-Cy3; mY-FITC; Cambio Ltd., Cambridge, UK; http://www.cambio.co.uk). Microscopy of slides was performed using an Olympus AH-2 photomicroscope equipped with Quips XL Genetics Workstation system including a Photometrics KAF 1400-G2 CCD camera (Abbott Laboratories, Abbott Park, IL, USA; http://www.abbottmolecular.com).
Colony-forming assay
Single-cell suspensions of cells were plated on gelatinized 10-cm gridded tissue culture dishes (Greiner Bio-One GmbH, Frickenhausen, Germany; http://www.greinerbioone.com) using 5×103 cell density to determine ESCs colony-forming unit (CFU) (Freshney, 2000). Colonies were microscopically enumerated after 4 days of culture. Before calculating the CFU values, plates were stained for ALP activity, and the numbers of colonies that were mostly positive, mixed, and unstained colonies was determined. A colony with greater than 90% staining was considered as “ALP+” (undifferentiated), 20–90% was called “ALP+ mixed,” and less than 20% called “ALP−” (differentiated). Three independent experiments were performed with four parallel replicates. The colony-forming efficiency percentage was calculated using the formula of colony forming efficiency (%)=(number of ALP-positive colonies/number of cells seeded)×100.
Growth efficiency
Cells were dispersed to single-cell suspension using 0.25% trypsin–EDTA, counted using a hemocytometer, and plated on mitomycin-C inactivated primary mouse embryonic fibroblast (MEF) covered six-well plates (Greiner Bio-One GmbH) at 105 cell density/per plate. Each trial was plated in duplicate. Population doubling times (PDT) were calculated after trypsinization and haemocytometer counting performed every 12 h over a 72-h culture period by using the online software of Doubling Time–Several Time Points calculator (http://www.doubling-time.com).
In vitro differentiation
For in vitro differentiation of ESCs, the hanging drop method (Doetschman et al., 1985) was used (20 μL/drops, 4×104 cells/mL). The basic differentiation medium consisted of high glucose Dulbecco-modified Eagle medium (DMEM), supplemented with penicillin (50 U/mL), streptomycin (50 μg/mL), Na-pyruvate (0.11% w/v), 0.1 mM 2-mercaptoethanol, nonessential amino acids (NEAA; 100×), FBS; 10% v/v; Hyclone, Logan, UT, USA; Waltham; Thermo Fisher Scientific Inc., Wilmington, DE, USA; http://www.hyclone.com). After 2 days of culture (day 2) in hanging drops, the formed EBs were collected and put into suspension culture in 10-cm bacterial Petri dish (Greiner Bio-One GmbH), where the medium was changed daily. Neural differentiation was induced supplementing the media with 10−6 M all-trans retinoic acid (RA) from day 4 until day 8. After day 8 the EBs were transferred to gelatine-coated 24-well dishes individually, and cultured further in basic media containing no RA. In case of cardiac muscle differentiation the media was supplemented with 1% dimethyl sulfoxide (DMSO) on day 2 for 2 days; thereafter, the EBs were cultured in suspension in basic media. On day 10 the EBs were plated individually into gelatinized 24-well plates (Greiner Bio-One GmbH), and cultured for further analysis. The length of beating periods were compared based on their frequency distribution.
Reverse transcription-PCR (RT-PCR)
Total RNA was isolated from the EB lysates using RNeasy Mini kit (Qiagen, Düsseldorf, Germany; http://www.qiagen.com) with an on-column DNase digestion step, following the manufacturer's procedures. The total RNA concentration and the quality of all samples were evaluated using NanoDrop (Thermo Fisher Scientific Inc.; http://www.nanodrop.com). One microgram total RNA from each EB samples was reverse transcribed with MMLV Reverse Transcriptase and oligodT primers, using the manufacturer's protocol. RT-PCR was performed in a Perkin-Elmer 9600 thermocycler (Applied Biosystems Inc., Foster City, CA, USA; www.appliedbiosystems.com). The reaction mixture consisted of JumpStart™ REDTaq™ ReadyMix™ (Sigma), 100 mM of each primer (see details in Supplementary Table S2), and 5 μL cDNA in a final volume of 50 μL. The reaction conditions were template denaturation and polymerase activation at 95°C for 2 min followed by 26–34 cycles of 95°C denaturation for 30 sec, 60°C annealing and extension for 45 sec at each cycle. For final extension, one cycle at 72°C for 10 min was applied. The cycle number of the amplification process was determined experimentally to produce the most sensitive results. For control, the Hprt1 was used. The PCR products were visualized on 1.5% agarose gel electrophoresis.
Statistical analysis
The chi-square test was used to compare the data obtained from the experiments. Values of p<0.05 were considered statistically significant. Immunoassay results were confirmed in at least three independent experiments.
All results of the FACS analysis, the colony-forming assay, and growth efficiency experiments were analyzed by one-way analysis of variance (ANOVA). A level of p<0.05 was considered statistically significant.
Results
Description of the cell lines analyzed
In the current study, we compared mouse ntESCs of different donor cell origins from two genetic backgrounds (126SV and B6D2) with their genotype control ESCs (derived from fertilized embryos) and a parthenogenetic ESC line (as a recipient oocyte control), which were established in our laboratory earlier (Kobolak et al., 2010). Furthermore, two NT methods [zona-free (ZF) and piezoelectric microinjection (PEM) technology] (Kobolak et al., 2010) were also compared by using the same nuclear donor cell type, that is, cumulus cells, to establish ntESCs and compare their performance in in vitro studies. Therefore, these cell lines are referred to as NT ZF and NT PEM to describe the technique used in their production, or MEF NT or CUM NT accordingly, to describe their donor cell background, herein. Details of NT, ntESC establishment, and primary characterization of the established cell lines were published recently (Kobolak et al., 2010) and summarized in Table 1.
