Abstract
In H295R human adrenocortical cells, ACTH rapidly activates ceramide (Cer) and sphingosine (SPH) turnover with a concomitant increase in SPH-1-phosphate secretion. These bioactive lipids modulate adrenocortical steroidogenesis, primarily by acting as second messengers in the protein kinase A/cAMP-dependent pathway. Acid ceramidase (ASAH1) directly regulates the intracellular balance of Cer, SPH, and SPH-1-phosphate by catalyzing the hydrolysis of Cer into SPH. ACTH/cAMP signaling stimulates ASAH1 transcription and activity, supporting a role for this enzyme in glucocorticoid production. Here, the role of ASAH1 in regulating steroidogenic capacity was examined using a tetracycline-inducible ASAH1 short hairpin RNA H295R human adrenocortical stable cell line. We show that ASAH1 suppression increases the transcription of multiple steroidogenic genes, including Cytochrome P450 monooxygenase (CYP)17A1, CYP11B1/2, CYP21A2, steroidogenic acute regulatory protein, hormone-sensitive lipase, 18-kDa translocator protein, and the melanocortin-2 receptor. Induced gene expression positively correlated with enhanced histone H3 acetylation at target promoters. Repression of ASAH1 expression also induced the expression of members of the nuclear receptor nuclear receptor subfamily 4 (NR4A) family while concomitantly suppressing the expression of dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1. ASAH1 knockdown altered the expression of genes involved in sphingolipid metabolism and changed the cellular amounts of distinct sphingolipid species. Finally, ASAH1 silencing increased basal and cAMP-dependent cortisol and dehydroepiandrosterone secretion, establishing ASAH1 as a pivotal regulator of steroidogenic capacity in the human adrenal cortex.
In the human adrenal cortex, cortisol is synthesized from cholesterol by cytochrome P450 monooxygenase (CYP)11A1, CYP17A1, CYP11B1/2, and CYP21A2 and 3β-hydroxysteroid dehydrogenase (3β-HSD) type II enzymes in a process primarily regulated by the peptide hormone ACTH (1–4). In the zonae fasciculata and reticularis, ACTH increases steroid hydroxylase gene expression by activating adenylyl cyclase and consequently increasing intracellular cAMP (4). This second messenger activates protein kinase A, which acutely promotes cholesterol mobilization to the inner mitochondrial membrane and chronically induces the transcription of genes required for steroid hormone production (3, 5–7). The transcription of most steroidogenic genes is regulated by the nuclear receptor steroidogenic factor 1 (SF-1) nuclear receptor (NR)5A1, which in response to ACTH signaling binds to target promoters and facilitates the recruitment of coactivator proteins (3, 4, 8–11). Further, additional transcription regulators, including β-catenin (12, 13), dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 (DAX-1) (NR0B1) (14, 15), and the NR4A family of transcription factors (16–19), are equally important for maintaining optimal transcriptional output.
Sphingolipids have emerged as important second messengers in various signaling transduction pathways (20–27). In steroidogenesis, sphingosine (SPH) modulates steroidogenic gene transcription by serving as an antagonist for SF-1 (28). We have previously demonstrated that SPH is bound to SF-1 under basal conditions and that cAMP stimulation promotes SPH displacement from the receptor's ligand-binding pocket. SPH binding to SF-1 antagonizes the ability of cAMP to activate CYP17A1 gene transcription and stimulate dehydroepiandrosterone (DHEA) production. Silencing the expression of the SPH-generating enzyme acid ceramidase (ASAH1) mimics cAMP-stimulated CYP17A1 transcription (28), which supports a role for this enzyme in regulating SF-1 function and steroidogenic gene transcription. In many respects, steroid hormone biosynthesis and sphingolipid metabolism have a reciprocal relationship (29). In H295R cells, ACTH stimulates sphingolipid metabolism by rapidly promoting the catabolism of sphingomyelin (SM), ceramide (Cer), and SPH (30). ACTH/cAMP signaling acutely increases the enzymatic activities of SPH kinase (SK) (30, 31) and ASAH1 (32) in H295R cells. Further, we have recently established that cAMP-responsive element-binding protein is an essential transcriptional regulator of the ASAH1 gene in H295R cells (32).
