Abstract
Vascular inflammation is initiated by stimuli acting on endothelial cells. A clinical feature of vascular inflammation is increased circulating interleukin 6 (IL-6) type cytokines such as leukemia inhibitory factor (LIF), but their role in vascular inflammation is not fully defined. IL-6 type cytokines activate transcription factor signal transducer and activator of transcription 3 (STAT3), which has a key role in inflammation and the innate immune response. Canonical STAT3 gene induction is due to phosphorylation of (1) Y705, leading to STAT3 dimerization and DNA binding and (2) S727, enhancing homodimerization and DNA binding by recruiting p300/CBP. We asked whether enhancing S727 STAT3 phosphorylation using the protein phosphatase 1 (PP1) inhibitor, calyculin A, would enhance LIF-induced gene expression in human microvascular endothelial cells (HMEC-1). Cotreatment with calyculin A and LIF markedly increased STAT3 S727 phosphorylation, without affecting the increase in the nuclear fraction of STAT3 phosphorylated on Y705. PP2A inhibitors, okadaic acid and fostriecin, did not enhance STAT3 S727 phosphorylation. Surprisingly, calyculin A eliminated LIF-induced gene expression: (1) calyculin A reduced binding of nuclear extracts to a STAT3 consensus site, thereby reducing the overall level of binding observed with LIF; and (2) calyculin A caused p300/CBP phosphorylation, thus resulting in reduced acetylation activity and degradation. Together, these findings reveal a pivotal role of a protein serine/threonine phosphatases that is likely PP1 in HMEC in controlling STAT3 transcriptional activity.
Introduction
Vascular inflammation occurs in coronary heart disease, myocardial infarction, arteriosclerosis, atherosclerosis, systolic/diastolic heart failure, metabolic syndrome, diabetes, and hypertension (López Farré and Casado 2001; Yung and others 2006; Coccheri 2007; Ganne and Winer 2008; Dawood and Schlaich 2009; Lakshmi and others 2009). Inflammation is often initiated by stimuli, such as the interleukin 6 (IL-6) type cytokines, acting on endothelial cells to enhance reactive oxygen species (ROS) generation, as well as leukocyte chemotaxis and adherence (Nian and others 2004; Hou and others 2008; Brasier 2010). The IL-6 type cytokines include IL-6, IL-11, leukemia inhibitory factor (LIF), cardiotrophin 1, oncostatin M, ciliary neurotrophic factor, and cardiotrophin-like cytokine (Kurdi and Booz 2007). On binding to their cell surface receptors, these cytokines activate several intracellular signaling events, notably the Janus kinase 1 (JAK1)-signal transducer and activator of transcription 3 (STAT3) pathway. STAT3 is a transcription factor that is activated by phosphorylation of tyrosine residue 705 (Y705). After phosphorylation, STAT3 forms homodimers or heterodimers with other STAT family members that bind specific promoters to induce target gene expression (Kurdi and Booz 2007). STAT3 is also phosphorylated by various kinases on serine residue 727 (S727) within the C-terminus transcription activation domain. Previous reports have shown that S727 phosphorylation is required for maximal transcriptional activity and DNA binding of STAT3, as well as STAT3 homodimerization (Zhang and others 1995; Kurdi and Booz 2007).
Others have reported that treatment of ALK+ TCL cells, glioblastoma multiforme cells, 293T cells, human antigen-specific CD4+ T cell lines, and cutaneous T cell lymphoma lines with the PP1/PP2A inhibitor calyculin A caused a marked increase in STAT3 S727 phosphorylation (Woetmann and others 1999; Zhang and others 2002a; Ghosh and others 2005). In this study, we tested the hypothesis that by simultaneously increasing nuclear STAT3 S727 and Y705 phosphorylation with calyculin A and LIF, we could enhance STAT3-related gene expression in human microvascular endothelial cells (HMEC). Unexpectedly, we observed contrary findings that reveal a novel point of control for STAT3-mediated gene response which has significance for understanding the inflammatory process.
Materials and Methods
Materials
Tissue culture reagents were from Invitrogen. Fetal bovine serum (FBS, SH30070.03) was from Thermo Scientific. Okadaic acid, xanthine, and protease inhibitor cocktail for use with mammalian cell and tissue extracts were from Sigma-Aldrich. Antibodies for STAT3, STAT3 pY705, histone H4, and LSD1 were from Cell Signaling Technology. The antibody against pS727 STAT3 was from Millipore. Fostriecin and antibodies for Ac-histone H4 K5, histone H1, p300, phospho-p300 S89, and GAPDH were from Santa Cruz Biotechnology. RIPA-based kinase extraction buffer and activated vanadate were from Boston Bioproducts. Calyculin A was from Santa Cruz Biotechnology, and Sigma-Aldrich. Xanthine oxidase from buttermilk was obtained from EMD Chemicals. Binding of nuclear extracts to a STAT3 consensus oligonucleotide was measured using the TransAM STAT3 kit from Active Motif. Nuclear extraction kits were from Active Motif (STAT3 oligonucleotide binding) and Thermo Scientific (Westerns). RNA was extracted with the RNAqueous kit from Ambion.