Comparison of the pluripotency of ntESCs and control ESCs
In order to compare the stem cell characteristics of ntESCs and their control ESC counterparts (Table 1), common pluripotency markers were analyzed. An analysis of ALP, using an ALP enzymatic assay, was performed for all ntESCs and their control ESCs, which revealed all cell lines were positive for ALP activity (Fig. 1). In addition, the cell lines were characterized in vitro, by immunocytochemistry (ICC), using conventional pluripotency markers such as SSEA-1, POU5F1 (Nichols et al., 1998; Pesce et al., 1999), NANOG (Mitsui et al., 2003), SOX2 (Avilion et al., 2003), and a regulatory factor of early embryonic differentiation, FGF4 (Ambrosetti et al., 1997; Avilion et al., 2003). Representative ICC images, justifying the expression of these proteins of pluripotency markers in the cell populations examined, are shown in Supplementary Figure S1. Furthermore, the HM1 and B6D2 control cell lines have been tested independently from the current work, in tetraploid chimera experiments, in our own laboratory. These lines were able to support germline transmission (unpublished data), indicating that these are indeed true pluripotent ESC lines. In summary, the ICC results demonstrated that all cell lines have similar pluripotency characteristics.
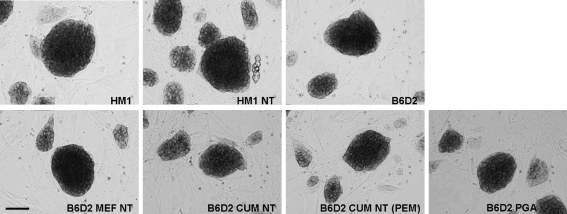
FIG. 1.
Alkaline phosphatase (ALP) activity of ESCs. Representative pictures of ALP enzymatic assay of ntESCs and control ESCs. Names of the cell lines are given on each picture. Scale bar represents 100 μm.
Furthermore, flow cytometry [fluorescence-activated cell sorting (FACS)] was used to measure the percentage of cells that express the same set of markers as used in the ICC experiments. By comparing the cell lines, HM1 and HM1 NT did not differ in any of the examined markers. Furthermore, the B6D2 ESC line (the genotype control) did not differ significantly from HM1, or from HM1 NT, with the exception of SOX2 (Table 2A). The B6D2 PGA cell line showed the most significant differences from both the controls and from the same genotype (B6D2) counterparts.
Table 2A.
FACS Analysis of ntESCs: Single Staining
| ESCs | POU5F1 | SSEA-1 | NANOG | FGF4 | SOX2 |
|---|---|---|---|---|---|
| HM1 | 75%(±4.6) | 68%(±5.9) | 67%(±4.4) | 47%(±6.1) | 81%(±3.9) |
| HM1 NT | 73%(±3.2) | 62%(±4.0) | 64%(±3.6) | 45%(±5.5) | 87%(±3.6) |
| B6D2 | 72%(±4.5) | 65%(±5.1) | 61%(±4.1) | 52%(±6.1) | 65%(±7.1)a,b |
| B6D2 MEF NT | 66%(±2.9)a | 71%(±5.3) | 59%(±6.8) | 67%(±5.3)a,b,c | 77%(±9.7)c |
| B6D2 CUM NT | 67%(±6.8)a | 62%(±6.6) | 73%(±2.1)b,c,d | 64%(±6.0)a,b | 80%(±8.6)c |
| B6D2 CUM NT (PEM) | 64%(±3.1)a,b,c | 58%(±3.6)a,d | 57%(±4.3)a,d,e | 60%(±9.7)a,b | 78%(±8.1)c |
| B6D2 PGA | 74%(±4.7)d,f | 47%(±7.5)1 | 41%(±6.8)1 | 58%(±8.5)b | 72%(±4.0)b |
The data are presented as the mean±SD of three independent samples; significant differences (ANOVA) are labeled with uppercase letters as follows: adata were significantly different from the mean value of HM1; bdata were significantly different from the mean value of HM1 NT; cdata were significantly different from the mean value of B6D2; ddata were significantly different from the value of B6D2 MEF NT; edata were significantly different from the value of B6D2 CUM NT; fdata were significantly different from the value of B6D2 CUM NT (PEM).
Data were significantly different from all other values; genotype controls are bolded.
The percentage of POU5F1-positive cells varied from 64% (±3.1) to 75% (±4.6) in the studied cell lines. Significant differences (p<0.05) were observed between the HM1 (129/Ola genotype) and B6D2 genotype ntESCs: B6D2 MEF NT (66%±2.9), B6D2 CUM NT (67%±6.8) and B6D2 CUM NT (PEM) (64%±3.1, Table 2A).
An evaluation of SSEA-1 expression revealed that the highest percentage of positive cells were detected in the B6D2 MEF NT (71%±5.3) cell line, which did not significantly differ from the HM1 cell line (68%±5.9, Table 2A). The B6D2 CUM NT (PEM) (58%±3.6) and the parthenogenetic cell line, B6D2 PGA (47%±7.5), had significantly (p<0.05) lower expression.
NANOG expression in the cell lines was compared to the HM1 control. The B6D2 CUM NT showed higher NANOG expression levels (73%±2.1), whereas the B6D2 CUM NT (PEM) and the B6D2 PGA ntESCs had lower expression levels (57%±4.3 and 41%±6.8, respectively; Table 2A). Furthermore, both CUM ntESCs were significantly different from the B6D2 MEF NT cell line (Table 2A).
Generally, FGF4 expression was low, when compared to all the other examined proteins in this study (Table 2A). The three B6D2 ntESC lines expressed significantly higher FGF4 levels (p<0.05), compared to the HM1 control or HM1 ntESC line (Table 2A). However, only B6D2 MEF NT (67%±5.3) differed significantly from the B6D2 control (52%±6.1).
