Abstract
Humans are not (and have never been) alone. From the moment we are born, millions of micro-organisms populate our bodies and coexist with us rather peacefully for the rest of our lives. This microbiome represents the totality of micro-organisms (and their genomes) that we necessarily acquire from the environment. Micro-organisms living in or on us have evolved to extract the energy they require to survive, and in exchange they support the physiological, metabolic and immune capacities that have contributed to our evolutionary success. Although currently categorized as an autoimmune disorder and regarded as a complex genetic disease, the ultimate cause of rheumatoid arthritis (RA) remains elusive. It seems that interplay between predisposing genetic factors and environmental triggers is required for disease manifestation. New insights from DNA sequence-based analyses of gut microbial communities and a renewed interest in mucosal immunology suggest that the microbiome represents an important environmental factor that can influence autoimmune disease manifestation. This Review summarizes the historical clues that suggest a possible role for the microbiota in the pathogenesis of RA, and will focus on new technologies that might provide scientific evidence to support this hypothesis.
Introduction
Since the invention of the microscope by Antony van Leeuwenhoek in the 17th century, it has become evident that humans have never lived alone. While describing his observations of what he called oral plaque “animalcules” to the Royal British Society, van Leeuwenhoek noted, “…that the people living in our United Netherlands are not as many as the living animals that I carry in my own mouth this very day.” He was not far off. Indeed, this geo-arithmetical relationship has now been calculated, and the micro-organisms sharing our body spaces total around 100 trillion, outnumbering human cells by a factor of ten.1 The human gut alone harbors roughly 3 lbs of bacteria, whose collective genome encodes around 3 million different genes—100 times more than that of its human host.2
The term microbiome—as coined by Joshua Lederberg four centuries after the first description of van Leeuwenhoek’s animalcules—defines the ecological communities of commensal, symbiotic, and pathogenic micro-organisms that literally share our body space.3 Most of these micro-organisms have been all but ignored as determinants of health and disease.4 Through this extended view of self, humans can be regarded as a ‘superorganism’, composed of an ensemble of human and nonhuman cells (and their genomes) that constitute our body. The human microbiome has coevolved with us in a mutually beneficial relationship. These microbes—through the cellular constituents encoded by their genomes—provide us with physiological, metabolic and immune capacities in exchange for nutrients extracted from our body sites.
In 2007, the NIH launched the Human Microbiome Project (HMP), to gain a better understanding of the complex biological interactions between humans and the micro-organisms they harbor.5 Through the utilization of revolutionary culture-independent techniques, investigators in the HMP hope to fulfill two main aims: to characterize the microbial communities found at several different sites on the human body; and to analyze the role of these microbes in human health and disease. Autoimmune disorders occupy a prominent position among diseases that have long been thought to be triggered by micro-organisms.6 In particular, accumulating evidence suggests that the oral and intestinal microbiomes have a role in the development of rheumatoid arthritis (RA). Here, we summarize the historical, anthropological and epidemiological clues supporting this idea and describe the technological (massive parallel DNA sequencing) and scientific (mucosal immunology and host–microbe interactions) advances in micro-biomics, which shed new light on the part played by micro-organisms in the pathogenesis of RA.
An old hypothesis for a new disease
RA is a chronic, disabling and incurable disease characterized as a complex genetic autoimmune disorder.7 However, evidence as to how human genes contribute to the development of RA is inconclusive, as the presence of susceptibility genes identified to date is neither necessary nor sufficient for the development of disease. Current genetic discoveries resulting from genome-wide associations studies (GWAS) explain only 16% of disease variance (including an estimated 12% for the MHC class II region alone).8 Although many more common risk alleles with modest effect size are likely to be discovered in the future,9 enough data now exist to indicate that genetic predisposition to RA is not a guarantee of developing this disease. Most importantly, the prevalence of RA concordance in monozygotic twins in European studies is ≤15%,10,11 and it is unclear whether this level actually exceeds the risk of developing RA that exists in the general population.12
Although genes contribute to RA susceptibility, interaction between genetic effects and environmental factors is required to explain the observed differences in incidence of the disease, such as those mentioned above. Environmental risk factors associated with RA development include hormones, smoking and infection.13
It has been hypothesized that RA originated in the New World and has ‘spread’ to the rest of the planet as a result of 16th and 17th century expeditions by Europeans. The geographical dissemination of RA could have been brought about through contact with environmental factors (such as pathogens or dietary components that altered the microbiome), mixture of genes (through breeding), or a combination of genetic and environmental factors. Evidence to support this theory comes from different sources. Firstly—whereas gout, spondylo-arthropathies and osteoarthritis have all been accurately and extensively described in historical texts and paleopathological studies,14–18 no mention of RA exists in any medical or general literature until the 19th century (Figure 1). Furthermore, inconclusive evidence of RA can be drawn from the history of fine arts before the paintings of Rubens (early 17th century). Himself afflicted by a severe form of RA, Rubens clearly reflected his condition in his later works.19 More interesting, although debated, are the paleopathological studies by Rothschild et al.20 describing RA in six skeletons dating from 3,000–6,000 B.C. and found in northwestern Alabama, USA. These remains had clear evidence of symmetric, erosive, peripheral polyarthritis without axial disease or distal interphalangeal involvement. As no indication of RA was found in Old World skeletons,21 Rothschild and colleagues concluded that RA might actually be a transmissible disease caused by a pathogen contracted in the New World. Epidemiological evidence supports this notion. The highest prevalence of RA is found among Amerindian communities, such as the Chippewa, Pima and Tiglit.22 Conversely, studies conducted in Sub-Saharan Africa and Asia—the populations that most recently gained contact with European conquerors—have described a much lower frequency of RA in the populations inhabiting these areas,23,24 with white and European rates seeming to fall somewhere in between.25 Finally, although recently stabilized, overall RA incidence has decreased proportionally to the use of antibiotics since the widespread introduction of these agents to the clinic in the first half of the 20th century.26
Figure 1.