Cell culture
HMEC-1 were obtained from the Centers for Disease Control and Prevention. Cells were cultured in MCDB 131 medium with 15% FBS, 10 ng/mL epidermal growth factor, 10 mM glutamine, 1 μg/mL hydrocortisone, and antibiotic-antimycotic. For experiments, cells were grown to near confluency on 60 or 100 mm diameter culture dishes. Twelve to 15 h beforehand, growth medium was replaced with medium containing 0.5% FBS.
Western blots
Whole-cell lysates were prepared by scraping cells into ice-cold RIPA-based buffer containing 100 mM vanadate and protease inhibitor cocktail and were cleared by centrifugation at 100,000 g for 20 min at 4°C. Equal amounts of protein in Laemmli's-sodium dodecyl sulfate (SDS) reducing buffer were separated by SDS-PAGE. Separated proteins were blotted onto nitrocellulose membranes, and the immunoreactive bands were quantified using the Li-COR Odyssey infrared imaging system.
Real-time polymerase chain reaction
cDNA was prepared using SuperScript VILO cDNA synthesis Kit from Invitrogen. Gene amplification was performed using TaqMan Gene Expression Master Mix and TaqMan Gene expression assays from Applied Biosystems. TaqMan Gene Expression primers used were suppressor of cytokine signaling 3 (SOCS3) (Hs02330328_s1), chemokine (C-C motif) ligand 2 (CCL2) (Hs00234140_m1), and GAPDH (Hs99999905_m1). Real-time polymerase chain reaction was carried out in a BioRad iQ5 multicolor machine. Normalization of gene expression was carried out with housekeeping gene GAPDH. Data were analyzed using the iQTM 5 optical system software provided by Biorad.
Statistical analysis
Results are expressed as mean±SEM for n number of independent experiments. The data in Figs. 2 and 8 were analyzed by 2-way analysis of variance (ANOVA) and paired t-test, respectively. In all other cases, statistical significance was assessed by one-way ANOVA followed by an appropriate post hoc test. P≤0.05 was taken as significant.
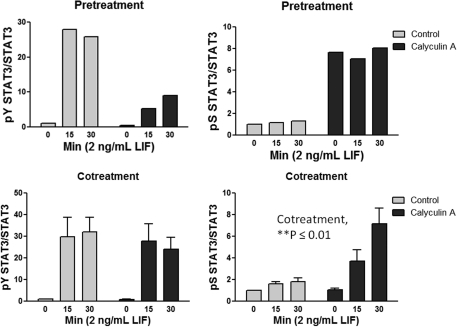
FIG. 2.
Pretreatment versus cotreatment with CA on LIF-induced STAT3 signaling. Results from the experiments described in Fig. 1 were quantified using the LiCor Odyssey system. Values shown are the mean of 2 independent experiments (Pretreatment) or mean±SEM for 3 independent experiments (Cotreatment). The results for Cotreatment were analyzed by 2-way analysis of variance with CA treatment and time as variables: For pY STAT3, time was significant as source of variation (P≤0.001), whereas cotreatment (CA) and cotreatment-time interaction were not, thus indicating that CA cotreatment did not affect LIF-induced STAT3 Y705 phosphorylation. For pS STAT3, cotreatment (CA), time, and cotreatment-time interaction as sources of variation were significant at P≤0.01, P≤0.01, and P≤0.05, respectively, thus indicating that cayculin A cotreatment resulted in greater STAT3 S727 phosphorylation in response to LIF stimulation in a time-dependent manner.
FIG. 8.
Oxidative stress increases STAT3 and p300 phosphorylation. HMEC-1 cells were incubated for 6 h in the absence or presence of 20 U/L xanthine oxidase and 500 μM xanthine. Cell extracts were prepared and analyzed for STAT3 Y705 and S727 phosphorylation and p300 S89 phosphorylation. (A) Representative blots from 3 independent experiments. (B) The fraction of phosphorylated to total protein (pY705/STAT3, pS727/STAT3, and pS89/p300) was determined using the LiCor Odyssey detection system and for each experiment normalized to the control. Results are mean±SEM of 3 independent experiments. ***P<0.001 and *P<0.05 versus control.
