Abstract
Cloning mammals by somatic cell nuclear transfer (SCNT) has become an established procedure, but the success rate remains low and gene expression abnormalities are also observed. In addition, SCNT pups exhibited an abnormal gene expression profile with a high degree of heterogeneity among individuals. Recently, we reported that somatic clones treated with trichostatin A (TSA) exhibited a significantly improved success rate, probably due to its effects on chromatin remodeling and histone modification in early embryos. Here we show that the TSA treatment also improves the long-term consistency of genome-wide gene expression regulation: the total number of genes commonly exhibiting up- or downregulation in the TSA clone pups decreased to half of the conventional SCNT pups, and the variation among individuals observed in the SCNT pups was also reduced to the level of the pups produced by the intracytoplasmic sperm injection (ICSI) method. Interestingly, the total gene expression profile of the TSA clones came to resemble that of the ICSI pups.
Introduction
The success of somatic cell cloning has demonstrated that the somatic cell nuclei can be reprogrammed so as to acquire totipotency. This technique has the distinct characteristic of reprogramming over the whole range of the establishment of induced pluripotent stem (iPS) cells, because somatic cells can be directly reprogrammed to the totipotent state of the zygote in the course of cloning, during which time they are gradually reprogrammed to take on the pluripotent embryonic stem cell-like state in the process of iPS cell establishment. However, the reprogramming molecular mechanism in the somatic cell nuclear transfer (SCNT) procedure has not been elucidated and the somatic cell cloning success rate remains very low. We have reported that the treatment of SCNT embryo with trichostatin A (TSA) results in a significant improvement in the success rate (Kishigami et al., 2006, 2007). Subsequently, significant improvement in cloning efficiency resulting from TSA treatment has also reported in other mammals, such as porcine, bovine, and rabbit models (Iager et al., 2008; Li et al., 2008; Shi et al., 2008). Recently, we reported that TSA enhances the reprogramming of somatic nuclei chromatin remodeling and histone modification in two-cell stage embryos (Bui et al., 2010), whereas the effect on long-term transcriptional regulation has still not been determined. Previously, we demonstrated that, despite their normal appearance, somatic cell-cloned mice exhibited a significantly abnormal gene expression profile in neonatal tissues and that the genes dysregulated in the clones by SCNT varies among the cloned individuals (Kohda et al., 2005). Therefore, to elucidate the long-term effects of TSA treatment on transcriptome regulation in somatic cell cloning, we examined the gene expression profile in the neonatal tissues of TSA clone mice using a DNA microarray method.
Materials and Methods
Ethics statement
All procedures described here were reviewed and approved by the Animal Experimentation Committee at RIKEN and were performed in accordance with the RIKEN Guiding Principles for the Care and Use of Laboratory Animals.
Mice
All mice analyzed in this study were produced by genetically identical crosses, for example, C57BL/6×DBA/2 (BDF1). All neonatal mice were delivered by Cesarean section. After confirmation of the start of respiration at the time of Cesarean section, the pups were sacrificed, and then the liver and brain were collected and immediately frozen in liquid nitrogen and stored at −80°C until use.
Somatic cell cloning
Cumulus cell donor nuclear transfer was performed as described elsewhere (Wakayama et al., 1998). TSA treatment was applied as described previously (Kishigami et al., 2006, 2007). Briefly, 50 nM TSA was added to the activation media for 6 h and then the cells were washed and transferred to KSOM containing 50 nM TSA for 4 h. Following complete washing, zygotes were cultured so as to permit development. Each group of embryos was transferred into pseudopregnant female mice. The mice conceived by in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI) used as the control were produced in the same way as previously described (Bui et al., 2010).
RNA preparation
Total RNA was prepared from neonatal tissues using ISOGEN (Nippon Gene Co. Ltd., Tokyo, Japan), as described previously (Kohda et al., 2005). Total RNA was further purified by the RNeasy Mini kit according to the manufacturer's protocol (Qiagen, Chatsworth, CA, USA).
DNA microarray
Analyses using the DNA microarray were performed with an Agilent system (G4122F; Whole Genome (4×44K) Oligo Microarray). The probes for the microarray were prepared and labeled by Cy3 according to the manufacturer's protocol (Agilent Technologies, Englewood, CO, USA). Arrays were scanned with a G2565BA Microarray Scanner System (Agilent Technologies). The expression levels of the different genes were assessed using “Feature Extract (Agilent Technologies).” The signals were normalized using the qspline algorithm implemented in the Bioconductor package of the statistics program R. To extract transcripts commonly exhibiting a more than twofold upregulation in the four clone pups, the fold change of each signal in the individuals within one group (SCNT and TSA) was calculated against the average of the IVF control and then the mean difference was checked with the Tukey test (p<0.05) using R.
Results
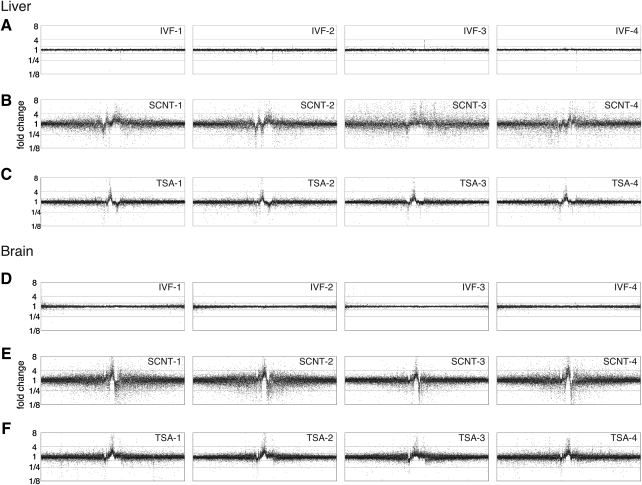
We analyzed the gene expression profile in the neonatal liver of four control pups produced by IVF (Fig. 1A) and four cumulus cell clones (Fig. 1B) using a DNA microarray with 41,174 probes. In the conventional cumulus cell clone neonate, the gene expression profile was substantially changed. The number of the transcripts commonly exhibiting a more than twofold upregulation in the four clone pups was 885, whereas the number exhibiting down regulation was 1550. At the same time, the number of transcripts exhibiting more than twofold up- or downregulation in at least one of the four pups was 4146 and 4212, respectively.