Ceramidases (N-acylsphingosine amidohydrolase, ASAH) are a family of hydrolases that catalyze the degradation of Cer into SPH and a free fatty acid. Five human ceramidases have been cloned and are categorized by the pH at which they exhibit optimal in vitro activity as acid (ASAH1), neutral (ASAH2), and three isoforms of alkaline [alkaline ceramidase (ACER)1–ACER3] (33). ASAH1 is a glycoprotein processed from a 55-kDa precursor via autoproteolytic cleavage (34) into a mature heterodimeric enzyme formed by an α-subunit (13 kDa) and a β-subunit (40 kDa) (35). Because Cer degradation is the only source of cellular SPH (36), these enzymes are not only essential for limiting Cer-mediated signaling but also for controlling the cellular functions of SPH and SPH-1-phosphate (S1P) (37–39). In mouse, ASAH1 is expressed early during embryogenesis, with targeted disruption of the ASAH1 gene resulting in embryonic lethality (40). Moreover, ASAH1 overexpression has been reported in various human cancers (41–44), and a genetic deficiency in ASAH1 catalytic activity causes the lysosomal sphingolipid storage disorder, Farber's disease (45).
To determine the functional significance of Cer metabolism in adrenocortical steroidogenesis, we generated an H295R stable cell line that expresses ASAH1 short hairpin RNA (shRNA) in a tetracycline (tet)-regulated manner. We show here that suppression of ASAH1 protein expression results in global changes in gene expression, including the induction of the steroidogenic genes CYP17A1, CYP11A1, CYP21A2, CYP11B1/2, steroidogenic acute regulatory protein (StAR), 18-kDa translocator protein (TSPO), hormone-sensitive lipase (HSL), and melanocortin-2 receptor (MC2R). Consistent with increased steroidogenic gene expression, these cells exhibit a higher capacity to secrete cortisol and DHEA, both basally and in response to activation of the ACTH signaling pathway. Further, suppression of ASAH1 alters the amount of several sphingolipid species, including a decrease in C16-sphingolipids and a shift in the acyl-chain composition of Cer, SM, and glycosphingolipid subspecies. Intriguingly, we show that ASAH1 knockdown (ASAH1KD) leads to a decrease in cell proliferation with a concomitant reduction in the protein levels of β-catenin, proliferating cell nuclear antigen (PCNA), and cyclin B2.
Results
Characterization of the H295R ASAH1KD stable cell line
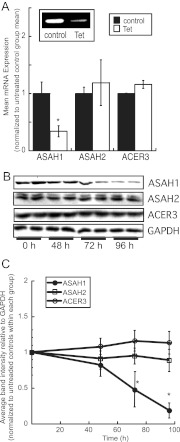
To characterize the functional significance of ASAH1 expression in regulating the steroidogenic capacity of H295R cells, we stably transfected H295R cells with the pENTR/H1/TO vector containing a sequence for an ASAH1 shRNA. As shown in Fig. 1A, ASAH1 mRNA levels were reduced by 68% in tet-treated cells (5 μg/ml tet for 48 h). Importantly, ASAH2 and ACER3 mRNA expression remained unchanged. ASAH1 protein levels are decreased after 72 and 96 h of tet treatment (Fig. 1, B and C). Similar levels of ASAH1 suppression were observed in another clone (data not shown).
Fig. 1.
Characterization of the H295R ASAH1KD cell line. A, ASAH1KD cells were treated with 5 μg/ml tet for 48 h, total RNA was harvested, and ASAH1, ASAH2, and ACER3 mRNA levels were quantified by qRT-PCR and normalized to the mRNA expression of β-actin. Inset, Representative agarose gel of qRT-PCR products from ASAH1 mRNA transcripts. Data are graphed as mean ± sem of three separate experiments, each done in triplicate. Asterisks indicate statistically significant differences from untreated controls (P < 0.05). B, ASAH1KD cells were treated with 5 μg/ml tet for 48, 72, or 96 h, cell lysates were harvested and separated by SDS-PAGE followed by Western blotting using antibodies against ASAH1, ASAH2, ACER3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). C, Densitometric analysis of Western blots of ASAH1, ASAH2, and ACER3 protein expression in, normalized to GAPDH protein content in ASAH1KD cells treated for 0–96 h with 5 μg/ml tet. Data graphed represent the mean ± sd of three separate experiments, each done in duplicate. Asterisks denote statistically significant difference (P < 0.05) when compared to untreated control groups.