Results
LIF-induced STAT3 phosphorylation in endothelial cells
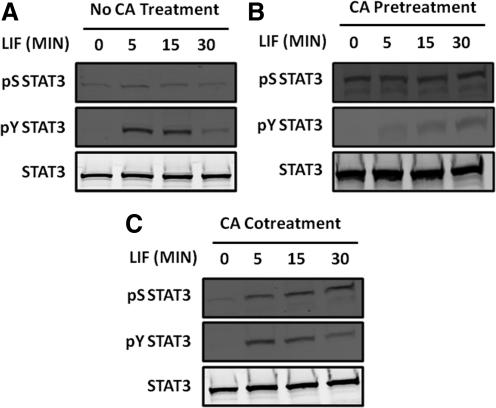
Initial studies were performed to determine the appropriate dosing regimen with calyculin A to assess the impact of enhanced S727 phosphorylation on LIF-induced STAT3 activation. Treatment of HMEC-1 with LIF (2 ng/mL) produced a time-dependent increase in phosphorylation levels of both Y705 and S727 STAT3 with the latter being less intense of the 2 (Fig. 1A). Pretreatment with calyculin A enhanced basal levels of STAT3 S727 phosphorylation, but LIF-induced Y705 phosphorylation was markedly attenuated under this condition (Fig. 1B). We had previously made a similar observation using cardiac myocytes, and provided evidence that S727 STAT3 phosphorylation serves to recruit a tyrosine phosphatase (Booz and others 2003). However, when cells were co-treated with LIF and calyculin A, we were able to produce an increase in STAT3 S727 phosphorylation without any reduction in LIF-induced Y705 phosphorylation (Fig. 1C). For that reason, cotreatment with calyculin A and LIF was performed in subsequent experiments. These findings are quantified in Fig. 2.
FIG. 1.
Differential effect of pretreatment versus cotreatment with calyculin A (CA) on LIF-induced STAT3 signaling. HMEC-1 were (A) treated with 2 ng/mL LIF for various times, (B) pretreated for 60 min with 100 nM CA and then treated with 2 ng/mL LIF, or (C) treated with 2 ng/mL LIF for various times along with 100 nM CA. Proteins were extracted, and levels of pY705 STAT3, pS727 STAT3, and STAT3 were evaluated by Western blotting. Blots shown are representative of (A) 5, (B) 2, and (C) 3 independent experiments. STAT3, signal transducer and activator of transcription 3; HMEC, human microvascular endothelial cells; LIF, leukemia inhibitory factor; DMSO, dimethyl sulfoxide.
Calyculin A inhibits the activity of both PP1 and PP2A (Ishihara and others 1989; Resjö and others 1999). To address which of these PP was involved in STAT3 S727 dephosphorylation, we used okadaic acid, which inhibits PP2A activity but has little effect on PP1 activity (Resjö and others 1999). As seen from Fig. 3A, treatment with okadaic acid (1 μM) failed to further increase STAT3 S727 phosphorylation when cotreated with LIF. We also tested the effect of fostriecin on STAT3 S727 phosphorylation. Fostriecin is a selective inhibitor of PP2A (IC50=3.2 nM) with little effect on PP1 (IC50=131 μM) (Walsh and others 1997) and was reported to inhibit PP2A in human endothelial cells (Grethe and Pörn-Ares 2006). As seen from Fig. 3B and C, cotreatment with fostriecin had no effect on STAT3 Y705 or S727 phosphorylation.
FIG. 3.
PP2A inhibitor cotreatment did not enhance STAT3 S727 phosphorylation. (A) HMEC-1 were treated with 2 ng/mL LIF for various times along with 1 μM okadaic acid (OA) or vehicle (0.04% v/v DMSO). Proteins were extracted, and levels of pY705 STAT3, pS727 STAT3, and STAT3 were evaluated by Western blotting. Results shown are representative of 2 independent experiments. (B) HMEC-1 were treated with vehicle or 2 ng/mL LIF in the presence or absence of 1 μM fostriecin for 2 h. Proteins were extracted, and levels of pY705 STAT3, pS727 STAT3, and STAT3 were evaluated by Western blotting. (C) Levels of pS727 STAT3 and pY705 STAT3 were normalized to total STAT3 using the LiCor Odyssey system. Values are mean±SEM for 3 independent experiments. ***P≤0.001 versus control or fostriecin. PP2A, protein phosphatase 2A.
Increased STAT3 S727 phosphorylation with calyculin A cotreatment did not enhance gene expression
To assess the effect of enhancing STAT3 S727 phosphorylation on LIF-induced gene expression, we selected 2 genes that are activated by LIF in HMEC-1 cells and which contain a STAF_HOXF_STAT framework in their promoters (unpublished observation), namely, SOCS3 and CCL2, also known as monocyte chemotactic protein-1 (MCP-1). As seen from Fig. 4, contrary to our expectations, enhancing STAT3 S727 phosphorylation by cotreatment with calyculin A did not enhance LIF-induced expression of these genes, but rather a marked inhibition was observed.
FIG. 4.
CA inhibited LIF-induced SOCS3 and CCL2 gene expression. HMEC-1 were treated for 1 h with nothing (Control), 0.04% v/v DMSO (vehicle control), 100 nM CA, 2 ng/mL LIF, or LIF with DMSO or with CA. RNA was extracted, reverse transcribed, and analyzed by real-time polymerase chain reaction for SOCS3 and CCL2 expression. Results were normalized to GAPDH and expressed as the fold increase over control levels. Values are mean±SEM for 4 independent experiments. ***P≤0.001 versus LIF or LIF + DMSO.