FIG. 1.
Gene expression in the mice conceived by conventional SCNT or SCNT with TSA treatment. The intensity of each signal from the DNA microarray was normalized by dividing it by the mean signal intensity of the IVF samples. If the value exceeded 1, it was used as the fold increase; if the value was less than 1, the inverse was used as the fold decrease. Genes with similar expression patterns among the ICSI individuals were clustered using Cluster 3.0, and the fold change was plotted in the gene order resulting from the cluster analysis. The gene expression profile of neonatal liver (A–C) and brain (D–F) in four samples of the control IVF (A, D), conventional SCNT (B, E), and SCNT with TSA (C, F) were plotted.
Next, we analyzed the cumulus cell clones treated with TSA in the same manner (Fig. 1C), and found that the number of the transcripts that commonly exhibited a more than twofold upregulation in the four clone pups was 781 and for twofold downregulation was 612. The genes that exhibited aberrant upregulation in the conventional clones were unchanged in number, whereas the genes exhibiting downregulation in the SCNT clones was reduced by the TSA treatment. The transcripts with and without the normalization resulting from the TSA treatment are summarized in Tables 1 and 2.
Table 1.
Genes Exhibiting Increase or Decrease in SCNT Pup Liver and Normalized in TSA
| |
Fold change |
|
Fold change |
||
|---|---|---|---|---|---|
| Gene name | SCNT | TSA | Gene name | SCNT | TSA |
| U20264 | 0.10 | 0.76 | NAP124059-1 | 5.53 | 1.47 |
| AK087779 | 0.16 | 1.05 | Hist1h3aa | 4.96 | 0.84 |
| AK051522 | 0.16 | 0.61 | 1700031C06Rik | 4.61 | 1.80 |
| AK040092 | 0.18 | 0.73 | Calca | 4.36 | 1.09 |
| 2610027H17Rik | 0.19 | 1.08 | Serpina6 | 3.99 | 0.83 |
| AK038045 | 0.19 | 0.59 | Ifi27a | 3.78 | 1.43 |
| AK050807 | 0.19 | 0.66 | A_52_P1101503 | 3.77 | 1.39 |
| AK050242 | 0.20 | 0.54 | Rmrp | 3.73 | 1.72 |
| NM_001013789 | 0.20 | 0.69 | G0s2 | 3.70 | 1.74 |
| Chka | 0.21 | 0.61 | Gata3 | 3.68 | 1.15 |
| Stra6 | 0.21 | 0.97 | Sox9 | 3.67 | 1.94 |
| AK046412 | 0.21 | 0.60 | Guca1a | 3.64 | 1.74 |
| AK051621 | 0.21 | 0.60 | Pfkp | 3.58 | 1.61 |
| AK084873 | 0.21 | 1.11 | Tnf | 3.30 | 1.28 |
| Epas1 | 0.21 | 0.55 | Ldhc | 3.26 | 1.15 |
| Tanc1 | 0.22 | 0.60 | Oosp1 | 3.23 | 1.49 |
| AK083952 | 0.22 | 0.70 | AV231787 | 3.22 | 1.33 |
| AK076939 | 0.22 | 0.55 | Tnnc1 | 3.19 | 1.31 |
| AK085204 | 0.22 | 1.01 | C130050O18Rik | 3.15 | 1.22 |
| 4930588G05Rik | 0.23 | 0.98 | Bmp10 | 3.15 | 1.21 |
| AK033340 | 0.23 | 0.85 | 2610035D17Rik | 3.15 | 0.75 |
| AK048657 | 0.23 | 0.55 | Smpdl3b | 3.14 | 1.39 |
| AK079436 | 0.23 | 0.70 | Olfr74 | 3.12 | 1.25 |
| AK079230 | 0.23 | 0.84 | B230317C12Rik | 3.11 | 1.01 |
| AK035046a | 0.23 | 0.88 | Ahnak | 3.09 | 1.23 |
| TC1469196 | 0.23 | 0.77 | Aqp7 | 3.07 | 1.24 |
| AK038328a | 0.23 | 0.93 | AK014119 | 3.05 | 1.25 |
| Tns1 | 0.24 | 0.64 | Tcrg | 3.03 | 0.88 |
| Gfod1 | 0.24 | 0.55 | Ksr1 | 3.02 | 1.11 |
| Nrip1 | 0.24 | 0.50 | Cst8 | 3.00 | 1.56 |
| AK050555 | 0.24 | 0.65 | 2310043J07Rik | 2.99 | 0.97 |
| AK031434 | 0.24 | 0.77 | Mras | 2.98 | 1.58 |
| Mtss1 | 0.24 | 1.11 | Vdr | 2.94 | 0.79 |
| AK047526 | 0.24 | 0.78 | Stambpl1 | 2.92 | 1.67 |
| AK047848 | 0.24 | 0.66 | Oosp1 | 2.91 | 0.80 |
| BC010335 | 0.25 | 0.56 | Ccl4 | 2.90 | 1.41 |
| AK078994 | 0.25 | 0.55 | Osm | 2.89 | 1.51 |
| AK047019 | 0.25 | 1.81 | Tbx21 | 2.88 | 1.33 |
| A630081D01Rika | 0.25 | 0.55 | Pdcd1 | 2.88 | 1.84 |
| TC1434196 | 0.25 | 0.97 | ENSMUST00000065731 | 2.87 | 1.74 |
| Eml5 | 0.25 | 2.00 | Shbg | 2.85 | 1.02 |
| Sap30bp | 0.26 | 0.55 | LOC628211 | 2.84 | 1.00 |
| Smg6a | 0.26 | 1.04 | Lgals1a | 2.83 | 1.44 |
| E130120C16Rika | 0.26 | 0.93 | Aqp7 | 2.82 | 1.22 |
| AK048969 | 0.26 | 0.53 | Mras | 2.81 | 1.70 |
| AK036907 | 0.26 | 0.85 | TC1514546 | 2.79 | 1.50 |
| Centg2 | 0.26 | 0.63 | Hist1h1e | 2.76 | 1.15 |
| 9530091C08Rik | 0.26 | 0.81 | 1700088E04Rik | 2.76 | 0.98 |
| 8030498J20Rik | 0.26 | 0.63 | Ifitm1 | 2.76 | 0.14 |
| AK054507 | 0.26 | 0.55 | Tdgf1a | 2.75 | 0.93 |
Commonly up- or down-regulated in liver and brain.