A larger variation in the level of SOX2 expression was observed among the cell lines. Although the HM1 NT cell line exhibited the highest positive cell number (87%±2.6), a significant (p>0.05) reduction of SOX2 expression was observed for the B6D2 (65%±7.1) and the B6D2 PGA (72%±4.0) cell lines (Table 2A). Furthermore, B6D2 was significantly different (p>0.05) from all B6D2 ntESCs (Table 2A).
Loss of POU5F1 expression correlates with a loss of pluripotency (Pesce and Scholer, 2001); therefore, double and triple immunolabeling with other pluripotency markers was performed. We examined whether the POU5F1-positive cells were also positive for other pluripotency markers, such as SSEA-1, NANOG, SOX2, and the early embryonic differentiation factor, FGF4. In these experiments the POU5F1-positive cells were considered as 100% and the ratio of cell population expressing both markers were calculated as the percentage of POU5F1 positives (Table 2B).
Table 2B.
FACS Analysis of ntESCs: double and Triple Staining
| ESCs | POU5F1/SSEA-1 | POU5F1/NANOG | POU5F1/FGF4 | POU5F1/SOX2 | POU5F1/SSEA-1/NANOG | POU5F1/FGF4/SOX2 |
|---|---|---|---|---|---|---|
| HM1 | 82%(±5.1) | 83%(±4.8) | 70%(±7.1) | 98%(±1.8) | 73%(±6.1) | 68%(±3.8) |
| HM1 NT | 75%(±4.9) | 80%(±3.0) | 65%(±5.2) | 96%(±3.0) | 64%(±3.0) | 62%(±3.9) |
| B6D2 | 81%(±4.1) | 76%(±7.1) | 79%(±4.6)b | 97%(±4.6) | 72%(±4.9) | 75%(±7.6)b |
| B6D2 MEF NT | 87%(±3.6)b | 78%(±5.3) | 78%(±5.0)b | 87%(±4.5) | 74%(±4.8)b | 72%(±5.3)b |
| B6D2 CUM NT | 74%(±3.2)d | 76%(±4.0) | 87%(±4.9)a,b | 91%(±7.6) | 71%(±4.9) | 84%(±3.8)a,b,c,d |
| B6D2 CUM NT (PEM) | 77%(±4.8)d | 76%(±2.2) | 83%(±5.1)a,b | 89%(±9.1) | 65%(±7.7) | 79%(±4.1)a,b |
| B6D2 PGA | 61%(±6.1)1 | 55%(±4.6)1 | 87%(±3.5)a,b | 88%(±3.2) | 49%(±5.8)1 | 81%(±3.0)a,b,d |
The data are presented as the mean±SD of three independent samples; significant differences (ANOVA) are labeled with uppercase letters as follows: adata were significantly different from the mean value of HM1; bdata were significantly different from the mean value of HM1 NT; cdata were significantly different from the mean value of B6D2; ddata were significantly different from the value of B6D2 MEF NT.
Data were significantly different from all other values; genotype controls are bolded.
The POU5F1/SSEA-1 double labeling indicated the B6D2 PGA cell line had a significantly (p<0.05) lower percentage of double positive cells (61%±6.1) compared to the HM1 (82%±5.1). Furthermore, this cell line differed significantly with the other cell lines as well. The two CUM NT ESCs (74%±3.2 and 77%±4.8, respectively; Table 2B) showed a significant difference from the B6D2 MEF NT cell line (87%±3.6). However, the double labeling of POU5F1/NANOG revealed that only the pESC (B6D2 PGA) line contained a significantly lower percentage of double-positive cells (55%±4.6) compared to the other cell lines. When POU5F1/FGF4 double-positive cells were measured and compared, the two genotypes showed a difference. The two CUM NT cell lines and the pESC were significantly (p<0.05) different from the HM1 cell line. Furthermore, the B6D2 genotype cell lines were significantly different (p<0.05) from the HM1 NT cell line (Table 2B). In the case of POU5F1/SOX2 double labelling none of the cell lines showed a significant difference compared to the controls (HM1 or B6D2) or each other (Table 2B).
In addition, two different combinations of triple labeling (i.e., POU5F1/SSEA-1/NANOG and POU5F1/FGF4/SOX2) were also performed and expression in ntESCs and control ESCs was compared (Table 2B). In the POU5F1/SSEA-1/NANOG triple-positive cell staining the lowest percentage of triple positive cells were found for the HM1 NT (64%±3.0) and the B6D2 PGA (49%±5.8) cell lines, compared to the HM1 (73%±6.1). However, when comparing the POU5F1/FGF4/SOX2 triple-positive subpopulations, both CUM NT ESCs and the pESC line showed significantly higher (p<0.05) percentages compared to the HM1 control (Table 2B). Furthermore, all B6D2 genotype cell lines were significantly different from the HM1 ntESC. In summary, the FACS analysis revealed significant variations among the cell lines, mainly due to differences in genetic background.
Growth efficiency
The ability to grow and multiply rapidly is another important feature of ESCs. To measure the growth efficiency a plating efficiency assay was performed. In general, the CFU varied from 4.4–9.8% (Table 3). The B6D2 ESCs had the highest CFU (9.8%±1.14), which was significantly higher (p<0.05) than the other cell lines, with the exception of the two cumulus cell-derived ntESCs (B6D2 CUM NT 7.1±1.3 and B6D2 CUM NT (PEM) 7.2±1.87). Although all other cell lines had a lower CFU compared to the HM1 line, only B6D2 PGA differed significantly (4.4±2.06; Table 3).
Table 3.