Increased S727-STAT3 phosphorylation did not impact STAT3 nuclear localization
To address the possibility that STAT3 S727 phosphorylation caused nuclear export of STAT3 as reported by others (Woetmann and others 1999), we isolated nuclei after LIF and/or calyculin A treatment. Notably, calyculin A co-treatment resulted in a marked increase in the proportion of nuclear STAT3 that was phosphorylated on S727 without changing the proportion which was phosphorylated on Y705 in response to LIF stimulation (Fig. 5A, B). In addition, as Fig. 5C shows, there was no significant difference between LIF and LIF-calyculin A treatments in nuclear STAT3 levels. By itself, calyculin A did not significantly affect nuclear STAT3 levels. Nuclear STAT3 levels assessed at 60 min did show a trend toward being higher with LIF treatment, but this did not reach statistical significance.
FIG. 5.
CA did not affect nuclear STAT3 levels. Cells were treated with vehicle, 100 nM CA, 2 ng/mL LIF (with vehicle), or LIF + CA. (A) After 1 h, nuclear extracts were prepared and analyzed by Western blotting for STAT3 pS727, STAT3 pY705, STAT3, and LSD1 or histone H1 (not shown) as a loading control. Results were quantified using the LiCor Odyssey system and for each experiment normalized to the control. (B) pS727 STAT3 and pY705 to total STAT3 ratios. For pS STAT3/STAT3: *P≤0.05 versus control or LIF, **P<0.01 versus control or LIF. For pY STAT3/STAT3: *P<0.05 versus control or CA, ***P<0.001 versus control or CA. (C) Nuclear STAT3 levels were normalized to histone H1 or LSD1 levels as a loading control. Values are mean±SEM of 9 independent experiments.
PP1 inhibition decreased STAT3 DNA binding ability
Since calyculin A did not have any significant effect on STAT3 nuclear levels, we considered the possibility that the decrease in LIF-induced SOCS3 and CCL2 gene expression was due to impaired DNA binding of STAT3. To address that possibility, nuclear extracts were assessed for their ability to bind an oligonucleotide containing STAT3 consensus binding sites. Figure 6 shows that calyculin A treatment alone produced a significant decrease in STAT3 nuclear binding. As expected, LIF treatment enhanced STAT3 binding. Binding resulting from co-treatment, although markedly enhanced compared with calyculin A treatment alone, was no different from vehicle control. These results indicate that calyculin A causes changes in STAT3 that attenuate DNA binding.
FIG. 6.
CA cotreatment inhibited LIF-induced binding of nuclear extracts to a STAT3 consensus binding sequence. Cells were treated for 1 h with 0.04% DMSO (control), 100 nM CA, 2 ng/mL LIF with DMSO, or LIF with 100 nM CA. Nuclear extracts (20 μg protein) were assayed for binding to a STAT3 consensus binding site using the TransAM STAT3 kit from Active Motif. Values are mean±SEM of 4 independent experiments. Values labeled with the same letter are significantly different from one another. a,b,cP<0.05, dP<0.001, and eP<0.01.
Calyculin A causes p300 inhibitory phosphorylation and degradation
The first function identified for the phosphorylation of the S727 residue in STAT3 was the recruitment of transcriptional co-activating proteins p300/CBP (Schuringa and others 2001; Ray and others 2002). We, thus, asked whether the complete block by calyculin A of LIF-induced gene expression could be due, in part, to an inhibitory action on p300/CBP. As Fig. 7A and B show, treatment of HMEC-1 with calyculin A caused a marked increase in the phosphorylation of p300/CBP on S89, which others have reported to be associated with inhibition of acetyltransferase activity (Yuan and Gambee 2000). Fostriecin (1 μM) by itself or with LIF had no effect on p300 S89 phosphorylation or degradation (data not shown). Moreover, as can be seen from Fig. 7A and C, calyculin A caused a remarkable decrease in the protein levels of p300/CBP. To further establish that calyculin A was targeting p300/CBP, we looked at the effect of this inhibitor on histone H4 acetylation at K5, as p300/CBP preferentially acetylates Histone H4 at this site and at K8 (Schiltz and others 1999). As Fig. 7D shows, calyculin A markedly reduced the fraction of histone H4 acetylated on K5.
FIG. 7.
CA causes phosphorylation of p300/CBP on S89 and p300/CBP degradation. (A) Cells were treated for 1 h with 0.04% DMSO (control), 100 nM CA, 2 ng/mL LIF with DMSO, or LIF with 100 nM CA. Cell extracts were analyzed by Western immunoblotting for p300 pS89, p300, and GAPDH (loading control). Results were quantified using the LiCor Odyssey system and show that (B) CA caused p300 phosphorylation on p89 and (C) p300 degradation regardless of the presence or absence of LIF. (D) Cell extracts were analyzed by Western immunoblotting for Ac-histone H4 K5 and histone H4. Values are mean±SEM of 3 independent experiments. ***P<0.001 versus control or LIF, *P<0.05 versus control.