Table 2.
Genes Exhibiting Increase or Decrease in SCNT and TSA in the Liver
| |
Fold change |
|
Fold change |
||
|---|---|---|---|---|---|
| Gene name | SCNT | TSA | Gene name | SCNT | TSA |
| AK078857 | 0.05 | 0.13 | Zfp748 | 7.13 | 8.46 |
| Ptbp2a | 0.08 | 0.08 | 6030458C11Rik | 6.58 | 7.90 |
| 2010107C10Rik | 0.09 | 0.03 | 1700065O13Rik | 4.88 | 2.51 |
| AK145625 | 0.09 | 0.17 | 2310047L11Rik | 4.85 | 7.99 |
| 8030488J09Rik | 0.12 | 0.17 | D330038O06Rik | 4.79 | 3.69 |
| Ifrd1 | 0.12 | 0.18 | Chac1 | 4.78 | 3.52 |
| Slc25a25 | 0.13 | 0.14 | Sh3rf1 | 4.77 | 4.75 |
| AK031320 | 0.14 | 0.34 | Zbtb3 | 4.71 | 2.38 |
| TC1467208a | 0.15 | 0.09 | TC1498661 | 4.63 | 2.05 |
| AK045690 | 0.15 | 0.26 | LOC639396 | 4.49 | 8.30 |
| Xlr4ba | 0.15 | 0.36 | BC050092 | 4.32 | 4.02 |
| AK048349 | 0.16 | 0.19 | TC1434594 | 4.07 | 8.96 |
| Slc25a25 | 0.16 | 0.12 | Gpr84 | 4.06 | 2.00 |
| ENSMUST00000094393 | 0.16 | 0.30 | 5930430L01Rik | 4.01 | 7.21 |
| AK172662 | 0.18 | 0.47 | AK011803 | 3.98 | 6.37 |
| AK039978 | 0.18 | 0.20 | AK052902 | 3.96 | 9.25 |
| Slc25a25 | 0.18 | 0.21 | 4930481A15Rik | 3.86 | 2.17 |
| 4631416L12Rik | 0.18 | 0.18 | Zfp418 | 3.82 | 3.90 |
| AK030494 | 0.19 | 0.43 | Zfp84 | 3.81 | 2.23 |
| Txnip | 0.19 | 0.22 | AK042809 | 3.68 | 3.12 |
| 2610207I05Rik | 0.19 | 0.45 | Ptger2 | 3.66 | 2.87 |
| Gadd45b | 0.19 | 0.33 | Zfp87 | 3.63 | 2.62 |
| Cry2 | 0.19 | 0.28 | Runx1 | 3.56 | 2.85 |
| AK086877 | 0.19 | 0.30 | Thoc7 | 3.56 | 4.79 |
| AK033329 | 0.19 | 0.38 | TC1410447 | 3.55 | 2.04 |
| Nfe2l2 | 0.19 | 0.15 | AK079452 | 3.49 | 3.21 |
| Zbtb20 | 0.20 | 0.23 | BB114266 | 3.47 | 2.66 |
| 5830410F13Rika | 0.20 | 0.38 | Rag1 | 3.46 | 2.79 |
| TC1507087 | 0.20 | 0.21 | Cpn1 | 3.33 | 5.91 |
| Wfikkn1 | 0.20 | 0.18 | A630031M23Rik | 3.30 | 6.24 |
| 9430098F02Rik | 0.20 | 0.27 | Pctk2 | 3.24 | 3.18 |
| Trim27 | 0.20 | 0.15 | Dars | 3.21 | 4.27 |
| 5930436O19Rik | 0.21 | 0.36 | Eif3s8 | 3.19 | 2.72 |
| Crtam | 0.21 | 0.22 | R3hdm1 | 3.18 | 3.52 |
| D130076A03Rika | 0.21 | 0.21 | 1810047C23Rik | 3.17 | 2.61 |
| AK028471 | 0.21 | 0.44 | AK051618 | 3.16 | 2.25 |
| AK085960 | 0.21 | 0.29 | Taz | 3.16 | 4.05 |
| Chchd7 | 0.21 | 0.22 | Rab5b | 3.14 | 3.07 |
| Fgd4 | 0.22 | 0.25 | Pde5a | 3.12 | 2.26 |
| AK047137 | 0.22 | 0.19 | AK044799 | 3.07 | 5.04 |
| AK085234 | 0.22 | 0.47 | Clec2d | 3.03 | 2.84 |
| C79248a | 0.22 | 0.26 | C3ar1 | 3.02 | 2.06 |
| AK033846 | 0.22 | 0.47 | Tnrc15 | 3.01 | 2.97 |
| D18Ertd232e | 0.22 | 0.29 | Kalrn | 2.99 | 3.62 |
| E430022E14Rik | 0.22 | 0.22 | ENSMUST00000014957 | 2.98 | 2.10 |
| Erbb2ip | 0.22 | 0.31 | AK079215 | 2.95 | 4.95 |
| Pard3 | 0.22 | 0.36 | Zfp11 | 2.92 | 2.66 |
| Arnt | 0.22 | 0.48 | 6030458C11Rik | 2.87 | 4.90 |
| TC1541797 | 0.23 | 0.32 | Phka2 | 2.86 | 4.03 |
| AK089858 | 0.27 | 0.06 | 5830417I10Rik | 2.83 | 2.31 |
Commonly up- or downregulated in liver and brain.