Historical, literary, artistic and paleopathological evidence of RA as a New World disease that has ‘spread’ to the rest of the world. Paleopathological evidence of RA exists only in skeletal remains from New World populations and RA was not documented in the Old World until the late 18th century, whereas other rheumatic diseases have been well described in biblical and ancient texts. Although debated, RA is thought to have spread to Europe after the beginning of trading with the Americas. The first medical literary evidence comes from a paper from 1800 by Landré-Beauvais, who reported his findings in “La goutte asthénique primitive”. In 1859, Alfred Garrod coined the term ‘Rheumatoid Arthritis’. Rubens seems to be the first painter to depict what seems to be RA of the hands in the mid 17th century. Paintings such as The three graces and The miracle of St Ignatius, show ulnar deviation, buttoniere deformities and MCP swelling. Epidemiological evidence supports the notion that RA is a disease of the New World. Amerindians and Eskimos have the highest prevalence of RA, followed by white populations. In keeping with this theory, African and Far East populations, being the latest to be in contact with European conquistadors, have a strikingly low prevalence of RA. Abbreviations: MCP, metacarpophalangeal; RA, rheumatoid arthritis. Permission to use image ‘Sir Alfred Baring Garrod. Photograph by Elliot and Fry’ obtained from the Wellcome Library, London ©
The idea that oral or intestinal micro-organisms are associated with the development of RA is not novel. For more than a century, several investigators have subscribed to this theory and many have sought to link intestinal microbiota to the etiology of RA: from the albuminous putrefaction therapies of Andrews and Hoke, to the toxemic factor hypothesis of Carl Warden,27 and recent attempts by Toivanen et al. using gas chromatography.28 Similar efforts by other groups pointed towards periodontal disease and its polymicrobial burden as the cause of RA. The oral sepsis hypothesis,29 which became widespread in the 1900s, led to the use of teeth extraction as a prevalent treatment of disease and was used very intensively in the treatment of RA for several decades. A vast body of literature has now been published showing epidemiological associations between the presence of periodontal disease and RA. Indirect evidence (mostly from nonspecific serological analyses) also suggests that certain bacterial phylotypes are involved in the pathogenesis of RA. Most recently, Porphyromonas gingivalis, a periodontopathic bacterium, has been recognized as a possible link between periodontitis, peptide citrullination, autoantibody formation and joint inflammation.30,31 The RA–periodontitis connection has been extensively reviewed elsewhere32,33 and, therefore, in this Review we will focus on recent findings relating to the intestinal microbiome and RA.
The DNA sequencing revolution
Since Louis Pasteur and Robert Koch redefined modern microbiology, the prevailing paradigm has stated that in order to be considered the cause of disease, a micro-organism must: a) be found in abundance in organisms with the disease but not in healthy controls; b) be isolated and cultured from an organism with the disease; c) cause disease when inoculated into healthy controls; and d) be reisolated in identical form and shape from the inoculated organism. However, from their very beginnings the Koch and Loffler postulates lacked,34 for a number of important reasons, a universal application. Firstly, many bacteria, such as Vibrio cholerae and Salmonella, are found in asymptomatic individuals. Secondly, not all subjects exposed to an infectious agent, Mycobacterium tuberculosis for example, will acquire the infection, as host factors are undoubtedly primordial and natural selection might have led to differences in genetic susceptibility to pathogens (immune competency). Finally and most recently, with the advance in DNA sequencing technologies, it became clear that the vast majority (as much as 80%) of bacteria living in our body cavities have never been cultured,35 largely because the nutrients and conditions commonly used in the laboratory do not favor the growth of most micro-organisms (particularly the anaerobes). Therefore, the scientific community has taken advantage of culture-independent DNA sequencing to perform taxonomic identification and to elucidate bacterial enzymatic function.
The 16S ribosomal RNA (16S rRNA) is a component of the 30S subunit of prokaryotic ribosomes, regions of which are highly conserved between different species of bacteria. The 16S rRNA gene is, therefore, useful for phylogenetic studies36 because universal PCR primers targeting these conserved regions can be used to amplify the gene in parts to provide the complete nucleotide sequence of the 16S rRNA without prior knowledge of which bacterial species are present.36 Concomitantly, the 16S rRNA gene also contains hypervariable regions that provide species-specific signature sequences and enable unbiased bacterial identification utilizing next-generation sequencing platforms.37,38
This approach answers the question of bacterial identity in a given ecological niche (‘who are they?’) and, thus, can be used to characterize the complexity of microbial communities present at different sites in the human body (Figure 2). However, 16S rRNA gene sequencing does not provide insight into the capacities provided by the molecular machinery encoded by the genes of the human microbiome, considered by many as our second genome.39 Therefore, whole-genome shotgun sequencing is used to enable the identification of metabolic and enzymatic pathways present in the microbial communities, and to answer the question, ‘what are they doing?’40 (Figure 2).
Figure 2.
Culture-independent genomic analysis of the human microbiome. Culture-independent techniques have advanced our capacity to survey complex microbial communities in human samples. Well-characterized individuals (healthy and diseased) are asked to donate samples for microbiome analyses. Two metagenomic sequencing approaches are utilized. Conserved and variable 16S rRNA genomic regions are amplified and subjected to pyrosequencing. The resulting sequences are then aligned, filtered and compared to publicly available databases of 16S rRNA sequences, enabling taxonomic classification of bacteria present or absent in a given sample. Whole genome shotgun sequencing provides information that enables identification of genes present and allows for subsequent comparison of enzymatic pathways and functions represented among different samples. Enzymatic databases are also available to assist in the identification of protein function, enabling the richness and diversity of functional capacities provided by the microbiome to be assessed. Abbreviations: PCR, polymerase chain reaction; rRNA, ribosomal RNA.
The international scientific community has recently established roadmaps for the HMP41 and the European Commission’s MetaHit consortium.42 Their common mission, as a complement to the Human Genome Project, is to generate resources enabling comprehensive characterization of the human microbiome and analysis of its role in human health and disease.
The microbiome and its host
Development of the human microbiome
Humans and other mammals undergo embryonic maturation protected by a closed, sterile, micro-organism-free environment. However, soon after birth, the neonate’s gut is colonized by bacteria derived from the surfaces contacted during, or soon after, delivery (mainly vaginal flora in vaginally delivered infants or skin-associated bacteria in the case of cesarean section).43 After a short period of relative instability, in which a few bacterial species compete for survival and alternate their dominance, phylogenetic richness and species diversity increase over time.44,45 By the end of the first year of life, a typical adult microbiota profile emerges, which becomes increasingly stable with age. At this stage, more than 1,000 different species from a dozen different divisions colonize the gastrointestinal tract.42,46 However, the human intestine is dominated by just two divisions of bacteria (Bacteroidetes and Firmicutes), which combined represent more than two-thirds of the total gut microbiota.35 Astoundingly, the composition of the adult human gut microbiome can be classified into just three distinct and stable combinations (enterotypes) that appear across populations from a variety of backgrounds.47 These enterotypes are dictated by the bacterial species present and are independent of individual host properties.