Oxidative stress increases STAT3 and p300 phosphorylation
We next sought to investigate whether phosphorylation of STAT3 S727 and p300 S89 that is associated with PP1 inhibition might have pathological or physiological relevance. Others have reported that PP1 is inhibited by oxidative stress (O'Loghlen and others 2003). We, thus, asked whether long-term exposure to oxidative stress would affect STAT3 and p300 phosphorylation. HMEC-1 were incubated with xanthine oxidase and xanthine to generate ROS. As Fig. 8 shows, exposure of HMEC-1 to oxidative stress produced a modest but significant increase in both STAT3 S727 and p300 p89 phosphorylation. Notable oxidative stress produced a marked increase in STAT3 Y705 phosphorylation, which is in keeping with the well-established observation that protein tyrosine phosphatases are inhibited in cellular systems by ROS (Winterbourn and Hampton 2008).
Discussion
We report here that the PP1 inhibitor, calyculin A, caused a marked increase in phosphorylation of STAT3 on S727, which had been previously linked to enhanced gene expression by STAT3 at certain promoters (Zhang and others 1995; Schuringa and others 2001; Ray and others 2002; Kurdi and Booz 2007). Okadaic acid and fostriecin, which exhibit almost an exclusively inhibitory effect on PP2A, did not cause a similar increase in STAT3 S727 phosphorylation, thus showing that PP1 or PP1-related protein is the phosphatase responsible for STAT3 S727 dephosphorylation in HMEC-1. Although several studies have implicated PP2A in dephosphorylating STAT3 S727 (Liang and others 1999; Woetmann and other 1999; Zhang and others 2002a; Togi and others 2009), a recent study found evidence that PP1 and not PP2A is responsible for STAT3 S727 dephosphorylation in a variety of human tumor cell lines (Haridas and others 2009).
The role of STAT3 S727 that lies within the transcription activation domain is complicated and best described as myriad. Evidence has been reported supporting the conclusion that phosphorylation of this residue regulates STAT3 subcellular distribution (Woetmann and others 1999), tyrosine phosphorylation (possibly through recruitment of a phosphatase) (Zhang and others 2002a; Booz and others 2003), cofactor recruitment (e.g., histone deacetylase p300 and Sp1) (Schuringa and others 2001; Yang and others 2005), maximal transcriptional activity (Zhang and others 1995), receptor binding (Zhang and others 2002b), and homodimerization (Zhang and others 1995).
Based on what has been reported, one might expect that simultaneous Y705 and S727 phosphorylation would enhance STAT3-related gene expression. However, we observed just the opposite, which was due in part to the inhibition and degradation of p300/CBP seen with calyculin A. Most likely, these actions occurred indirectly as the result of enhanced (stress) kinase signaling that would occur with inhibition of the repressive phosphatase actions of PP1 on these signaling pathways. Factors that control the stability of p300/CBP are poorly understood. In cardiac myocytes, the anticancer agent doxorubicin was found to cause p300 hyperphosphorylation and degradation via the p38 mitogen-activated protein kinases (Poizat and others 2005). Protein kinase Cδ, AMP-activated protein kinase, and salt inducible kinase 2 were reported to phosphorylate p300 on S89, thereby attenuating its ability to function as an acetylase (Yang and others 2001; Yuan and others 2002; Bricambert and others 2010). Whether p300/CBP is a direct target of PP1 is a question for future investigation.
Inhibition of DNA binding also partially explains the effect of calyculin A on STAT3-related gene expression. We observed that treatment with calyculin A alone strongly reduced binding of nuclear extracts to a STAT3 consensus binding site (Fig. 6), although nuclear levels of STAT3 were not affected (Fig. 5C). Others reported that calyculin A reduced binding of STAT3 from cutaneous T lymphoma cells to various STAT3 binding elements (Woetmann and others 1999). The explanation suggested was that increased serine phosphorylation of STAT3 caused decreased STAT3 tyrosine phosphorylation and consequently reduced STAT3 DNA binding. However, we did not observe a marked decrease in STAT3 phosphorylated on Y705 in response to calyculin A (Fig. 5B). An alternative explanation may be increased phosphorylation of STAT3 at 2 other sites, namely S691 or T714, which are known to be affected by DNA damage or the cell cycle, respectively. In fact, calyculin A was reported to cause increased threonine phosphorylation of STAT3 (Woetmann and others 1999). Although S691 and T714 phosphorylation likely have physiological relevance, to the best of our knowledge, the effect of phosphorylation of these sites on STAT3 DNA binding has not been reported and warrants investigation. In any case, the results shown in Fig. 6 indicate that the positive effect of STAT3 Y705 phosphorylation on DNA binding can offset any potential diminution in DNA binding produced by phosphorylation at another site.