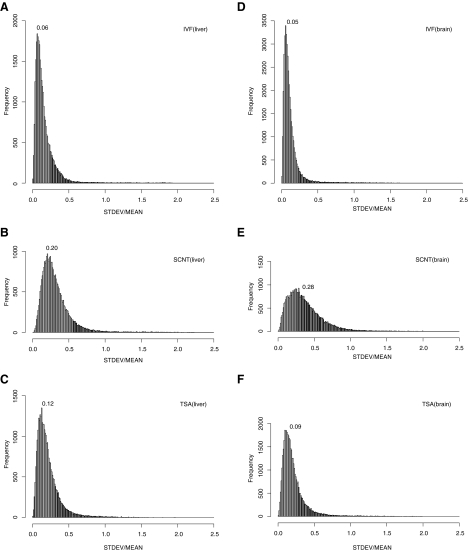
As previously reported, conventional cumulus clone pups displayed abnormal gene expression, with a wide degree of variation across the clones. In the case of the TSA-treated clones, the number of the transcripts in at least one of the four pup groups with more than twofold up- or downregulation was 2174 and 1512 in the neonatal liver, respectively. As shown in Figure 2, the standard deviation in the microarray signals among the four pups in each of the experimental groups was calculated, and a histogram drawn. The most frequent standard deviation for a conventional clone was 0.2, whereas that for an IVF control was 0.06. The standard deviation peak for a TSA-treated clone was 0.12. This means TSA treatment not only normalize gene expression level but also reduce the variation of gene expression.
FIG. 2.
Gene expression variation in SCNT with and without TSA treatment. Gene expression variation among the individuals in each group is represented as the value for the standard deviation in the microarray signals divided by the mean signal. These values of the liver (A–C) and brain (D–F) for the IVF control (A, D), SCNT (B, E) and SCNT with TSA treatment (C, F) were plotted as histograms (each break was 0.01). The most frequent values are also presented in terms of “mode.”
The gene expression changes in SCNT and their normalization were also observed in neonatal brain in the same individuals (Fig. 1D–F), but the commonly affected genes in the liver and brain comprised approximately 10% (Tables 3 and 4). The standard deviation in the gene expression was also larger in SCNT in the neonatal brain (mode=0.05 in IVF, whereas the mode=0.28 in SCNT), and TSA treatment also reduced the variation in the gene expression (mode=0.09 in SCNT with TSA) in the brain (Fig. 2D–F). These data indicated that the TSA treatment effects were not restricted to a single organ.
Table 3.
Genes Exhibiting Increase or Decrease in SCNT Pup Brain and Normalized in TSA
| |
Fold change |
|
Fold change |
||
|---|---|---|---|---|---|
| Gene name | SCNT | TSA | Gene name | SCNT | TSA |
| AK082373 | 0.05 | 0.79 | Igl-V1 | 17.00 | 0.93 |
| 2900027M19Rik | 0.05 | 0.86 | Chi3l3 | 8.18 | 1.25 |
| AK038604 | 0.06 | 1.36 | Tdgf1a | 7.06 | 1.94 |
| AK028486 | 0.07 | 0.61 | Ifi27a | 7.01 | 1.44 |
| AK047779 | 0.07 | 0.78 | D630033O11Rik | 6.74 | 1.53 |
| AK042559 | 0.07 | 0.78 | Hsd3b1 | 6.65 | 1.63 |
| AK035046a | 0.08 | 1.20 | NAP107172-1 | 6.53 | 1.70 |
| AK042974 | 0.08 | 1.23 | 1190007F08Rik | 6.21 | 1.59 |
| AK042794 | 0.08 | 1.20 | Cmtm2b | 6.21 | 1.75 |
| AK039039 | 0.08 | 0.73 | Zfp87 | 6.16 | 1.66 |
| AK029415 | 0.08 | 0.99 | BC006965 | 6.04 | 1.92 |
| AK043453 | 0.09 | 1.06 | Ryr1 | 5.91 | 0.70 |
| AK039932 | 0.09 | 0.55 | Pex11c | 5.76 | 1.53 |
| AK039246 | 0.09 | 1.06 | Gata1 | 5.42 | 1.76 |
| Opcml | 0.09 | 0.76 | Dusp23 | 5.38 | 1.62 |
| AK045769 | 0.09 | 0.84 | Hist1h1b | 5.37 | 0.78 |
| AK032764 | 0.09 | 0.73 | Irf6 | 5.36 | 1.08 |
| AK048842 | 0.09 | 0.80 | Acta2 | 5.36 | 1.22 |
| AK082198 | 0.10 | 0.59 | 4930504H06Rik | 5.14 | 1.23 |
| AK080985 | 0.10 | 0.96 | Cd52 | 5.02 | 1.12 |
| AK047123 | 0.10 | 0.64 | Olfr446 | 4.98 | 1.96 |
| Cdh10 | 0.10 | 1.05 | Lst1 | 4.95 | 1.98 |
| 2900022M07Rik | 0.10 | 1.05 | Olfr60 | 4.91 | 1.87 |
| AK086620 | 0.10 | 0.92 | Hist4h4 | 4.86 | 0.66 |
| Copg2as2 | 0.10 | 0.78 | Prlpo | 4.77 | 1.03 |
| AK090149 | 0.10 | 0.73 | Tmc1 | 4.69 | 1.59 |
| AK077975 | 0.10 | 0.65 | Hist1h3aa | 4.61 | 0.57 |
| A830054O04Rik | 0.10 | 1.06 | BC019731 | 4.57 | 1.89 |
| Sorbs2 | 0.10 | 0.90 | Zic4 | 4.52 | 1.65 |
| AK045636 | 0.11 | 1.13 | Olfr1443 | 4.36 | 1.58 |
| AK047993 | 0.11 | 0.91 | 1700016G05Rik | 4.36 | 1.64 |
| 9030425P06Rik | 0.11 | 0.70 | AK031219 | 4.36 | 1.86 |
| 6720482D04 | 0.11 | 0.81 | Tgm3 | 4.31 | 1.90 |
| AK053189 | 0.11 | 0.62 | 4833421E05Rik | 4.29 | 0.85 |
| AK089705 | 0.11 | 0.78 | Etv3 | 4.21 | 1.45 |
| AK048858 | 0.12 | 0.73 | AY344585 | 4.17 | 0.84 |
| A630081D01Rika | 0.12 | 0.69 | Lemd3 | 4.14 | 1.50 |
| Gria4 | 0.12 | 0.81 | B2m | 4.09 | 1.85 |
| Sox5 | 0.12 | 0.86 | 4930481A15Rik | 4.08 | 1.44 |
| AK038647 | 0.12 | 0.76 | 4933413N12Rik | 4.07 | 1.31 |
| AK040469 | 0.12 | 0.70 | NP063118 | 4.06 | 1.85 |
| Smg6a | 0.12 | 1.19 | 2010107E04Rik | 4.05 | 1.71 |
| 9630039A02Rik | 0.12 | 0.82 | Spink2 | 4.04 | 1.32 |
| AK047015 | 0.12 | 1.25 | Lgals1a | 4.03 | 1.43 |
| AK080560 | 0.12 | 1.05 | AK040677 | 4.03 | 1.92 |
| E130120C16Rika | 0.12 | 1.13 | Gng13 | 4.02 | 1.54 |
| A930035E12Rik | 0.12 | 0.93 | 1110054P19Rik | 4.00 | 1.84 |
| Arpp21 | 0.13 | 1.51 | 1600014C23Rik | 3.91 | 1.18 |
| AK053316 | 0.13 | 0.72 | 4930572J05Rik | 3.85 | 0.89 |
| AK038328a | 0.13 | 0.77 | 2900053A13Rik | 3.83 | 1.06 |
Commonly up- or downregulated in brain and liver.