ASAH1 suppression alters steroidogenic gene expression
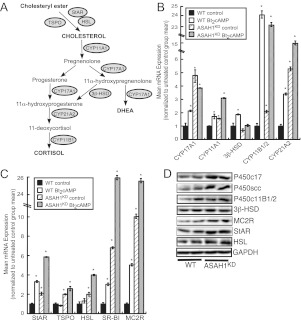
Because SPH is an antagonist for SF-1 and transient ASAH1 silencing induces CYP17A1 transcription (28), we determined the effect of reduced ASAH1 expression on global gene expression. ASAH1KD altered the expression of multiple classes of genes, including cell cycle and transcriptional regulators (Supplementals Fig. 1, published on The Endocrine Society's Journals Online web site at http://mend.endojournals.org). However, as shown in Fig. 2, reduction of ASAH1 expression markedly changed the expression of genes required for steroid hormone metabolism. Real-time RT-PCR revealed that ASAH1KD induced CYP11B1/2 and CYP21A2 transcription by 2.1- and 5.6-fold, respectively, while increasing expression of CYP17A1 by 4.8-fold (Fig. 2B). In addition to increasing the mRNA expression of CYP11A1 by 1.6-fold, ASAH1 suppression also potentiated dibutyryl (Bt2)cAMP-induced CYP11A1 mRNA expression (Fig. 2B). Intriguingly, ASAH1KD cells displayed significantly increased expression of genes involved in the uptake, deesterification, and transport of cholesterol. As shown in Fig. 2C, StAR, TSPO, and HSL expressions were induced by 2.0-fold in ASAH1KD cells. Moreover, mRNA expression of scavenger receptor type B class I (SR-BI), which mediates high-density lipoprotein cholesteryl ester delivery into adrenal cells (46), was also up-regulated by 6.8-fold (Fig. 2C). Notably, suppression of ASAH1 also enhanced Bt2cAMP-stimulated transcription of StAR, HSL, and SR-BI genes. Finally, mRNA expression of the MC2R gene, which encodes the cognate receptor for ACTH, was induced by 10.1-fold in ASAH1KD cells (Fig. 2C). Induced mRNA expression was concomitant with an increase in cellular protein levels of these steroidogenic genes (Fig. 2D). Significantly, similar to H295R cells, ASAH1KD in mouse Y-1 adrenocortical cells (Supplemental Fig. 2A) led to increased mRNA expression of StAR, CYP11A1, CYP21A2, and MC2R genes by at least 2-fold (Supplemental Fig. 2B), which was concomitant with an increase in cellular protein levels (Supplemental Fig. 2C). In contrast to the stimulatory effect of knocking down ASAH1 on adrenocortical steroidogenesis, silencing ASAH1 expression in MA-10 murine Leydig cells did not affect basal CYP17A1 or CYP11A1 mRNA expression but attenuated the Bt2cAMP-dependent expression of these genes (data not shown), suggesting differential roles for ASAH1 in adrenocortical vs. gonadal steroid hormone biosynthesis.
Fig. 2.
ASAH1KD increases the transcription of multiple steroidogenic genes. A, Diagram of the adrenocortical steroid hormone biosynthetic pathway. Circles represent genes that encode proteins that are involved in each step. B and C, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 72 h) cells were treated with 0.4 mm Bt2cAMP for 18 h. Total RNA was isolated, and the mRNA levels of various steroidogenic genes were quantified by qRT-PCR and normalized to the mRNA expression of β-actin. Data are graphed as mean ± sem of three separate experiments, each done in triplicate. Asterisks indicate statistically significant differences from untreated WT cells (P < 0.05). D, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 120 h) cell lysates were separated by SDS-PAGE, and the protein expression of P450c17 (encoded by CYP17A1), P450scc (CYP11A1), P450c11B (CYP11B1/2), 3β-HSD, MC2R, StAR, HSL, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was quantified by Western blotting.
Suppression of ASAH1 leads to increased cortisol and DHEA secretion
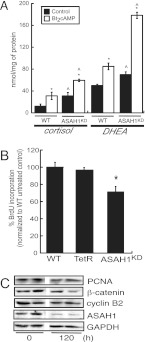
Because we observed increased expression of most genes required for steroid hormone production (Fig. 2), we postulated that ASAH1KD cells have increased steroidogenic capacity. To test this hypothesis, we quantified the levels of cortisol and DHEA secreted into the media from controls and Bt2cAMP-treated wild-type (WT) and ASAH1KD cells. As shown in Fig. 3A, ASAH1 suppression increased basal cortisol and DHEA secretion by 2.4- and 1.5-fold, respectively, compared with WT cells. Further, down-regulation of ASAH1 potentiated the stimulatory effect of Bt2cAMP on steroid hormone secretion (Fig. 3A). Similar results were obtained in mouse Y-1 cells that were transfected with 50 nm ASAH1 small interfering RNA (siRNA) oligonucleotides (Supplemental Fig. 2D). ASAH1KD increased both basal and Bt2cAMP-stimulated corticosterone secretion by 1.5- and 2.2-fold, respectively, compared with scrambled siRNA-transfected controls (Supplemental Fig. 2D).
Fig. 3.