Whereas the highly specific and refined selection process involved in establishment and maintenance of the microbiome remains poorly understood, the symbiotic processes and bidirectional cross-talk between microbiome and host immune system are increasingly appreciated. Indeed, the idea that commensal bacteria are a mere group of passive organisms that obtain nutritional benefit at the host’s expense has become untenable. Compelling evidence obtained over the past 3 years demonstrates that the intestinal microbiota is able to shape the immune system to maintain homeostasis in healthy states or promote inflammation when the composition of the microbial community becomes imbalanced (dysbiosis).48
Keeping the microbiome in check
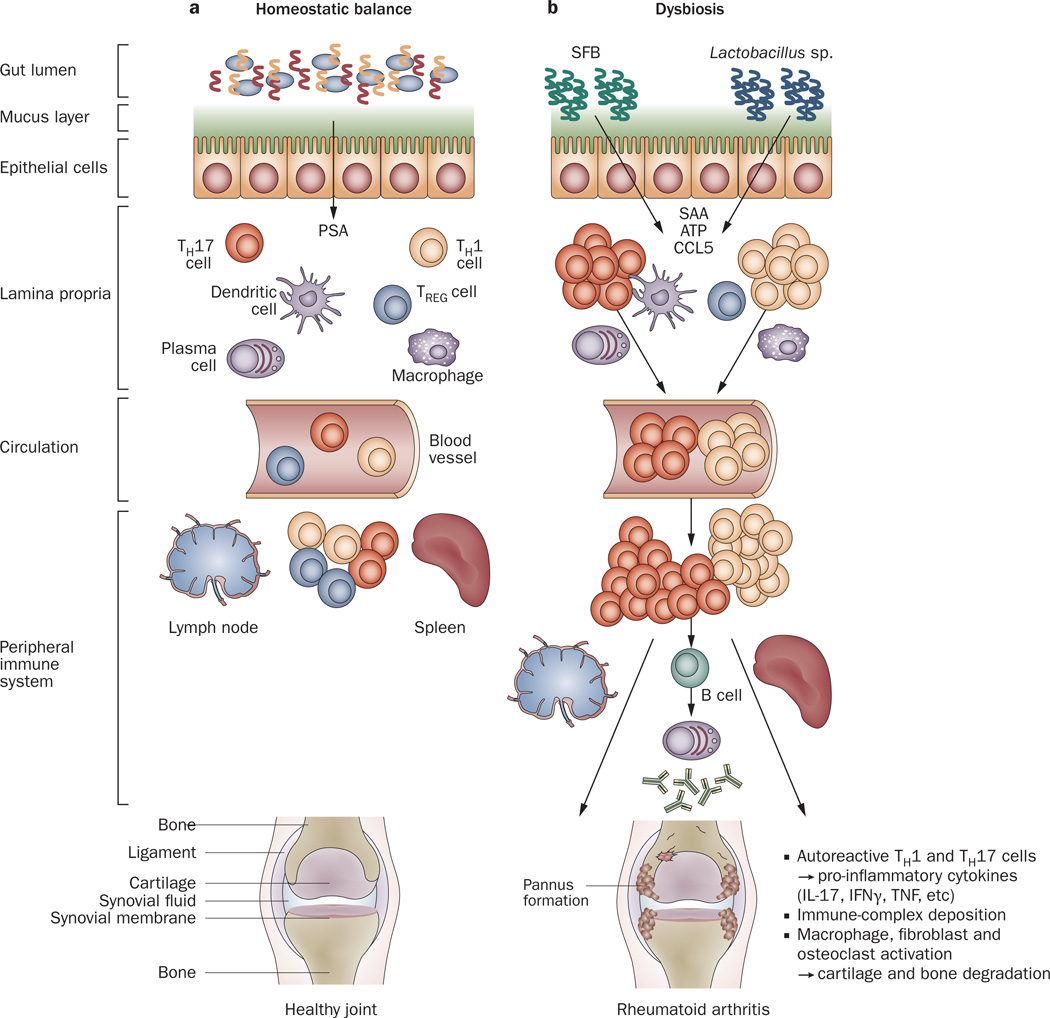
Multiple protective mechanisms prevent the gut microbiota from accessing the intestinal lamina propria and the systemic compartment (Figure 3a). A physicochemical barrier composed of a thick mucus layer,49 antimicrobial proteins,50 and secretory IgA antibodies51 coalesces to minimize the contact between the commensal microbes in the gut lumen and intestinal epithelial cells that line the gut wall. Bacteria escaping this initial ‘buffer zone’ encounter a second defense strategy, a physical boundary provided by the tight junctions formed between the intestinal epithelial cells.52 These cells are not only an anatomical boundary, but also exhibit active antibacterial properties. For example, epithelial cells produce a variety of bactericidal proteins such as defensins, cathelicidins and C-type lectins. Furthermore, epithelial cells present Toll-like receptors (TLRs) in their cellular membrane, which allow for the recognition of PAMPs (pathogen-associated molecular patterns), activation of the signaling adaptor molecule MyD88 (myeloid differentiation primary-response protein 88), and induction of downstream inflammatory responses. The lamina propria innate immune cells, constantly surveying the contents of the gut lumen in search of undesirable antigens, constitute another defense mechanism.53 Intestinal macrophages phagocytose micro-organisms and kill them within intracellular organelles containing digesting enzymes and reactive oxygen species. Concomitantly, lamina propria dendritic cells (DCs) behave as APCs (antigen-presenting cells) by presenting foreign peptides loaded into their MHC class II molecules; engagement of these molecules by B-cell or T-cell receptors primes these cells to initiate an adaptive immune response. Most recently, the influence of the gut microbiota on CD4+ T-cell differentiation and the subsequent nature of adaptive immune response has been clarified.48 The existence of a complex network of receptors, cytokines and transcription factors in naive T helper cells allows for their biological plasticity, enabling them to differentiate into several proinflammatory or anti-inflammatory subsets. Type 1 T helper (TH1) cells, for example, develop in response to detection of intracellular pathogens. By contrast, type 2 T helper (TH2) and type 17 T helper (TH17) cells are predominantly stimulated after identification of extracellular organisms. Regulatory T (TREG) cells counterbalance these proinflammatory factors by actively downregulating inflammation through activation of their specific transcription receptor factor, forkhead box protein P3 (FoxP3), and the subsequent generation of anti-inflammatory cytokines, such as IL-10. Therefore, a delicate equilibrium is in place to maintain a basal state of physiological gut mucosal inflammation. In a healthy gut environment, TH1, TH17, and TREG cells all act in concert with B cells, DCs, epithelial cells and phagocytic cells to prevent commensal organisms from causing damage to the host.54
Figure 3.