Table 4.
Genes Exhibiting Increase or Decrease in SCNT and TSA in the Brain
| |
Fold change |
|
Fold change |
||
|---|---|---|---|---|---|
| Gene name | SCNT | TSA | Gene name | SCNT | TSA |
| AK047096 | 0.05 | 0.31 | Mt4 | 15.62 | 6.42 |
| AK053534 | 0.06 | 0.47 | A_51_P445924 | 14.39 | 8.08 |
| AK034993 | 0.06 | 0.49 | ENSMUST00000081163 | 12.26 | 7.33 |
| AK043296 | 0.06 | 0.22 | AK006604 | 11.93 | 8.39 |
| Xlr4ba | 0.06 | 0.10 | LOC546361 | 11.20 | 8.78 |
| AK048863 | 0.07 | 0.46 | NAP070138-1 | 11.13 | 6.95 |
| AK045894 | 0.08 | 0.31 | NAP020861-001 | 10.96 | 7.40 |
| Fbxl5 | 0.09 | 0.09 | AV094648 | 10.62 | 7.23 |
| TC1479736 | 0.09 | 0.21 | D5Ertd135e | 10.58 | 9.84 |
| Cntn4 | 0.09 | 0.46 | NAP113002-1 | 10.20 | 7.71 |
| AK080684 | 0.09 | 0.45 | Pex1 | 10.07 | 5.55 |
| TC1413093 | 0.10 | 0.15 | NAP028543-1 | 9.66 | 7.58 |
| Xlr3b | 0.10 | 0.10 | LOC627004 | 9.24 | 7.12 |
| AK042781 | 0.10 | 0.16 | NAP101638-1 | 9.06 | 6.15 |
| 5830410F13Rika | 0.11 | 0.47 | 0610009B10Rik | 8.70 | 4.28 |
| Xpo1 | 0.11 | 0.30 | NAP057018-1 | 8.63 | 7.43 |
| AK085907 | 0.11 | 0.38 | NAP045499-1 | 8.59 | 6.37 |
| Dhx9 | 0.11 | 0.27 | Olfr144 | 8.57 | 4.09 |
| AK043796 | 0.11 | 0.43 | AK136065 | 8.34 | 3.93 |
| AK085847 | 0.11 | 0.39 | Smyd1 | 8.14 | 4.54 |
| 8430419K02Rik | 0.11 | 0.12 | NAP059572-1 | 7.79 | 6.10 |
| Ptbp2a | 0.11 | 0.03 | 4930544G11Rik | 7.67 | 5.67 |
| LOC664628 | 0.11 | 0.32 | LOC544881 | 7.01 | 6.63 |
| AK036012 | 0.11 | 0.48 | 4930571C24Rik | 6.96 | 4.71 |
| AK044191 | 0.11 | 0.26 | NAP061671-1 | 6.91 | 5.38 |
| AK037089 | 0.11 | 0.39 | BC036642 | 6.77 | 5.57 |
| TC1467208a | 0.12 | 0.09 | 4921534H16Rik | 6.76 | 3.21 |
| C79248a | 0.12 | 0.16 | 4933440N22Rik | 6.62 | 3.43 |
| Grid2 | 0.12 | 0.32 | LOC433882 | 6.54 | 7.56 |
| 4930546H06Rik | 0.13 | 0.35 | NAP070792-1 | 6.42 | 4.64 |
| D130076A03Rika | 0.13 | 0.18 | NAP113376-1 | 6.38 | 5.37 |
| 4732465E10Rik | 0.13 | 0.41 | NAP028025-1 | 5.91 | 4.89 |
| 3110068A07Rik | 0.14 | 0.41 | NAP062835-1 | 5.87 | 5.15 |
| AK038629 | 0.14 | 0.48 | MGC107702 | 5.85 | 3.75 |
| 2310035P21Rik | 0.14 | 0.45 | TC1454836 | 5.85 | 4.49 |
| Prim2 | 0.14 | 0.43 | Rps21 | 5.81 | 4.25 |
| C030038I04Rik | 0.14 | 0.35 | A630049P17 | 5.70 | 2.50 |
| Skiv2l2 | 0.14 | 0.21 | NAP028748-1 | 5.66 | 9.74 |
| AK029965 | 0.15 | 0.34 | Tmc6 | 5.58 | 3.99 |
| AK047189 | 0.15 | 0.44 | Gm1499 | 5.53 | 2.95 |
| AK033876 | 0.15 | 0.22 | Il17rd | 5.31 | 2.98 |
| AK043448 | 0.15 | 0.35 | NAP061630-1 | 5.30 | 3.32 |
| AK086440 | 0.15 | 0.44 | ENSMUST00000088062 | 5.18 | 3.07 |
| 2810055G20Rik | 0.16 | 0.39 | NAP070880-1 | 5.17 | 4.91 |
| Zfp91 | 0.16 | 0.30 | LOC673028 | 5.17 | 3.31 |
| 4831440D22Rik | 0.16 | 0.50 | V1ri6 | 5.09 | 3.01 |
| Taf15 | 0.16 | 0.33 | Cyp17a1 | 5.06 | 3.38 |
| D230015P20Rik | 0.16 | 0.10 | RP23-3N21.1 | 5.02 | 5.40 |
| Pcdhgb4 | 0.16 | 0.27 | Hes2 | 4.99 | 3.74 |
| AK037099 | 0.17 | 0.39 | Zc3hav1 | 4.98 | 2.53 |
Commonly up- or downregulated in brain and liver.