LIF-induced STAT3 nuclear levels at 60 min tended to be higher in the absence of calyculin A (Fig. 5C). However, the increase did not reach statistical significance. Although STAT3 was originally identified as a latent cytoplasmic transcription factor that translocates to the nucleus on cytokine-induced Y705 phosphorylation, STAT3 is now known to constitutively shuttle between the cytoplasm and nucleus independent of its phosphorylation state (Sehgal 2008). Thus, subcellular distribution is unlikely to explain our findings on impaired LIF-induced SOCS3 and CCL2 gene expression on cotreatment with calyculin A (Fig. 4). Both SOCS3 and CCL2 contain a STAF_HOXF_STAT framework within their promoters, and their induction is commonly associated with an inflammatory state. SOCS3 acts as a classic negative inhibitor of JAK-STAT signaling, whereas CCL2 is important for the recruitments of lymphocytes to sites of tissue injury and inflammation. Our results raise the possibility that expression of CCL2 and SOCS3 may be tamped or kept under control with increasing stress by attenuation of PP1 activity.
Although we demonstrated that oxidative stress mimicked the effects of calyculin A on STAT3 and p300 phosphorylation, other stress stimuli may be involved as well. Notably, PP1 inhibition by calyculin A in lung microvascular cells has been linked to disruption of barrier function, thus suggesting a role for this phosphatase in cell architecture (Kelly and others 1998). Consistent with that possibility, we observed that HMEC-1 seemed to adopt a more rounded appearance in response to treatment with calyculin A (unpublished observation). The regulatory mechanism described here may come into play in attenuating certain aspects of IL-6 type cytokine inflammatory signaling when other stresses are present in the cell.
In conclusion, our findings highlight the pivotal role of a serine threonine phosphatase that is most likely PP1 in controlling the nuclear actions of STAT3 by balancing opposing signaling processes (Fig. 9). On one hand, PP1 exerts positive actions on the transcriptional activity of STAT3 by preventing p300/CBP phosphorylation and degradation, and potentially opposing phosphorylation of STAT3 at 1 or 2 inhibitory sites. On the other hand, PP1 serves to attenuate STAT3 transcriptional activity by dephosphorylating the site responsible for p300/CBP recruitment. p300/CBP enhances STAT3 transcriptional activity by allowing for the acetylation of histone at certain promoters containing STAT3 responsive elements (Ray and others 2002; Ray and others 2008). Understanding how these opposing signaling events are coordinated and affected by stress could lead to novel therapeutic strategies to control the impact of inflammation on microvascular endothelial cells.
FIG. 9.
Scheme illustrating the pivotal role of PP1 in controlling the actions of STAT3 on gene expression in HMEC-1 by exerting both positive and negative actions. STAT3 transcriptional activation by the IL-6 type cytokines is due to enhanced phosphorylation of S727 and Y705. PP1 attenuates STAT3 transcriptional activity by dephosphorylating the site (S727) responsible for p300/CBP recruitment, but potentiates STAT3 DNA binding by dephosphorylating other sites (S691 and T714) in the C-terminus of STAT3. PP1 also favors STAT3-mediated transcription by opposing phosphorylation and subsequent inhibition and degradation of p300/CBP. Under conditions of PP1 inhibition (occurring perhaps with oxidative stress), increased STAT3 DNA binding due to S727 phosphorylation would be offset by p300 phosphorylation and degradation due to stress kinase activation. In the absence of an additional (stress) signal, IL-6 type cytokine-induced STAT3 activation occurs in the absence of p300 S89 phosphorylation. IL, interleukin.
Acknowledgments
The authors gratefully acknowledge the technical assistance of Mr. Hani Jamal Alturkmani with some of the experiments. This work was supported by grants from the National Heart, Lung, and Blood Institute (5R01HL088101-03 and 3R01HL088101-02S1) to GWB and from the Lebanese University, the Lebanese National Council for Scientific Research (CNRS # 05-10-09), and the COMSTECH-TWAS (09-122 RG/PHA/AF/AC_C) to M.K.
Author Disclosure Statement
No competing financial interests exist.