Comparison of Colony-Forming Efficiencies (CFU) and Population Doubling Time (PDT) of ntESCs
| ESCs | ALP+ colonies | ALP+ mixed colonies | ALP− colonies | CFU of ALP+ colonies | PDT in hours |
|---|---|---|---|---|---|
| HM1 | 288 (±29.8) | 182 (±23.7) | 24 (±8.5) | 7.2 (±1.18) | 12.76 (±0.29) |
| HM1 NT | 240 (±36.9) | 176 (±31.2) | 22 (±10.5) | 6.0 (±1.71) | 13.33 (±0.35)b |
| B6D2 | 293 (±29.0) | 207 (±25.1) | 18 (±12.3) | 9.8 (±1.14)a | 12.07 (±0.41) |
| B6D2 MEF NT | 271 (±29.2) | 134 (±17.3) | 16 (±9.4) | 6.8 (±1.46)b | 13.23 (±0.64)b |
| B6D2 CUM NT | 285 (±17.3) | 130 (±19.7) | 17 (±9.7) | 7.1 (±1.30) | 13.57 (±0.46)b |
| B6D2 CUM NT (PEM) | 289 (±38.1) | 168 (±23.6) | 29 (±7.5) | 7.2 (±1.87) | 12.93 (±0.65) |
| B6D2 PGA | 174 (±32.6)1 | 169 (±35.8) | 26 (±8.6) | 4.4 (±2.06)a,b | 14.36 (±1.03)1 |
The data are presented as the mean±SD of three independent experiments with four parallel replicates; significant differences (ANOVA) are labeled with uppercase letters as follows: adata were significantly different from the mean value of HM1, bdata were significantly different from the mean value of B6D2.
Data werer significantly different from all other values; genotype controls are bolded.
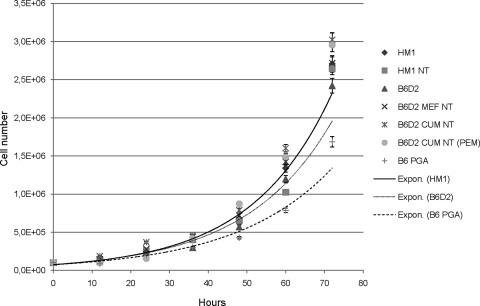
Additionally, we assessed the growth efficiency of the cell lines by calculating the PDT. Cell lines showed very similar growth curves and rates, with the doubling times ranging from 12.07 (±0.41) h (B6D2) to 14.36 (±1.03) h (B6D2 PGA). Of interest, the PDT of the two genotype controls did not differ significantly, with the doubling time of the B6D2 found to be shorter than that of the HM1 (12.07 (±0.41) and 12.76 (±0.29), respectively. The PDT of the ntESCs was very similar to each other. Here, the B6D2 MEF NT, the B6D2 CUM NT, and the B6D2 PGA were different from their genotype control (Table 3 and Fig. 2).
FIG. 2.
Comparison of the growth efficiencies of ntESCs and control ESCs. Population doubling times (PDT) were calculated after trypsinization and hemocytometer counting performed every 12 h over a 72-h culture period by using the online software of Doubling Time–Several Time Points calculator. Exponential curves were adapted to the data of both control (HM1 and B6D2) and the PGA ESC line to make visible the tendency of growth efficiencies.
Karyotype analysis
Cells were karyotyped by using FISH analysis and euploidy of each cell line was calculated (Table 4). The karyotype analysis revealed that the parthenogenetic and cumulus cell-derived ntESCs were female, whereas all other ESC lines were male. A lower level of euploidy (44%) was found for the B6D2 PGA ESC compared to all other examined cell lines showing an euploidy ratio of 50% or higher. The cell lines with the highest values were B6D2, HM1 NT, and B6D2 CUM NT (PEM) (76, 73, and 72%, respectively). The FISH analysis identified frequently noneuploid cells containing 39 or 41 chromosomes instead of 40 (Table 4). Either the Y chromosome or one of the X chromosomes were often lost and the karyogram revealed an XO genotype. In some cases, when 41 chromosomes were found, a translocation or deletion was detected on chromosome X. However, no chromosome specific deletions, translocations, or fusions were observed in the ntESCs lines, which might potentially be linked to the nuclear transfer process.
Table 4.
Karyotype of ntESCs
| ESCs | Passage number | Gender | Karyotype (euploidy %) | Notes |
|---|---|---|---|---|
| HM1 | p23 | XY | 68 | |
| HM1 NT | p4 | XY | 73 | |
| B6D2 | p5 | XY | 76 | |
| B6D2 MEF NT | p5 | XY | 63 | X0 |
| B6D2 CUM NT | p9 | XX | 65 | X0 |
| B6D2 CUM NT (PEM) | p9 | XX | 72 | deletion on chromosome X |
| B6D2 PGA | p3 | XX | 44 | deletion on chromosome X |
In vitro differentiation
To compare the developmental potential of the ntESCs and control ESCs, an in vitro assay, determining the spontaneous differentiations of the cell lines was performed by leukemia inhibitory factor (LIF) withdrawal. The experiments identified the derivatives of all three germ layers within differentiated EBs with interclass correlation (ICC) (Kobolak et al., 2010). Further detailed examination of the cardiac and neural differentiation in correlation to the expression levels of differentiation-specific genes and proteins were performed.
Cardiac lineage
The analysis of the appearance of beating areas following plating revealed no significant differences between ntESC lines and the control HM1 ESC, neither in the appearance of the first beating cell clusters, nor in the number of beating EBs (Table 5). The first day of beating has occurred between day 5 and 7, and on the last day of the experiment (day 30) still few beating cell-clusters existed in most cell lines. Additionally, the frequency distribution of the beating period was determined occurring in highest frequent beating periods between day 13 to 17. The shortest (9 days long) beating period showed the B6D2 PGA cells (Table 5).
Table 5.
Comparison of the Beating Profile of ntESC Cardiac Cell Clusters
| ESCs | Average number of beating EBs | First day of beating | Last day of beating | Most frequent duration of beating period |
|---|---|---|---|---|
| HM1 | 43 (±4.2) | 6 | 30 | 17 days |
| HM1 NT | 38 (±8.4) | 5 | 26 | 14 days |
| B6D2 | 44 (±3.6) | 5 | 30 | 17 days |
| B6D2 MEF NT | 40 (±5.5) | 6 | 30 | 13 days |
| B6D2 CUM NT | 42 (±3.2) | 5 | 30 | 16 days |
| B6D2 CUM NT (PEM) | 43 (±4.9) | 6 | 30 | 15 days |
| B6D2 PGA | 35 (±7.3) | 7 | 25 | 9 daysa |
In one experiment 48 EBs were plated. The attached EBs were counted from each experiment and the average number of beating EBs were calculated. SD±values are given in brackets.