Host–microbiota interactions in health and inflammatory arthritis. a | In healthy individuals, a well balanced host–microbial cross-talk is essential for the maintenance of homeostasis. A thick mucus layer and epithelial cells prevent direct contact with the gut-associated immune cells, which constantly survey the contents of the intestinal lumen and eliminate undesired antigens. Commensal bacteria, such as Bacteroides fragilis, can activate pro-tolerogenic machinery. A specific cell wall component, PSA, is sufficient to induce TREG-cell activation, IL-10 production and TH17-cell repression to avoid uncontrolled inflammation. b | When either genetic or environmental factors alter the balance in the microbiota composition, dysbiosis ensues. Potentially harmful micro-organisms (such as SFB or Lactobacillus) predominate and local expansion of proinflammatory cells (TH17 cells, TH1 cells and others) occurs via different molecules (such as ATP, SAA or CCL5 signaling). These autoreactive T cells migrate to peripheral immune compartments and activate B cells to differentiate into autoantibody-producing plasma cells. These cells and antibodies then migrate to synovial tissue where the inflammatory cascade is amplified through the activation of effector components, including macrophages, fibroblasts, osteoclasts, cytokines and proteinases. If self-perpetuating, this process can lead to arthritis and pannus formation. Abbreviations: ATP, adenosine-5'-triphosphate; CCL5, CC-chemokine ligand 5; IFNγ, interferon γ; IL-17, interleukin-17; PSA, polysaccharide A; SAA, serum amyloid A; SFB, segmented filamentous bacteria; TH1, type 1 T helper cell; TH17, type 17 T helper cell; TREG, regulatory T cell.
Immune homeostasis—it takes two to tango
How did the human immune system learn to fight and eradicate pathogens while tolerating the massive antigenic burden of commensal–symbiotic microbiota? This question remains a matter of intense research. However, critical work suggests that even single gut bacterial species can be sufficient to tilt the homeostatic balance of the immune system in either direction. Bacteroides fragilis, a common culturable commensal, supports anti-inflammatory responses by activating IL-10-producing TREG cells through a specific capsular component, polysaccharide A (PSA). This molecule exploits the TLR2 signaling pathway to actively suppress immunity and, simultaneously, dampen TH17 responses.55 A defined set of indigenous intestinal Clostridia species demonstrated similar TREG-cell-inducing properties.56 On the other hand, commensal segmented filamentous bacteria (SFB) were shown to be sufficient to induce production and activation of lamina propria TH17 cells, which subsequently secrete their signature proinflammatory cytokine, IL-17.57
Dysbiosis and autoimmune disease
As introduced above, the gut microbiota has a considerable impact on the maintenance of a local homeostatic balance owing to its proximity to the host’s intestinal immunity and its ability to influence immune responses. Therefore, it is reasonable to expect that dysbiosis might lead to inflammatory bowel disease (IBD) and related conditions.58 Indeed, multiple lines of evidence suggest that an increase in pathobionts (microbial symbionts that can cause disease after alterations in gut environment) and/or a decrease in symbionts (commensal species with beneficial effects to the host) creates imbalance in the immune responses by the innate and adaptive systems,59 resulting in the initiation of an inflammatory cascade with subsequent local tissue disruption and clinical disease. The findings presented in a paper published in 2011 clearly exemplify this theory, by demonstrating that the inflammasome pathway—a key component of the innate immune response that ultimately leads to production of the proinflammatory IL-1 and IL-18 cytokines—is required to maintain health. Knockout of various inflammasome-related genes in mice (including deletion of the genes encoding NLRP6 [NACHT, LRR and PYD domains-containing protein 6], ASC1 [Asc-type amino acid transporter 1], caspase-1, or IL-18), results in proliferation of Prevotella and TM7 phylotypes, which leads to exacerbation of colitis.60 Remarkably, wild-type animals demonstrate a similar disease phenotype when co-housed with knockout animals. Until recently, however, the concept that gut microbiota could influence the peripheral immune system or cause extraintestinal immune-driven disease was difficult to imagine.54 Seminal independent studies, performed by different groups,61,62 have altered this notion through the demonstration that single commensal bacterial species can drive a local adaptive immune response, which subsequently triggers or prevents autoimmunity at distant tissue sites, such as the central nervous system (Figure 3b). For example, studies in experimental autoimmune encephalomyelitis (EAE), an animal model of human MS (multiple sclerosis), suggest that alteration of the intestinal microbiome can enhance disease penetrance and severity.63 Conversely, oral immunization with an attenuated Salmonella typhimurium expressing the colonization factor antigen I protects against both EAE and collagen-induced arthritis.64 In the collagen-induced arthritis model, IL-35 activates CD39+ TREG cells that, in turn, induce immune suppression and arthritis abrogation via IL-10 production.65
The microbiome and RA pathogenesis
Evidence from germ-free animal models
Several studies have established the importance of the gut microbiota in the proper immune system maturation and competency by taking advantage of experiments comparing animals raised under germ-free conditions (through the use of special isolators that preclude the presence of bacteria) with those housed in conventional cages. A similar rationale was also used to assess the influence of intestinal bacteria as triggers for inflammatory arthritis in various susceptible animal models (Figure 4). A rich literature exists in which the authors have sought to establish possible associations between microbes and arthritis.66 However, given the focus and space limitations for this Review, we will herein illustrate this area of study with the aid of a few notable examples.
Figure 4.