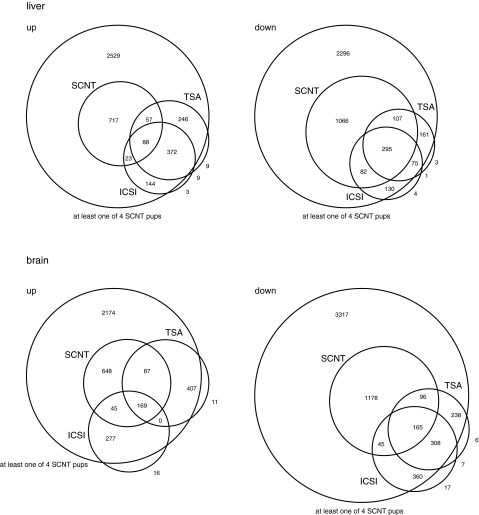
Recently, we reported that the process of ICSI also induces long-term transcriptome perturbations (Kohda et al., 2011). Interestingly, approximately 60 and 20% of the genes that were affected by ICSI treatment in terms of transcriptional regulation were also commonly affected by somatic cell cloning, with or without TSA treatment, respectively (Table 5). These overlapping findings were statistically significant (by chi-square test, p-value<2.2−16 both for SCNT with and without TSA) and this suggests there are common mechanisms in SCNT and ICSI that may contribute to the observed transcriptome perturbations. We have summarized the numbers of genes affected by SCNT, SCNT with TSA treatment, and ICSI in a Venn diagram (Fig. 3). As mentioned above, the number of commonly affected genes in SCNT pups was reduced in the TSA-treated SCNT pups, and there was a significant number of affected genes in SCNT with TSA that did not overlap with conventional SCNT (i.e., SCNT with TSA treatment-specific genes). However, almost all of the genes affected by the SCNT with TSA treatment overlapped with the genes that exhibited up- or downregulation in at least one of the four conventional SCNT clones. It is also evident that the affected genes in SCNT with TSA treatment overlapped well with ICSI pups.
Table 5.
Genes Exhibiting Increase or Decrease in SCNT and ICSI in the Liver
| |
Fold change |
|
Fold change |
||
|---|---|---|---|---|---|
| Gene name | SCNT | ICSI | Gene name | SCNT | ICSI |
| AK078857 | 0.06 | 0.14 | Zfp748 | 5.70 | 7.84 |
| Ptbp2 | 0.10 | 0.08 | 6030458C11Rik | 5.27 | 8.46 |
| 2010107C10Rik | 0.10 | 0.04 | 1700065O13Rik | 3.91 | 3.28 |
| AK145625 | 0.11 | 0.18 | 2310047L11Rik | 3.88 | 8.98 |
| 8030488J09Rik | 0.15 | 0.18 | D330038O06Rik | 3.83 | 3.87 |
| Ifrd1 | 0.15 | 0.19 | Chac1 | 3.83 | 3.04 |
| Slc25a25 | 0.16 | 0.14 | Sh3rf1 | 3.81 | 6.35 |
| AK031320 | 0.17 | 0.36 | Zbtb3 | 3.77 | 2.54 |
| TC1467208 | 0.18 | 0.09 | TC1498661 | 3.71 | 2.20 |
| AK045690 | 0.18 | 0.28 | LOC639396 | 3.59 | 11.41 |
| Ppp1r3g | 0.19 | 0.25 | BC050092 | 3.45 | 4.40 |
| AK048349 | 0.19 | 0.19 | TC1434594 | 3.25 | 9.84 |
| Slc25a25 | 0.19 | 0.12 | Gpr84 | 3.25 | 2.08 |
| ENSMUST00000094393 | 0.20 | 0.32 | 5930430L01Rik | 3.21 | 6.01 |
| AK172662 | 0.21 | 0.49 | AK011803 | 3.19 | 6.61 |
| AK039978 | 0.21 | 0.22 | AK052902 | 3.17 | 8.51 |
| Slc25a25 | 0.22 | 0.21 | 4930481A15Rik | 3.09 | 2.40 |
| 4631416L12Rik | 0.22 | 0.19 | Zfp418 | 3.05 | 2.77 |
| AK030494 | 0.22 | 0.44 | Zfp84 | 3.05 | 2.40 |
| Txnip | 0.22 | 0.22 | AK042809 | 2.95 | 3.53 |
| 2610207I05Rik | 0.22 | 0.49 | Sox9 | 2.94 | 2.03 |
| Gadd45b | 0.22 | 0.33 | Ptger2 | 2.93 | 3.26 |
| Cry2 | 0.22 | 0.31 | Zfp87 | 2.90 | 2.66 |
| AK086877 | 0.23 | 0.32 | Pfkp | 2.87 | 2.62 |
| AK033329 | 0.23 | 0.38 | Runx1 | 2.85 | 3.02 |
| Nfe2l2 | 0.23 | 0.16 | Thoc7 | 2.84 | 3.76 |
| Zbtb20 | 0.24 | 0.24 | TC1410447 | 2.84 | 2.08 |
| 5830410F13Rik | 0.24 | 0.40 | AK079452 | 2.79 | 3.48 |
| TC1507087 | 0.24 | 0.23 | BB114266 | 2.77 | 2.69 |
| Wfikkn1 | 0.24 | 0.20 | Rag1 | 2.77 | 2.41 |
| 9430098F02Rik | 0.24 | 0.29 | Cpn1 | 2.67 | 6.03 |
| Trim27 | 0.25 | 0.15 | A630031M23Rik | 2.64 | 7.91 |
| 5930436O19Rik | 0.25 | 0.36 | Pctk2 | 2.59 | 3.23 |
| Crtam | 0.25 | 0.23 | Dars | 2.57 | 4.91 |
| D130076A03Rik | 0.25 | 0.24 | Il1f9 | 2.56 | 2.45 |
| TC1525352 | 0.25 | 0.26 | Eif3s8 | 2.55 | 2.75 |