Value differs significantly from the others.
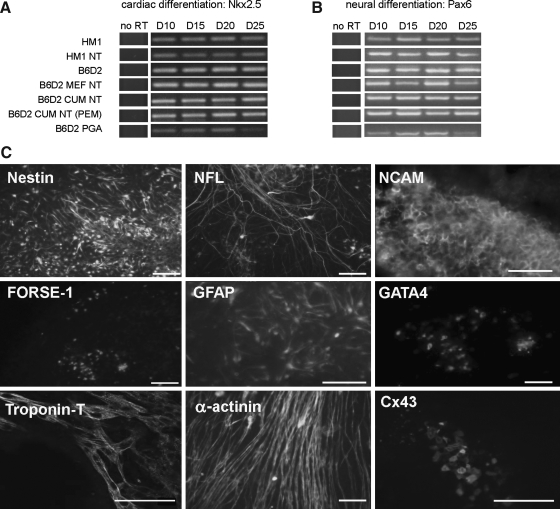
Gene expression was performed for cardiac differentiation marker genes (Gata4, Nkx2-5, Mef2c, Myl2, and Nppa). Samples were compared on days 5, 10, 15, 20, 25, and 30. No differences were observed in the expression patterns of the examined genes at any time point during the experiment. All cell lines expressed the chosen markers with the same dynamics during the monitored differentiation period (e.g., Nkx2.5) (Fig. 3A).
FIG. 3.
In vitro differentiation of ntESCs. Gene expression of (A) Nkx2.5 (cardiac lineage) and B) Pax6 (neural lineage) of in vitro differentiated ESCs with RT-PCR. no RT: pooled samples of a cell line, without reverse transcriptase. (C) Representative immunocytochemistry of HM1 ntESC during neural and cardiac differentiation. Images were taken at day 20 of differentiation, except for GATA-4 and FORSE-1 immunocytochemistry, which was performed on day 10 (nuclear staining), and Cx43 on day 15. Neural markers: NCAM, GFAP, NESTIN, FORSE-1, and NFL; Cardiac markers: GATA-4, Troponin T, α-actinin, Cx43. Scale bar represents 50 μm.
At day 10, 20 and 30, samples were analyzed by immunocytochemistry to detect the presence of GATA-4, alpha-cardiac actin (ACTC1), Troponin T (TNNT2), and Connexin 43 (GJA1), which are representative markers of late phase cardiac differentiation (see details in Fig. 3C). All cell lines were found to be positive for all the selected antibodies. The amount of positive EB cells differed within and among the cell lines. Therefore, no major differences were observed between the studied cell lines. On average, MEF-NT and PGA cell lines had the lowest attachment scores among the cell lines when plated on gelatinized dishes. In the PGA cell line, smaller areas were pulsing, and smaller clusters of cells gave positive signals with troponin-T or alfa-cardiac actin.
Neural differentiation
A classic neural differentiation approach, by using the 4−/4+ RA induction, was used on the ntESCs to study their capability to form neural cells. The experiment lasted 25 days and samples were collected at days 10, 15, 20, and 25 for RT-PCR and immunostaining.
All the examined neural markers (Pax6, Ncam1, Gfap, Nes, and Gbx) were expressed in each cell line. Alterations in the timing of expression were not found (e.g., Pax6) (Fig. 3B). By ICC, the examined markers (NCAM, GFAP, NESTIN, FORSE-1, and NFL) were detected in the differentiating EBs from all cell lines. To monitor the appearance of neural precursor cells an ICC against FORSE-1 was performed. Small cell populations, positive for FORSE-1, were detected in all cell lines at day 10; however, no positive cells were observed past this time point in any cell line. Representative ICC of the HM1 NT ESC is depicted in Figure 3C.
Discussion
Despite the potential of ESC use for tissue repair, these cells, if transplanted, would likely induce immunorejection upon their differentiation or neoplastic transformation. Nuclear transfer can overcome part of this difficulty by providing a donor-specific histocompatible cell source for cell therapy purposes. However, little is known about the biological performance of ntESC lines. For the generation of previous datasets, only a few cell types of nuclear donor cells from either the B6D2 or 129B6 genetic backgrounds have been used (Brambrink et al., 2006; Wakayama et al., 2006). Reproductive cloning (Wakayama et al., 2006) and/or tetraploid embryo complementation (Brambrink et al., 2006; Wakayama et al., 2006) has been performed to investigate the ultimate differentiation capacity of the ntESCs. These assays were able to only demonstrate major defects and minor differences between the cell lines due to donor cell source or NT technique have not yet been studied.
Recently, we have successfully established ntESCs derived from different nuclear donor cell types using either ZF or PEM technology (Kobolak et al., 2010). In this study the efficiency of ntESC derivation was not related to the NT method used. Furthermore, we demonstrated that the ZF NT technique resulted in cell lines with the same potential as ntESCs produced from PEM.
The major reason for undertaking the current study was to define the differences between ntESCs and control ESC lines on cellular and molecular level and gain further insight into the functional distinction between these cell lines. Our study focused on the transcriptional expression and in vitro differentiation potential of our previously established ntESC lines.