Multiple animal models of inflammatory arthritis have demonstrated that the gut microbiota is critical for the development of disease. The use of gnotobiotic experiments, in which animals are kept germ-free until specific microorganisms are introduced, have advanced our understanding of how local changes in the gut flora produce an imbalance in the proinflammatory and anti-inflammatory immune response and ultimately trigger autoimmunity at distal sites. SFB are sufficient to activate lamina propria TH17 cells in the K/BxN model of inflammatory arthritis. These cells migrate to the periphery, produce IL-17 (their signature cytokine) and stimulate plasma cells to produce arthritogenic autoantibodies. However, when kept in germ-free conditions, these animals do not develop arthritis. Lactobacillus is also capable of arthritis-induction in the Il1rn−/− model. Increase in TH17 cell activity and decrease in TREG cell function are key to the development of joint inflammation. Abbreviations: AA, adjuvant arthritis; CIA, collagen-induced arthritis; IL-17, interleukin-17; SFB, segmented filamentous bacteria; TH1, type 1 T helper cell; TH17, type 17 T helper cell; TLR, Toll-like receptor; TREG, regulatory T cell.
The first description of the possible involvement of bacterial flora in the pathology of arthritis came in the late 1970s when rats raised under germ-free conditions developed severe joint inflammation with 100% penetrance in an adjuvant-induced arthritis model, while conventionally raised controls showed only mild disease at a very low incidence.67 This finding suggests that, although a microbiota is not necessary for the development of arthritis, its presence has a potential suppressive effect through modulation of the immune response. The mechanism behind this suppressive effect remains unclear. The humoral immune system is not a prerequisite for arthritis, as germ-free rats do not produce specific autoantibodies to heat-shock protein (hsp) 65 and yet do develop clinical disease. This observation seems to indicate that immunity to hsp65 in experimental arthritis is unrelated to disease and may instead be regarded as an epiphenomenon dependent on the presence of gut flora.68 These findings were followed by several reports attributing either protective69,70 or proarthritogenic71,72 roles to certain Gram-negative enterobacteria (strains of Escherichia coli and Bacteroides species) when introduced into otherwise germ-free arthritis-prone rats. An important observation implicating commensal gut flora in a different model for arthritis was made by Taurog et al. in 1994, when they showed that HLA-B27 transgenic rats (a spontaneous model of spondyloarthropathy) raised in a germ-free environment did not develop inflammatory intestinal or peripheral joint disease, whereas inflammatory lesions in the skin and genitals were unaffected by the germ-free state.73 Another notable and classical example with similar findings was reported in a streptococcal cell wall-induced rat arthritis model. In this model, animals reared conventionally are resistant to joint inflammation, whereas germ-free rats become susceptible to arthritic disease, mainly through loss of T-cell tolerance.74 These findings support the concept that gut and joint inflammation are interconnected through the part played by the commensal gut flora in immune homeostasis. Whether flora is protective or deleterious for joint health might also be related to the host genetic background, as differing experimental results described above were conducted using alternate rat strains with various degrees of arthritis susceptibility.
Evidence from gnotobiotic animal studies
The use of gnotobiotic animals (germ-free mice that are colonized with defined microbiota at specific stages of their life) has enhanced our understanding of the role of gut microbial communities in systemic disease. IL-1 receptor antagonist-knockout (Il1rn−/−) mice, which spontaneously develop an autoimmune T-cell-mediated arthritis, did not develop disease when raised in a germfree environment. However, monocolonization of Il1rn−/− mice with the commensal Lactobacillus bifidus resulted in rapid disease onset, of comparable severity and incidence to the arthritis observed in non-germ-free mice.61 L. bifidus-triggered arthritis in this model is driven by an imbalance in TREG–TH17 cell homeostasis and mediated through TLR2–TLR4 signaling.
Gnotobiotic studies have also been performed in the K/BxN T-cell receptor transgenic model of inflammatory arthritis, which is caused by autoreactive T-cell-driven production of autoantibodies against glucose-6-phosphate isomerase. In this case, arthritis is attenuated in germ-free animals and this decrease in disease is linked to a deficiency of peripheral TH17 cells. Introduction of a single gut-residing commensal, SFB, restored the disease phenotype, while arthritis was strongly inhibited if K/BxN mice were treated from birth with antibiotics targeting SFB.62 Joint inflammation in SKG mice, another TH17-driven RA-like disease model, is also dependent on the presence of micro-organisms, although fungal phylotypes and cell wall components seem more relevant to pathogenesis in these animals.75
Taken together, these data suggest that a particular intestinal microbiota is required to trigger (if not drive) systemic autoimmunity leading to inflammatory arthritis in animal models of RA-like disease. These findings also support the notion that a state of dysbiosis might require genetic host susceptibility, as illustrated by the inability of wild-type animals to mount an inflammatory response even in the presence ‘proarthritogenic’ gut flora.
Insight from human arthritides
Proof-of-principle that the ‘gut–joint axis’ hypothesis is relevant to human rheumatic disease can be found in the pathogenesis of several arthritides. Spondyloarthropathies, particularly reactive arthritis and IBD-related arthritis, are the most prevalent examples.76,77 Jejunoileal-bypass-associated arthritis, which now occurs infrequently due to the use of bariatric banding alternatives to this procedure, can also be included in this category. Several reports have associated this condition with bacterial overgrowth and deposition of resultant immune complexes in the synovium.78 Arguably, however, the human model best fitting the gut–joint axis hypothesis is represented by classic Whipple disease, in which the presence of a single bacterium in the small intestine, Tropheryma whipplei, is sufficient for the development of joint inflammation in predisposed individuals.79
Long before the vastness of the human microbiome was unveiled, the rheumatology community embraced therapeutic regimens that targeted the entero–arthropathy connection; several have been classified as DMARDs and are still in use today. In the 1940s, sulfasalazine became the first rationally designed drug in the field of rheumatology. As RA was thought to be caused by streptococci found in milk,80 sulfasalazine was created as a deliberate attempt to combine a sulfonamide antibiotic with a salicylate anti-inflammatory agent through an azo bond. This ‘combination therapy’ showed good initial results in RA,81 before sulfasalazine went out of use for several decades (although it remained as standard of care for IBD). More rigorously designed trials rescued sulfasalazine from darkness, when it was shown to be superior to placebo82 and equivalent to methotrexate when used in combination with the antimalarial agent hydroxychloroquine.83 Today, the triple DMARD therapy (combination of sulfasalazine, hydroxychloroquine and methotrexate) remains one of the few first choices for all patients with RA who demonstrate poor prognostic features and moderate or high levels of disease activity, regardless of disease duration.84 Furthermore, the efficacy of triple DMARD therapy is equivalent to that observed for combined use of methotrexate and a biologic agent in early RA.85 In several studies, tetracycline antibiotics have also been proven efficacious in the treatment of early seropositive RA,86,87 leading to the approval of one of these, minocycline, as a DMARD. Despite encouraging clinical outcomes, the underlying mechanisms of action for all these drugs have never been completely elucidated.