References
- Booz GW. Day JN. Baker KM. Angiotensin II effects on STAT3 phosphorylation in cardiomyocytes: evidence for Erk-dependent Tyr705 dephosphorylation. Basic Res Cardiol. 2003;98:33–38. doi: 10.1007/s00395-003-0387-x. [DOI] [PubMed] [Google Scholar]
- Brasier AR. The nuclear factor-κB-interleukin-6 signalling pathway mediating vascular inflammation. Cardiovasc Res. 2010;86:211–218. doi: 10.1093/cvr/cvq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricambert J. Miranda J. Benhamed F. Girard J. Postic C. Dentin R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest. 2010;120:4316–4331. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccheri S. Approaches to prevention of cardiovascular complications and events in diabetes mellitus. Drugs. 2007;67:997–1026. doi: 10.2165/00003495-200767070-00005. [DOI] [PubMed] [Google Scholar]
- Dawood T. Schlaich MP. Mediators of target organ damage in hypertension: focus on obesity associated factors and inflammation. Minerva Cardioangiol. 2009;57:687–704. [PubMed] [Google Scholar]
- Ganne S. Winer N. Vascular compliance in the cardiometabolic syndrome. J Cardiometab Syndr. 2008;3:35–39. doi: 10.1111/j.1559-4572.2008.05630.x. [DOI] [PubMed] [Google Scholar]
- Ghosh MK. Sharma P. Harbor PC. Rahaman SO. Haque SJ. PI3K-AKT pathway negatively controls EGFR-dependent DNA-binding activity of Stat3 in glioblastoma multiforme cells. Oncogene. 2005;24:7290–7300. doi: 10.1038/sj.onc.1208894. [DOI] [PubMed] [Google Scholar]
- Grethe S. Pörn-Ares MI. p38 MAPK regulates phosphorylation of Bad via PP2A-dependent suppression of the MEK1/2-ERK1/2 survival pathway in TNF-α induced endothelial apoptosis. Cell Signal. 2006;18:531–540. doi: 10.1016/j.cellsig.2005.05.023. [DOI] [PubMed] [Google Scholar]
- Haridas V. Nishimura G. Xu ZX. Connolly F. Hanausek M. Walaszek Z. Zoltaszek R. Gutterman JU. Avicin D: a protein reactive plant isoprenoid dephosphorylates Stat 3 by regulating both kinase and phosphatase activities. PLoS One. 2009;4:e5578. doi: 10.1371/journal.pone.0005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T. Tieu BC. Ray S. Recinos Iii A. Cui R. Tilton RG. Brasier AR. Roles of IL-6-gp130 signaling in vascular inflammation. Curr Cardiol Rev. 2008;4:179–192. doi: 10.2174/157340308785160570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H. Martin BL. Brautigan DL. Karaki H. Ozaki H. Kato Y. Fusetani N. Watabe S. Hashimoto K. Uemura D. Hartshorne DJ. Calyculin A and okadaic acid: inhibitors of protein phosphatase activity. Biochem Biophys Res Commun. 1989;159:871–877. doi: 10.1016/0006-291x(89)92189-x. [DOI] [PubMed] [Google Scholar]
- Kelly JJ. Moore TM. Babal P. Diwan AH. Stevens T. Thompson WJ. Pulmonary microvascular and macrovascular endothelial cells: differential regulation of Ca2+ and permeability. Am J Physiol. 1998;274:L810–L819. doi: 10.1152/ajplung.1998.274.5.L810. [DOI] [PubMed] [Google Scholar]
- Kurdi M. Booz GW. Can the protective actions of JAK-STAT in the heart be exploited therapeutically? Parsing the regulation of interleukin-6-type cytokine signaling. J Cardiovasc Pharmacol. 2007;50:126–141. doi: 10.1097/FJC.0b013e318068dd49. [DOI] [PubMed] [Google Scholar]
- Lakshmi SV. Padmaja G. Kuppusamy P. Kutala VK. Oxidative stress in cardiovascular disease. Indian J Biochem Biophys. 2009;46:421–440. [PubMed] [Google Scholar]
- Liang H. Venema VJ. Wang X. Ju H. Venema RC. Marrero MB. Regulation of angiotensin II-induced phosphorylation of STAT3 in vascular smooth muscle cells. J Biol Chem. 1999;274:19846–19851. doi: 10.1074/jbc.274.28.19846. [DOI] [PubMed] [Google Scholar]
- López Farré A. Casado S. Heart failure, redox alterations, and endothelial dysfunction. Hypertension. 2001;38:1400–1405. doi: 10.1161/hy1201.099612. [DOI] [PubMed] [Google Scholar]
- Nian M. Lee P. Khaper N. Liu P. Inflammatory cytokines and postmyocardial infarction remodeling. Circ Res. 2004;94:1543–1553. doi: 10.1161/01.RES.0000130526.20854.fa. [DOI] [PubMed] [Google Scholar]
- O'Loghlen A. Pérez-Morgado MI. Salinas M. Martín ME. Reversible inhibition of the protein phosphatase 1 by hydrogen peroxide. Potential regulation of eIF2α phosphorylation in differentiated PC12 cells. Arch Biochem Biophys. 2003;417:194–202. doi: 10.1016/s0003-9861(03)00368-0. [DOI] [PubMed] [Google Scholar]