The accuracy and time-specific expression of the pluripotent transcriptional regulatory system is fundamental for the maintenance of ESC cell renewal and for their differentiation potential. To date, three transcription factors are known to play a critical role in the maintenance of ESC pluripotency: Pou5f1, Nanog, and Sox2. These factors comprise one characterized essential circuit for maintaining ESC pluripotency. It is generally considered that POU5F1 regulates Sox2, and additionally, the POU5F1–SOX2 protein complex activates Pou5f1 expression (Okumura-Nakanishi et al., 2005). Together with NANOG (Mitsui et al., 2003) these factors play an essential role in early development and are required for the propagation of undifferentiated ESCs in culture (Niwa et al., 2000). However, a number of other important factors (such as Tdgf1, Dnmt3b, Gabrb3, Gdf3, Utf1, and Zfp42) are expressed in undifferentiated ESC cells, and have been widely studied, although their role in maintaining pluripotency and self-renewal is more ambiguous (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Tanaka et al., 2002).
In our initial analyses, the most important in vitro pluripotency markers (POU5F1, NANOG, SSEA-1, SOX2, and ALP) and a regulator factor of early embryonic differentiation, FGF4 were analyzed at cellular level by use of immunocytochemistry and flow cytometry. In our ICC experiments, no major differences were observed among the control and ntESCs. However, B6D2 ntESCs, originating from both cumulus and MEF cells, and some cases the B6D2 control showed significant differences compared to the HM1 control in the flow cytometry experiments. Furthermore, the B6D2 ntESCs, independent from their nuclear cell origin, differed significantly from HM1 NT in several comparisons. These results indicate that the observed alterations may be more correlated with the genetic background of the ntESC than the type of the nuclear donor cell used.
The only exception found was the HM1 ESC-derived ntESC line, where a strong correlation was observed between the parental HM1 ESC and its NT derivative: no significant differences were detected in any of the examined pluripotency markers—eeither by single or multiple staining—at the protein level. This observation might indicate that ESCs could be reprogrammed more efficiently than somatic cells (Azuara et al., 2006).
A comparison of the two NT methods, using cumulus nuclear donor origin cell lines, revealed very similar results in flow cytometry and ICC experiment that might indicate a smaller distance between the two cell lines [B6D2 CUM NT and B6D2 CUM NT (PEM)] than the same genotype ESC lines (HM1 vs. HM1 NT) (Tong et al., 2007).
Immunocytochemistry performed using a single antibody may only partially validate cell pluripotency and could therefore lead to limited conclusions (Zangrossi et al., 2007). We expected that the ratios of double- or triple-positive cells (expressing two or three pluripotency markers) could characterize the pluripotency of a cell population more precisely. In our experiments, the double and triple staining revealed very similar tendencies among the cell lines, and also supported the results found from the single antibody labeling. The results were in accordance with previous observations of ntESCs. In a previous study, following analyzing of SSEA-1, SSEA-4, and PDGFR-α by cell sorting, only SSEA-1 was produced by all ntESCs examined (Wakayama et al., 2006). However, we examined other pluripotency markers, namely, POU5F1, NANOG, SOX2, and a regulator factor FGF4 in the ntESCs by flow cytometry. It should be noted, however, that these markers have also been measured in fertilized embryo-derived ESCs produced from different genotypes. The above-mentioned differences in the protein expression of ESCs might be in correlation with their in vivo developmental competence, the germline transmission, which known to be strongly influenced by the genetic background (Carstea et al., 2009).
ESCs have the potential to give rise to multiple cell lineages. ESCs also exhibit a very unusual cell cycle pattern, characterized by a short G1 phase and longer S-phase. This unique cell cycle pattern and the mechanisms underlying cell cycle control indicates that cell cycle machinery plays an important role in the maintenance of the stem cell state. For the first time, we describe the colony-forming capabilities and growth rates of ntESCs. The colony-forming assay revealed a significant difference between ntESCs and their fertilized embryo-derived counterparts in B6D2 genotype; however, both 129SV cell lines (HM1 and HM1 NT) also differed from the B6D2 ESC. Furthermore, the PDT of the cell lines significantly differed between the two genotypes 129/Ola (HM1 and HM1 NT) and B6D2. No difference was observed among the ntESCs, suggesting that their self-renewal capacity could be a sign of cell pluripotency.
Karyotype analyses of the ESCs allowed us to assess the gender of the cell lines and observe whether these lines contained any chromosome aberrations. From FISH analysis we could identify chromosome abnormalities in most of the cell lines. However, the specific chromosomal defects were not correlated with either the nuclear donor or the NT technique used. Further analysis and comparison of the euploidy ratios of these ESCs confirmed the observations of FISH results. Based on the literature, ESCs with more than 50% euploidy can contribute to in vivo development and colonize the germline successfully (Longo et al., 1997), thus karyotype results of the ntESCs, are in accordance with the published data on fertilized embryo-derived ESCs (Nagy, 2003; Suzuki et al., 1997).
The ESC lines were differentiated in vitro either spontaneously or by directed differentiation into cardiac and neural cell lineages. The spontaneous differentiation study revealed that the ntESC lines could form all three germ layers, thus proving their pluripotency. The directed in vitro differentiation studies revealed no major differences among the ntESC lines, which were analyzed by RT-PCR and immunostaining. In regard to the lower differentiation capacity observed in the parthenogenetic ESCs, it has been demonstrated earlier that their differentiation capacity into the mesodermal lineage is restricted (McKarney et al., 1997; Morali et al., 2000). Previous in vitro differentiation studies (Munsie et al., 2000) on ntESC support our findings concerning lack of differences observed in either neural or cardiac differentiation.
In conclusion, pluripotency marker analysis at the protein level revealed significant differences among the analysed ntESCs and their control counterparts, although, the observed differences had no effect on their in vitro cardiac and neural lineage differentiation potential. The observed differences should be examined further at the molecular level to conclude whether relevant differences among NT and control embryo-derived ESCs exists.