Future directions
Several studies addressing the role of the gut microbiota in RA pathogenesis are currently underway. Intriguingly, patients with RA have defective circulating TREG cell function88 and increased abundance of TH17 cells (and their signature cytokine, IL-17) in both plasma and the synovium. 89,90 Conceivably, intestinal dysbiosis could lead (in a predisposed host) to a break in immune tolerance, followed by a systemic immune disequilibrium that ultimately favors proinflammatory responses resulting in tissue damage in the periphery (for example, joints).
Given preliminary data,91 one can speculate that patients with RA carry a distinctive enterotype, which might either trigger or drive autoimmunity in individuals with genetic predisposing factors. Alternatively, different enterotypes might prove protective, even in genetically predisposed individuals. If either of these hypotheses can be established, it would open multiple avenues for the identification of potential therapeutic targets, biomarker discovery, or even the development of preventive approaches in RA. Mechanistic studies will then be required to establish causation, including gnotobiotic experiments in humanized mice, monocolonization of animal models with candidate organisms, and host immune response evaluation.
Conclusions
A fine equilibrium between ‘peace-keeping’ and potentially proinflammatory intestinal bacteria is necessary to keep gut immunity in check and prevent a state of dysbiosis, which might lead to local and distant deleterious consequences in the host. Impressive advances in sequencing technologies, compelling animal data and mounting human evidence suggest that gut microbiota indeed play a part in the pathogenesis of diseases such as autoimmune arthritis.
Well-defined human studies using 16S rRNA pyrosequencing methods, coupled with shotgun analyses and gnotobiotic experiments, are needed to better understand if and when the intestinal community composition in patients with RA differs from that in healthy populations. Prospective studies evaluating the microbiome–host relationship in RA are also necessary to establish not only potential causation, but also the effects of immunosuppressive therapies on the microbiota, which might subsequently impact on the host’s well-being. A deeper understanding of the biological complexities of our ‘two genomes’ will help in the elucidation of possible triggering factors in RA and close the gap in our knowledge regarding the role of gene–environment interactions in other autoimmune processes involved in disease pathogenesis.
Key points.
-
▪
In rheumatoid arthritis (RA)—a complex, polygenic, autoimmune disorder with a major impact on individuals and society—genes have a role, but environmental factors are required for disease manifestation
-
▪
Multiple lines of epidemiological and clinical investigation have implicated several micro-organisms in RA pathogenesis; however, causation could not be established
-
▪
The microbiome is defined as the totality of micro-organisms and their genes inhabiting a unique environment; the human microbiome outnumbers human genes by several orders of magnitude
-
▪
Understanding of the role of micro-organisms in modulating health and disease has been greatly advanced by culture-independent DNA sequencing technologies and novel insights into mucosal immunology
-
▪
Germ-free and gnotobiotic experiments have provided a deeper understanding of host–microbial interactions and have shown that gut bacteria can induce autoimmunity in genetically predisposed animal models
-
▪
Studies are underway to assess the role of the microbiome in human RA and related diseases in the hope that disease mechanisms will be elucidated and therapeutic targets identified
Review criteria.
We searched for original articles and abstracts focusing on the microbiome in MEDLINE, PubMed, the Google Scholar search engine and the American College of Rheumatology website with no restriction on publication date. The search terms used in various combinations were “microbiome”, “rheumatoid”, “arthritis”, “infection”, “gnotobiotic” and “germ-free”. All papers identified were English-language full-text papers and abstracts. We also searched the reference lists of identified articles for further papers of relevance.
Acknowledgments
The writing of this manuscript has been supported in part by Grant No. RC2 AR05898 to S. B. Abramson from the US NIH through the American Recovery and Reinvestment Act (ARRA) of 2009, and by KL2 Program in Translational Research to J. U. Scher, Grant No. 1 UL1 RR029893 from the National Center for Research Resources, NIH. The authors thank Ms. Ann Rupel for assistance in preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