- Poizat C. Puri PL. Bai Y. Kedes L. Phosphorylation-dependent degradation of p300 by doxorubicin-activated p38 mitogen-activated protein kinase in cardiac cells. Mol Cell Biol. 2005;25:2673–2687. doi: 10.1128/MCB.25.7.2673-2687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S. Lee C. Hou T. Boldogh I. Brasier AR. Requirement of histone deacetylase1 (HDAC1) in signal transducer and activator of transcription 3 (STAT3) nucleocytoplasmic distribution. Nucleic Acids. 2008;36:4510–4520. doi: 10.1093/nar/gkn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S. Sherman CT. Lu M. Brasier AR. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol Endocrinol. 2002;16:824–836. doi: 10.1210/mend.16.4.0811. [DOI] [PubMed] [Google Scholar]
- Resjö S. Oknianska A. Zolnierowicz S. Manganiello V. Degerman E. Phosphorylation and activation of phosphodiesterase type 3B (PDE3B) in adipocytes in response to serine/threonine phosphatase inhibitors: deactivation of PDE3B in vitro by protein phosphatase type 2A. Biochem J. 1999;341:839–845. [PMC free article] [PubMed] [Google Scholar]
- Schiltz RL. Mizzen CA. Vassilev A. Cook RG. Allis CD. Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Schuringa JJ. Schepers H. Vellenga E. Kruijer W. Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett. 2001;495:71–76. doi: 10.1016/s0014-5793(01)02354-7. [DOI] [PubMed] [Google Scholar]
- Sehgal PB. Paradigm shifts in the cell biology of STAT signaling. Semin Cell Dev Biol. 2008;19:329–340. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togi S. Kamitani S. Kawakami S. Ikeda O. Muromoto R. Nanbo A. Matsuda T. HDAC3 influences phosphorylation of STAT3 at serine 727 by interacting with PP2A. Biochem Biophys Res Commun. 2009;379:616–620. doi: 10.1016/j.bbrc.2008.12.132. [DOI] [PubMed] [Google Scholar]
- Walsh AH. Cheng A. Honkanen RE. Fostriecin, an antitumor antibiotic with inhibitory activity against serine/threonine protein phosphatases types 1 (PP1) and 2A (PP2A), is highly selective for PP2A. FEBS Lett. 1997;416:230–234. doi: 10.1016/s0014-5793(97)01210-6. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC. Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Woetmann A. Nielsen M. Christensen ST. Brockdorff J. Kaltoft K. Engel AM. Skov S. Brender C. Geisler C. Svejgaard A. Rygaard J. Leick V. Odum N. Inhibition of protein phosphatase 2A induces serine/threonine phosphorylation, subcellular redistribution, and functional inhibition of STAT3. Proc Natl Acad Sci USA. 1999;96:10620–10625. doi: 10.1073/pnas.96.19.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Hong YH. Shen XQ. Frankowski C. Camp HS. Leff T. Regulation of transcription by AMP-activated protein kinase: phosphorylation of p300 blocks its interaction with nuclear receptors. J Biol Chem. 2001;276:38341–38344. doi: 10.1074/jbc.C100316200. [DOI] [PubMed] [Google Scholar]
- Yang XP. Irani K. Mattagajasingh S. Dipaula A. Khanday F. Ozaki M. Fox-Talbot K. Baldwin WM., 3rd Becker LC. Signal transducer and activator of transcription 3α and specificity protein 1 interact to upregulate intercellular adhesion molecule-1 in ischemic-reperfused myocardium and vascular endothelium. Arterioscler Thromb Vasc Biol. 2005;25:1395–1400. doi: 10.1161/01.ATV.0000168428.96177.24. [DOI] [PubMed] [Google Scholar]
- Yuan LW. Gambee JE. Phosphorylation of p300 at serine 89 by protein kinase C. J Biol Chem. 2000;275:40946–40951. doi: 10.1074/jbc.M007832200. [DOI] [PubMed] [Google Scholar]
- Yuan LW. Soh JW. Weinstein IB. Inhibition of histone acetyltransferase function of p300 by PKCδ. Biochim Biophys Acta. 2002;1592:205–211. doi: 10.1016/s0167-4889(02)00327-0. [DOI] [PubMed] [Google Scholar]
- Yung LM. Leung FP. Yao X. Chen ZY. Huang Y. Reactive oxygen species in vascular wall. Cardiovasc Hematol Disord Drug Targets. 2006;6:1–19. doi: 10.2174/187152906776092659. [DOI] [PubMed] [Google Scholar]
- Zhang Q. Raghunath PN. Xue L. Majewski M. Carpentieri DF. Odum N. Morris S. Skorski T. Wasik MA. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J Immunol. 2002a;168:466–474. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- Zhang T. Seow KT. Ong CT. Cao X. Interdomain interaction of Stat3 regulates its Src homology 2 domain-mediated receptor binding activity. J Biol Chem. 2002b;277:17556–17563. doi: 10.1074/jbc.M105525200. [DOI] [PubMed] [Google Scholar]
- Zhang X. Blenis J. Li HC. Schindler C. Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–1994. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]