Supplementary Material
Acknowledgments
The authors thank Dr. Melinda Pirity for the critical reading and discussion of the manuscript. We are grateful to Mrs Hajnalka Csákány Tolnainé and Györgyi Kungl for their excellent technical assistance during the experiment. Supporting grants: Wellcome Trust (Grant No. 070246), EU FP6 (“TEAMOHOLIC” MEXT-CT-2003-509582; “MEDRAT” LSHG-CT-2006-518240; “CLONET” MRTN-CT-2006-035468), EU FP7 (“PLURISYS” HEALTH-2007-B-223485; PartnErS, PIAP-GA-2008-218205; EpiHealth, FP7-HEALTH-2011-278418; InduVir, PEOPLE-IRG-2009-245808), and NKFP_07_1-ES2HEART-HU; BONUS HU_08/2-2009-0008.
Author Disclosure Statement
The authors declare that no competing interests exist.
References
- Ambrosetti D.C. Basilico C. Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein–protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion A.A. Nicolis S.K. Pevny L.H., et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V. Perry P. Sauer S., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barberi T. Klivenyi P. Calingasan N.Y., et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat. Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- Blelloch R. Wang Z. Meissner A., et al. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T. Hochedlinger K. Bell G., et al. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc. Natl. Acad. Sci. USA. 2006;103:933–938. doi: 10.1073/pnas.0510485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne J.A. Pedersen D.A. Clepper L.L., et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Carstea A.C. Pirity M.K. Dinnyes A. Germline competence of mouse ES and iPS cell lines: Chimera technologies and genetic background. World J. Stem Cells. 2009;1:22–29. doi: 10.4252/wjsc.v1.i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibelli J.B. Stice S.L. Golueke P.J., et al. Transgenic bovine chimeric offspring produced from somatic cell-derived stem-like cells. Nat. Biotechnol. 1998;16:642–646. doi: 10.1038/nbt0798-642. [DOI] [PubMed] [Google Scholar]
- Doetschman T.C. Eistetter H. Katz M., et al. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- Fan Y. Tong M. Zhao C., et al. Comparative pluripotency analysis of mouse embryonic stem cells derived from wild-type and infertile hermaphrodite somatic cell nuclear transfer blastocysts. Chin. Sci. Bull. Sci. 2008;53:3648–3655. [Google Scholar]
- Fang Z.F. Gai H. Huang Y.Z., et al. Rabbit embryonic stem cell lines derived from fertilized, parthenogenetic or somatic cell nuclear transfer embryos. Exp. Cell Res. 2006;312:3669–3682. doi: 10.1016/j.yexcr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Feng Q. Lu S.J. Klimanskaya I., et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Freshney R.I. Culture of Animal Cells: A Manual of Basic Technique. New York: Wiley; 2000. [Google Scholar]
- Gurer F. Ozden H. Muslumanoglu H., et al. Therapeutic use of cloning: Osmangazi turk identical embryonic stem cells and embryonic stem cell transfer to diabetic mice. J Health Sci. 2009;55:503–515. [Google Scholar]
- Hipp J. Atala A. Sources of stem cells for regenerative medicine. Stem Cell Rev. 2008;4:3–11. doi: 10.1007/s12015-008-9010-8. [DOI] [PubMed] [Google Scholar]
- Inoue K. Noda S. Ogonuki N., et al. Differential developmental ability of embryos cloned from tissue-specific stem cells. Stem Cells. 2007;25:1279–1285. doi: 10.1634/stemcells.2006-0747. [DOI] [PubMed] [Google Scholar]
- Ivanova N.B. Dimos J.T. Schaniel C., et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jiang W. Bai Z. Zhang D., et al. Differentiation of mouse nuclear transfer embryonic stem cells into functional pancreatic beta cells. Diabetologia. 2008;51:1671–1679. doi: 10.1007/s00125-008-1065-1. [DOI] [PubMed] [Google Scholar]
- Kaji K. Norrby K. Paca A., et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase E. Yamazaki Y. Yagi T., et al. Mouse embryonic stem (ES) cell lines established from neuronal cell-derived cloned blastocysts. Genesis. 2000;28:156–163. [PubMed] [Google Scholar]
- Kim D. Kim C.H. Moon J.I., et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S. Wakayama S. Thuan N.V., et al. Production of cloned mice by somatic cell nuclear transfer. Nat. Protoc. 2006;1:125–138. doi: 10.1038/nprot.2006.21. [DOI] [PubMed] [Google Scholar]
- Kobolak J. Bodo S. Rungsiwiwut R., et al. Generation of mouse embryonic stem cell lines from zona-free nuclear transfer embryos. Cell Reprogram. 2010;12:105–113. doi: 10.1089/cell.2009.0040. [DOI] [PubMed] [Google Scholar]
- Kou Z. Kang L. Yuan Y., et al. Mice cloned from induced pluripotent stem cells (iPSCs) Biol. Reprod. 2010;83:238–243. doi: 10.1095/biolreprod.110.084731. [DOI] [PubMed] [Google Scholar]
- Li J. Mombaerts P. Nuclear transfer-mediated rescue of the nuclear genome of nonviable mouse cells frozen without cryoprotectant. Biol. Reprod. 2008;79:588–593. doi: 10.1095/biolreprod.108.069583. [DOI] [PubMed] [Google Scholar]
- Longo L. Bygrave A. Grosveld F.G., et al. The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res. 1997;6:321–328. doi: 10.1023/a:1018418914106. [DOI] [PubMed] [Google Scholar]
- McKarney L.A. Overall M.L. Dziadek M. Myogenesis in cultures of uniparental mouse embryonic stem cells: differing patterns of expression of myogenic regulatory factors. Int. J. Dev. Biol. 1997;41:485–490. [PubMed] [Google Scholar]
- Mitsui K. Tokuzawa Y. Itoh H., et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morali O.G. Jouneau A. McLaughlin K.J., et al. IGF-II promotes mesoderm formation. Dev. Biol. 2000;227:133–145. doi: 10.1006/dbio.2000.9875. [DOI] [PubMed] [Google Scholar]