J. U. Scher and S. B. Abramson contributed equally to all aspects of the preparation of this manuscript.
References
- 1.Savage DC. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 3.Lederberg J. Infectious history. Science. 2000;288:287–293. doi: 10.1126/science.288.5464.287. [DOI] [PubMed] [Google Scholar]
- 4.Lederberg J, McCray AT. ‘Ome sweet ‘omics—A genealogical treasury of words. Scientist. 2001;15:8–9. [Google Scholar]
- 5.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chervonsky AV. Influence of microbial environment on autoimmunity. Nat. Immunol. 2010;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 7.Klareskog L, Catrina AI, Paget S. Rheumatoid arthritis. Lancet. 2009;373:659–672. doi: 10.1016/S0140-6736(09)60008-8. [DOI] [PubMed] [Google Scholar]
- 8.MacGregor AJ, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aho K, Koskenvuo M, Tuominen J, Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J. Rheumatol. 1986;13:899–902. [PubMed] [Google Scholar]
- 11.Silman AJ, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br. J. Rheumatol. 1993;32:903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- 12.Svendsen AJ, Holm NV, Kyvik K, Petersen PH, Junker P. Relative importance of genetic effects in rheumatoid arthritis: historical cohort study of Danish nationwide twin population. BMJ. 2002;324:264–266. [PMC free article] [PubMed] [Google Scholar]
- 13.Tobón GJ, Youinou P, Saraux A. The environment, geo-epidemiology, and autoimmune disease: Rheumatoid arthritis. J. Autoimmun. 2010;35:10–14. doi: 10.1016/j.jaut.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 14.Short CL. The antiquity of rheumatoid arthritis. Arthritis Rheum. 1974;17:193–205. doi: 10.1002/art.1780170302. [DOI] [PubMed] [Google Scholar]
- 15.Ruffer MA, Rietti A. On osseous lesions in ancient Egyptians. J. Pathol. Bacteriol. 1912;16:439–465. [Google Scholar]
- 16.Bourke JB. A review of the paleopathology of arthritic diseases. In: Brothwell D, Sandison AT, editors. Diseases in Antiquity. Springfield, IL, USA: Thomas; 1967. pp. 352–369. [Google Scholar]
- 17.Zorab PA. Historical and prehistorical background of ankylosing spondylitis. Proc. R. Soc. Med. 1961;54:415–420. [PMC free article] [PubMed] [Google Scholar]
- 18.Wells C. Joint pathology in ancient Anglo-Saxons. J. Bone Joint. Surg. 1962;44B:948–949. [Google Scholar]
- 19.Appelboom T. Hypothesis: Rubens—one of the first victims of an epidemic of rheumatoid arthritis that started in the 16th–17th century? Rheumatology (Oxford) 2005;44:681–683. doi: 10.1093/rheumatology/keh252. [DOI] [PubMed] [Google Scholar]
- 20.Rothschild BM, Turner KR, DeLuca MA. Symmetrical erosive peripheral polyarthritis in the Late Archaic Period of Alabama. Science. 1988;241:1498–1501. doi: 10.1126/science.3047874. [DOI] [PubMed] [Google Scholar]
- 21.Rothschild BM, Woods RJ, Rothschild C, Sebes JI. Geographic distribution of rheumatoid arthritis in ancient North America: implications for pathogenesis. Semin. Arthritis Rheum. 1992;22:181–187. doi: 10.1016/0049-0172(92)90018-9. [DOI] [PubMed] [Google Scholar]
- 22.Ferucci ED, Templin DW, Lanier AP. Rheumatoid arthritis in American Indians and Alaska Natives: a review of the literature. Semin. Arthritis Rheum. 2005;34:662–667. doi: 10.1016/j.semarthrit.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Zeng QY, et al. Rheumatic diseases in China. Arthritis Res. Ther. 2008;10:R17. doi: 10.1186/ar2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGill PE, Oyoo GO. Rheumatic disorders in Sub-saharan Africa. East Afr. Med. J. 2002;79:214–216. doi: 10.4314/eamj.v79i4.8882. [DOI] [PubMed] [Google Scholar]
- 25.Neovius M, Simard JF, Askling J. Nationwide prevalence of rheumatoid arthritis and penetration of disease-modifying drugs in Sweden. Ann. Rheum. Dis. 2011;70:624–629. doi: 10.1136/ard.2010.133371. [DOI] [PubMed] [Google Scholar]
- 26.Myasoedova E, Crowson CS, Kremers HM, Therneau TM, Gabriel SE. Is the incidence of rheumatoid arthritis rising?: results from Olmsted County, Minnesota, 1955–2007. Arthritis Rheum. 2010;62:1576–1582. doi: 10.1002/art.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warden CC. The toxemic factor in rheumatoid arthritis. Cal. State. J. Med. 1909;7:299–301. [PMC free article] [PubMed] [Google Scholar]
- 28.Eerola E, et al. Intestinal flora in early rheumatoid arthritis. Br. J. Rheumatol. 1994;33:1030–1038. doi: 10.1093/rheumatology/33.11.1030. [DOI] [PubMed] [Google Scholar]
- 29.Hunter W. Oral sepsis as a cause of disease. Br. Med. J. 1900;2:215–216. doi: 10.1136/bmj.2.2065.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikuls TR, et al. Antibody responses to Porphyromonas gingivalis (P. gingivalis) in subjects with rheumatoid arthritis and periodontitis. Int. Immunopharmacol. 2009;9:38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitchon CA, et al. Antibodies to Porphyromonas gingivalis are associated with anticitrullinated protein antibodies in patients with rheumatoid arthritis and their relatives. J. Rheumatol. 2010;37:1105–1112. doi: 10.3899/jrheum.091323. [DOI] [PubMed] [Google Scholar]
- 32.Loyola-Rodriguez JP, Martinez-Martinez RE, Abud-Mendoza C, Patino-Marin N, Seymour GJ. Rheumatoid arthritis and the role of oral bacteria. J. Oral Microbiol. 2010 doi: 10.3402/jom.v2i0.5784. http://dx.doi. org/10.3402/jom.v2i0.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA—the citrullinated enolase connection. Nat. Rev. Rheumatol. 2010;6:727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- 34.Koch R. An address on bacteriological research. Br. Med. J. 1890;2:380–383. doi: 10.1136/bmj.2.1546.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huse SM, et al. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000255. e1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L. Genomics: The tale of our other genome. Nature. 2010;465:879–880. doi: 10.1038/465879a. [DOI] [PubMed] [Google Scholar]
- 40.Nelson KE, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl Acad. Sci. USA. 2011;108 Suppl. 1:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johansson ME, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer-Hoffert U, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 51.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 52.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 53.Kelsall B. Recent progress in understanding the phenotype and function of intestinal dendritic cells and macrophages. Mucosal. Immunol. 2008;1:460–469. doi: 10.1038/mi.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat. Rev. Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 55.Round JL, et al. The Toll-Like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 60.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdollahi-Roodsaz S, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 2008;118:205–216. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ochoa-Reparaz J, Mielcarz DW, Begum-Haque S, Kasper LH. Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease. Ann. Neurol. 2011;69:240–247. doi: 10.1002/ana.22344. [DOI] [PubMed] [Google Scholar]