| AK028471 | 0.25 | 0.46 | R3hdm1 | 2.54 | 3.95 |
| AK085960 | 0.25 | 0.32 | 1810047C23Rik | 2.53 | 2.94 |
| 4921522E08Rik | 0.26 | 0.23 | AK051618 | 2.53 | 3.08 |
| Chchd7 | 0.26 | 0.24 | Taz | 2.52 | 3.90 |
| Fgd4 | 0.26 | 0.27 | NAP045909-1 | 2.52 | 2.22 |
| AK047137 | 0.26 | 0.19 | Rab5b | 2.51 | 3.44 |
| AK085234 | 0.26 | 0.50 | Pde5a | 2.50 | 2.40 |
| C79248 | 0.26 | 0.27 | Ahnak | 2.47 | 2.57 |
| D18Ertd232e | 0.27 | 0.30 | AK044799 | 2.45 | 4.61 |
| E430022E14Rik | 0.27 | 0.22 | Clec2d | 2.42 | 2.87 |
| Erbb2ip | 0.27 | 0.34 | C3ar1 | 2.42 | 2.37 |
| Pard3 | 0.27 | 0.38 | Tnrc15 | 2.41 | 3.04 |
| TC1541797 | 0.27 | 0.33 | Kalrn | 2.39 | 5.21 |
| Lrrc16 | 0.27 | 0.44 | ENSMUST00000014957 | 2.38 | 2.41 |
FIG. 3.
Summary of the number transcripts affected by SCNT or ICSI. The number of microarray probes corresponding to the affected transcripts by SCNT, SCNT with TSA treatment, and ICSI in the neonatal liver and brain are summarized in a Venn diagram. The total number of the microarray probes was 41,174.
Discussion
In this study, it is demonstrated that TSA treatment in SCNT not only increases the efficiency of cloning, but also improves the long-term transcriptome integrity (i.e., transcriptional regulation in neonatal tissues). Recently, we have reported that TSA treatment enhances the reprogramming of somatic nuclei in terms of chromatin remodeling, histone modification, DNA replication, and transcriptional activity in two-cell stage embryos (Bui et al., 2010). This evidence suggests that the TSA treatment improved the condition of the SCNT embryo nucleus in general at the two-cell stage. The reduction in the variance of the gene expression level in this study in the SCNT pups which resulted from TSA treatment is in good accord with these observations.
Almost all of the genes affected in both SCNT with TSA and ICSI were included in the genes affected in at least one of the four SCNT pups (Fig. 3). In other words, the alteration in the aberrant gene expression induced in the conventional SCNT procedure was significantly reduced to a smaller number of genes by TSA treatment. Furthermore, the gene profile shifted to one resembling that of the ICSI produced pups. The genes affected by SCNT, especially by SCNT with TSA treatment, overlapped with the ICSI affected genes (Fig. 3). This suggests that there may be common steps and/or reactions in SCNT and ICSI which induce this subgroup of affected genes. The close overlap of the SCNT- and ICSI-affected genes suggests that as a result of common technical steps or procedures, such as the injection of the nuclei with a glass pipette, bypassing the acrosome reaction at fertilization, there might be a slight difference of chromatin decondensation or calcium oscillation in SCNT and ICSI. It is important to identify this common event at an early stage of development and refine the technique to overcome the problem.
On the other hand, there remains a group of genes affected by SCNT but not by ICSI (Table 6). These genes are purely affected by the “reprogramming process” of SCNT and not by the manipulation of SCNT such as the injection step with a glass pipette. To understand the mechanism of the reprogramming by nuclear transfer, it is important to identify the first target genes of the “reprogramming process” of SCNT among the SCNT affected genes comparing ICSI and SCNT embryo in preimplantation stages.
Table 6.