- Munsie M.J. Michalska A.E. O'Brien C.M., et al. Isolation of pluripotent embryonic stem cells from reprogrammed adult mouse somatic cell nuclei. Curr. Biol. 2000;10:989–992. doi: 10.1016/s0960-9822(00)00648-5. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Nakagawa M. Koyanagi M. Tanabe K., et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nichols J. Zevnik B. Anastassiadis K., et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nishikawa S.-i. Goldstein R.A. Nierras C.R. The promise of human induced pluripotent stem cells for research and therapy. Nat. Rev. 2008;9:725–729. doi: 10.1038/nrm2466. [DOI] [PubMed] [Google Scholar]
- Niwa H. Miyazaki J. Smith A. G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Noggle S. Fung H.L. Gore A., et al. Human oocytes reprogram somatic cells to a pluripotent state. Nature. 2011;478:70–75. doi: 10.1038/nature10397. [DOI] [PubMed] [Google Scholar]
- Oback B. Wells D.N. Donor cell differentiation, reprogramming, and cloning efficiency: elusive or illusive correlation? Mol. Reprod. Dev. 2007;74:646–654. doi: 10.1002/mrd.20654. [DOI] [PubMed] [Google Scholar]
- Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K. Nakagawa M. Hyenjong H., et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Okumura-Nakanishi S. Saito M. Niwa H., et al. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- Park I.-H. Zhao R. West J.A., et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pesce M. Scholer H.R. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Pesce M. Anastassiadis K. Scholer H.R. Oct-4: lessons of totipotency from embryonic stem cells. Cells Tissues Organs. 1999;165:144–152. doi: 10.1159/000016694. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M. Yoon S. Matsuzaki Y., et al. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Ribas R. Oback B. Ritchie W., et al. Development of a zona-free method of nuclear transfer in the mouse. Cloning Stem Cells. 2005;7:126–138. doi: 10.1089/clo.2005.7.126. [DOI] [PubMed] [Google Scholar]
- Selfridge J. Pow A.M. McWhir J., et al. Gene targeting using a mouse HPRT minigene/HPRT-deficient embryonic stem cell system: inactivation of the mouse ERCC-1 gene. Somat. Cell Mol. Genet. 1992;18:325–336. doi: 10.1007/BF01235756. [DOI] [PubMed] [Google Scholar]
- Suzuki H. Kamada N. Ueda O., et al. Germ-line contribution of embryonic stem cells in chimeric mice: influence of karyotype and in vitro differentiation ability. Exp. Anim. 1997;46:17–23. doi: 10.1538/expanim.46.17. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tanaka T.S. Kunath T. Kimber W.L., et al. Gene expression profiling of embryo-derived stem cells reveals candidate genes associated with pluripotency and lineage specificity. Genome Res. 2002;12:1921–1928. doi: 10.1101/gr.670002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong G.Q. Heng B.C. Ng S.C. Cumulus-specific genes are transcriptionally silent following somatic cell nuclear transfer in a mouse model. J. Zhejiang Univ. Sci. B. 2007;8:533–539. doi: 10.1631/jzus.2007.B0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassiliev I. Vassilieva S. Truong K.P., et al. Isolation and in vitro characterization of putative porcine embryonic stem cells from cloned embryos treated with trichostatin A. Cell Reprogram. 2011 doi: 10.1089/cell.2010.0102. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Mizutani E. Kishigami S., et al. Mice cloned by nuclear transfer from somatic and ntES cells derived from the same individuals. J. Reprod. Dev. 2005a;51:765–772. doi: 10.1262/jrd.17061. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Ohta H. Kishigami S., et al. Establishment of male and female nuclear transfer embryonic stem cell lines from different mouse strains and tissues. Biol. Reprod. 2005b;72:932–936. doi: 10.1095/biolreprod.104.035105. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Jakt M.L. Suzuki M., et al. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastocysts. Stem Cells. 2006;24:2023–2033. doi: 10.1634/stemcells.2005-0537. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Cummins J.M. Wakayama T. Nuclear reprogramming to produce cloned mice and embryonic stem cells from somatic cells. Reprod. Biomed. Online. 2008a;16:545–552. doi: 10.1016/s1472-6483(10)60462-2. [DOI] [PubMed] [Google Scholar]
- Wakayama S. Ohta H. Hikichi T., et al. Production of healthy cloned mice from bodies frozen at −20°C for 16 years. Proc. Natl. Acad. Sci. USA. 2008b;105:17318–17322. doi: 10.1073/pnas.0806166105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama T. Production of cloned mice and ES cells from adult somatic cells by nuclear transfer: how to improve cloning efficiency? J. Reprod. Dev. 2007;53:13–26. doi: 10.1262/jrd.18120. [DOI] [PubMed] [Google Scholar]
- Wakayama T. Tabar V. Rodriguez I., et al. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science. 2001;292:740–743. doi: 10.1126/science.1059399. [DOI] [PubMed] [Google Scholar]
- Wang L. Duan E. Sung L.Y., et al. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol. Reprod. 2005;73:149–155. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- Wernig M. Meissner A. Foreman R., et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wun I.C. Dittman R.E. Human somatic cell nuclear transfer. Chin. J. Physiol. 2008;51:208–213. [PubMed] [Google Scholar]
- Yang X. Smith S.L. Tian X.C., et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Zangrossi S. Marabese M. Broggini M., et al. Oct-4 Expression in adult human differentiated cells challenges its role as a pure stem cell marker. Stem Cells. 2007;25:1675–1680. doi: 10.1634/stemcells.2006-0611. [DOI] [PubMed] [Google Scholar]
- Zhao C. Yao R. Hao J., et al. Establishment of customized mouse stem cell lines by sequential nuclear transfer. Cell Res. 2007;17:80–87. doi: 10.1038/sj.cr.7310139. [DOI] [PubMed] [Google Scholar]
- Zhao T. Zhang Z.N. Rong Z., et al. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.