- 64.Kochetkova I, Trunkle T, Callis G, Pascual DW. Vaccination without autoantigen protects against collagen II-induced arthritis via immune deviation and regulatory T cells. J. Immunol. 2008;181:2741–2752. doi: 10.4049/jimmunol.181.4.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kochetkova I, Golden S, Holderness K, Callis G, Pascual DW. IL-35 stimulation of CD39+ regulatory T cells confers protection against collagen II-induced arthritis via the production of IL-10. J. Immunol. 2010;184:7144–7153. doi: 10.4049/jimmunol.0902739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hyrich KL, Inman RD. Infectious agents in chronic rheumatic diseases. Curr. Opin. Rheumatol. 2001;13:300–304. doi: 10.1097/00002281-200107000-00010. [DOI] [PubMed] [Google Scholar]
- 67.Kohashi O, et al. Susceptibility to adjuvant-induced arthritis among germfree, specific-pathogen-free, and conventional rats. Infect. Immun. 1979;26:791–794. doi: 10.1128/iai.26.3.791-794.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bjork J, Kleinau S, Midtvedt T, Klareskog L, Smedegard G. Role of the bowel flora for development of immunity to hsp 65 and arthritis in three experimental models. Scand. J. Immunol. 1994;40:648–652. doi: 10.1111/j.1365-3083.1994.tb03518.x. [DOI] [PubMed] [Google Scholar]
- 69.Kohashi O, Kohashi Y, Takahashi T, Ozawa A, Shigematsu N. Reverse effect of gram-positive bacteria vs. gram-negative bacteria on adjuvant-induced arthritis in germfree rats. Microbiol. Immunol. 1985;29:487–497. doi: 10.1111/j.1348-0421.1985.tb00851.x. [DOI] [PubMed] [Google Scholar]
- 70.Kohashi O, Kohashi Y, Takahashi T, Ozawa A, Shigematsu N. Suppressive effect of Escherichia coli on adjuvant-induced arthritis in germ-free rats. Arthritis Rheum. 1986;29:547–553. doi: 10.1002/art.1780290413. [DOI] [PubMed] [Google Scholar]
- 71.Rath HC, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J. Clin. Invest. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sinkorova Z, Capkova J, Niederlova J, Stepankova R, Sinkora J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2(k)) male mice. Hum. Immunol. 2008;69:845–850. doi: 10.1016/j.humimm.2008.08.296. [DOI] [PubMed] [Google Scholar]
- 73.Taurog JD, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 1994;180:2359–2364. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van den Broek MF, van Bruggen MC, Koopman JP, Hazenberg MP, van den Berg WB. Gut flora induces and maintains resistance against streptococcal cell wall-induced arthritis in F344 rats. Clin. Exp. Immunol. 1992;88:313–317. doi: 10.1111/j.1365-2249.1992.tb03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshitomi H, et al. A role for fungal β-glucans and their receptor Dectin-1 in the induction of autoimmune arthritis in genetically susceptible mice. J. Exp. Med. 2005;201:949–960. doi: 10.1084/jem.20041758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodriguez-Reyna TS, Martinez-Reyes C, Yamamoto-Furusho JK. Rheumatic manifestations of inflammatory bowel disease. World. J. Gastroenterol. 2009;15:5517–5524. doi: 10.3748/wjg.15.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter JD, Hudson AP. Reactive arthritis: clinical aspects and medical management. Rheum. Dis. Clin. North Am. 2009;35:21–44. doi: 10.1016/j.rdc.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 78.Ross CB, Scott HW, Pincus T. Jejunoileal bypass arthritis. Baillieres Clin. Rheumatol. 1989;3:339–355. doi: 10.1016/s0950-3579(89)80025-1. [DOI] [PubMed] [Google Scholar]
- 79.Moos V, Schneider T. Changing paradigms in Whipple’s disease and infection with Tropheryma whipplei. Eur. J. Clin. Microbiol. Infect. Dis. doi: 10.1007/s10096-011-1209-y. http://dx.doi.org/10.1007/s10096-011-1209-y. [DOI] [PubMed] [Google Scholar]
- 80.Svartz N. The primary cause of rheumatoid arthritis is an infection—the infectious agent exists in milk. Acta Med. Scand. 1972;192:231–239. doi: 10.1111/j.0954-6820.1972.tb04807.x. [DOI] [PubMed] [Google Scholar]
- 81.Svartz N. The treatment of rheumatic polyarthritis with acid azo compounds. Rheumatism. 1948;4:180–185. [PubMed] [Google Scholar]
- 82.Hannonen P, Mottonen T, Hakola M, Oka M. Sulfasalazine in early rheumatoid arthritis. A 48-week double-blind, prospective, placebo-controlled study. Arthritis Rheum. 1993;36:1501–1509. doi: 10.1002/art.1780361104. [DOI] [PubMed] [Google Scholar]
- 83.O’Dell JR, et al. Treatment of rheumatoid arthritis with methotrexate alone, sulfasalazine and hydroxychloroquine, or a combination of all three medications. N. Engl. J. Med. 1996;334:1287–1291. doi: 10.1056/NEJM199605163342002. [DOI] [PubMed] [Google Scholar]
- 84.Saag KG, et al. American College of Rheumatology 2008 recommendations for the use of nonbiologic and biologic disease-modifying antirheumatic drugs in rheumatoid arthritis. Arthritis Rheum. 2008;59:762–784. doi: 10.1002/art.23721. [DOI] [PubMed] [Google Scholar]
- 85.Moreland LW, et al. TEAR: Treatment of Early Aggressive RA; a randomized, double-blind, 2-year trial comparing immediate triple DMARD versus MTX plus etanercept to step-up from initial MTX monotherapy. Arthritis Rheum. 2009;60 Suppl. 10:1895. [Google Scholar]
- 86.Tilley BC, et al. Minocycline in rheumatoid arthritis. A 48-week, double-blind, placebo-controlled trial. MIRA Trial Group. Ann. Intern. Med. 1995;122:81–89. doi: 10.7326/0003-4819-122-2-199501150-00001. [DOI] [PubMed] [Google Scholar]
- 87.O’Dell JR, et al. Treatment of early seropositive rheumatoid arthritis: doxycycline plus methotrexate versus methotrexate alone. Arthritis Rheum. 2006;54:621–627. doi: 10.1002/art.21620. [DOI] [PubMed] [Google Scholar]
- 88.Zanin-Zhorov A, et al. Protein kinase C-θ mediates negative feedback on regulatory T cell function. Science. 2010;328:372–376. doi: 10.1126/science.1186068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hot A, Miossec P. Effects of interleukin (IL)-17A and IL-17F in human rheumatoid arthritis synoviocytes. Ann. Rheum. Dis. 2011;70:727–732. doi: 10.1136/ard.2010.143768. [DOI] [PubMed] [Google Scholar]
- 90.Colin EM, et al. 1, 25-dihydroxyvitamin D3 modulates Th17 polarization and interleukin-22 expression by memory T cells from patients with early rheumatoid arthritis. Arthritis Rheum. 2010;62:132–142. doi: 10.1002/art.25043. [DOI] [PubMed] [Google Scholar]
- 91.Scher JU, et al. Characteristic oral and intestinal microbiota in rheumatoid arthritis (RA): a trigger for autoimmunity? Arthritis Rheum. 2010;62 suppl. 10 [Google Scholar]