Genes Exhibiting Increase or Decrease in SCNT But Not Affected in ICSI in the Liver
| |
Fold change |
|
Fold change |
||
|---|---|---|---|---|---|
| Gene name | SCNT | ICSI | Gene name | SCNT | ICSI |
| U20264 | 0.13 | 1.04 | NAP124059-1 | 4.42 | 1.58 |
| Xlr4b | 0.18 | 0.66 | Hist1h3a | 3.97 | 0.87 |
| AK087779 | 0.20 | 0.73 | Calca | 3.49 | 1.15 |
| AK051522 | 0.20 | 0.65 | Il1b | 3.42 | 2.00 |
| Xlr3b | 0.21 | 0.82 | Serpina6 | 3.19 | 1.06 |
| AK040092 | 0.22 | 0.75 | Ifi27 | 3.02 | 1.45 |
| 2610027H17Rik | 0.22 | 1.10 | A_52_P1101503 | 3.02 | 1.56 |
| AK038045 | 0.23 | 0.62 | Rmrp | 2.98 | 1.76 |
| AK050807 | 0.23 | 0.71 | G0s2 | 2.96 | 1.99 |
| AK050242 | 0.24 | 0.55 | Gata3 | 2.94 | 1.17 |
| NM_001013789 | 0.24 | 0.69 | Guca1a | 2.92 | 1.86 |
| Chka | 0.25 | 0.63 | Smyd2 | 2.81 | 1.61 |
| Stra6 | 0.25 | 0.98 | Lst1 | 2.73 | 1.44 |
| AK046412 | 0.25 | 0.64 | Ptgfr | 2.69 | 1.55 |
| AK051621 | 0.25 | 0.64 | Mt1 | 2.65 | 1.93 |
| AK084873 | 0.25 | 0.94 | Tnf | 2.64 | 1.39 |
| Epas1 | 0.26 | 0.55 | Ldhc | 2.61 | 1.00 |
| Tanc1 | 0.26 | 0.61 | Oosp1 | 2.59 | 1.53 |
| AK083952 | 0.26 | 0.73 | Tnnc1 | 2.55 | 1.30 |
| AK076939 | 0.26 | 0.59 | C130050O18Rik | 2.52 | 1.27 |
| AK033846 | 0.27 | 0.51 | Bmp10 | 2.52 | 1.73 |
| AK085204 | 0.27 | 1.10 | 2610035D17Rik | 2.52 | 0.82 |
| Arnt | 0.27 | 0.52 | Smpdl3b | 2.51 | 1.55 |
| 4930588G05Rik | 0.27 | 1.27 | Olfr74 | 2.50 | 1.43 |
| AK033340 | 0.27 | 0.88 | BF228116 | 2.49 | 1.85 |
| AK048657 | 0.27 | 0.55 | B230317C12Rik | 2.49 | 1.12 |
| AK079436 | 0.28 | 0.73 | Atp5k | 2.49 | 1.81 |
| C530043K16Rik | 0.28 | 0.51 | Drr1 | 2.48 | 1.61 |
| AK079230 | 0.28 | 0.87 | B3galt5 | 2.46 | 1.57 |
| AK035046 | 0.28 | 0.89 | Aqp7 | 2.46 | 1.37 |
| TC1469196 | 0.28 | 0.83 | Fcer1g | 2.44 | 1.22 |
| AK038328 | 0.28 | 0.93 | AK014119 | 2.44 | 1.38 |
| Tns1 | 0.28 | 0.66 | Tcrg | 2.42 | 0.99 |
| Gfod1 | 0.28 | 0.56 | Ksr1 | 2.42 | 1.25 |
| Bclaf1 | 0.28 | 0.50 | Cst8 | 2.40 | 0.99 |
| Nrip1 | 0.29 | 0.51 | TC1498165 | 2.39 | 1.74 |
| AK050555 | 0.29 | 0.66 | Pdik1l | 2.36 | 1.60 |
| AK031434 | 0.29 | 0.80 | Vdr | 2.35 | 1.04 |
| Mtss1 | 0.29 | 1.11 | Stambpl1 | 2.33 | 1.74 |
| AK047526 | 0.29 | 0.80 | Ccl4 | 2.32 | 0.75 |
| AK047848 | 0.29 | 0.72 | 1700026L06Rik | 2.32 | 1.21 |
| BC010335 | 0.30 | 0.57 | Osm | 2.31 | 1.62 |
| AK078994 | 0.30 | 0.56 | Tbx21 | 2.31 | 1.50 |
| A630081D01Rik | 0.30 | 0.60 | Pdcd1 | 2.30 | 1.51 |
| TC1434196 | 0.30 | 1.00 | ENSMUST00000065731 | 2.30 | 1.37 |
| Eml5 | 0.30 | 1.27 | Shbg | 2.28 | 1.05 |
| Sap30bp | 0.31 | 0.60 | D430015B01Rik | 2.28 | 1.46 |
| Smg6 | 0.31 | 1.11 | LOC628211 | 2.27 | 1.13 |
| E130120C16Rik | 0.31 | 0.95 | A_52_P1157520 | 2.26 | 1.60 |
| AK048969 | 0.31 | 0.56 | Lgals1 | 2.26 | 1.25 |
It is also reported that one of the major causes of SCNT embryo lethality at the preimplantation stage is ectopic expression of the Xist gene and aberrant X chromosome inactivation, and it was demonstrated that targeted disruption of the Xist gene significantly improves the efficiency of SCNT (Inoue et al., 2010). The combination of TSA treatment and suppression of ectopic Xist expression appears to more effectively normalize the long-term transcriptome integrity and success rate of SCNT.
Acknowledgments
This work was supported by grants from the JSPS (18510168).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bui H.T. Wakayama S. Kishigami S., et al. Effect of trichostatin A on chromatin remodeling, histone modifications, DNA replication, and transcriptional activity in cloned mouse embryos. Biol. Reprod. 2010;83:454–463. doi: 10.1095/biolreprod.109.083337. [DOI] [PubMed] [Google Scholar]
- Iager A.E. Ragina N.P. Ross P.J., et al. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells. 2008;10:371–379. doi: 10.1089/clo.2007.0002. [DOI] [PubMed] [Google Scholar]
- Inoue K. Kohda T. Sugimoto M., et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- Kishigami S. Mizutani E. Ohta H., et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Kishigami S. Bui H.T. Wakayama S., et al. Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J. Reprod. Dev. 2007;53:165–170. doi: 10.1262/jrd.18098. [DOI] [PubMed] [Google Scholar]
- Kohda T. Inoue K. Ogonuki N., et al. Variation in gene expression and aberrantly regulated chromosome regions in cloned mice. Biol. Reprod. 2005;73:1302–1311. doi: 10.1095/biolreprod.105.044958. [DOI] [PubMed] [Google Scholar]
- Kohda T. Ogonuki N. Inoue K., et al. Intracytoplasmic sperm injection induces transcriptome perturbation without any transgenerational effect. Biochem. Biophys. Res. Commun. 2011;410:282–288. doi: 10.1016/j.bbrc.2011.05.133. [DOI] [PubMed] [Google Scholar]
- Li J. Svarcova O. Villemoes K., et al. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology. 2008;70:800–808. doi: 10.1016/j.theriogenology.2008.05.046. [DOI] [PubMed] [Google Scholar]
- Shi L.H. Ai J.S. Ouyang Y.C., et al. Trichostatin A and nuclear reprogramming of cloned rabbit embryos. J. Anim. Sci. 2008;86:1106–1113. doi: 10.2527/jas.2007-0718. [DOI] [PubMed] [Google Scholar]
- Wakayama T. Perry A.C. Zuccotti M., et al. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]