ASAH1 repression increases steroidogenic capacity. A, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 48 h) cells were treated with 0.4 mm Bt2cAMP for 48 h. Cortisol and DHEA secreted into the media were quantified by EIA as described in Materials and Methods and normalized to the total cellular protein content of each sample. Asterisks and carats indicate statistically significant differences from the untreated controls within each group or from WT-untreated controls, respectively. B, H295R WT, H295R-TetR (TetR), and ASAH1KD cells were seeded in 96-well plates at 5 × 103 cells/well. H295R-TetR and ASAH1KD cells were then treated with 5 μg/ml tet for 96 h. Cell proliferation was measured with a BrdU incorporation ELISA kit. Data are expressed as mean percentage of control ± sem of three separate experiments, each done in quintuplicate. Asterisk denotes statistically significant difference from WT cells (P < 0.05). C, ASAH1KD cells were treated with 5 μg/ml tet for 120 h. Total cell lysates were harvested and separated by SDS-PAGE followed by Western blot analysis using anti-PCNA, anti-β-catenin, anticyclin B2, anti-ASAH1, or antiglyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies.
ASAH1KD decreases cellular proliferation
To further assess the role of ASAH1 in regulating H295R cell function, we determined the effect of ASAH1KD on cell proliferation. As shown in Fig. 3B, suppression of ASAH1 expression decreased cell proliferation by 29.1% compared with WT cells. In addition, analysis of various proliferative protein markers revealed that ASAH1 depletion decreased cellular levels of β-catenin, PCNA, and cyclin B2 (Fig. 3C). Significantly, no change in protein expression was observed in H295R-tet repressor (TetR) cells, which express the TetR protein but not ASAH1 shRNA (data not shown). Further, no significant difference in cell viability [determined by (3-4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide assays] was observed among WT, H295R-TetR, and H295R-ASAH1KD cells (data not shown).
ASAH1KD cells have altered sphingolipid gene expression and sphingolipid content
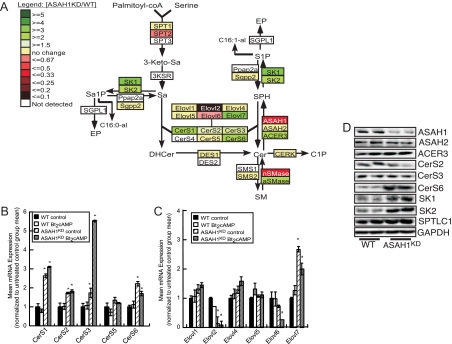
Next, we examined the effect of ASAH1 suppression on the transcription of sphingolipid genes in ASAH1KD cells. As shown in Fig. 4A, reduced ASAH1 expression increased the mRNA expression of acid sphingomyelinase 1 (aSMase1), SK1, and SK2 by 2.3-, 3.3-, and 2.1-fold, respectively. ACER3 transcription was also increased by 2.0-fold. Conversely, the expression of the catalytic subunit of the enzyme that catalyzes the rate-limiting step in de novo sphingolipid biosynthesis [serine palmitoyltransferase long-chain base subunit (SPTLC)2] was significantly decreased by 2.6-fold in ASAH1KD cells (Fig. 4A). ASAH1KD cells expressed higher levels of Cer synthase (CerS)1, CerS2, CerS3, and CerS6 mRNA transcripts (Fig. 4B). These genes encode the family of CerS enzymes that catalyze the N-acylation of sphinganine to form (dihydro)Cer (47). Because CerS enzymes use very long-chain fatty acyl-coenzyme A (CoA) that are made by the elongation of stearoyl- and palmitoyl-CoA (47), we analyzed the expression of the corresponding fatty acyl-CoA elongases (Elovl) (encoded by Elovl genes). The mRNA transcript level of Elovl2, which catalyzes the elongation of polyunsaturated fatty acyl-CoA of up to 24 carbons (48), was decreased by 7.6-fold in ASAH1KD cells (Fig. 4C). Conversely, mRNA expression of Elovl7, which encodes a saturated very long-chain fatty acid elongase (49), was induced by 2.7-fold (Fig. 4C). Protein expression levels of ASAH1, ASAH2, SK1, and SK2 mirrored the changes in mRNA expression (Fig. 4D). Although CerS transcripts were increased in ASAH1KD cells, CerS protein levels, with the exception of CerS6, remained unchanged (Fig. 4D), suggesting multiple levels of regulation of the CerS gene family. Of note, although ACER3 mRNA expression was induced in response to suppression of ASAH1 expression (Fig. 4A), no statistically significant change in protein level was observed (Fig. 4D).
Fig. 4.
ASAH1 suppression alters the transcription of multiple sphingolipid genes. A, Total RNA from H295R WT and ASAH1KD cells (pretreated with 5 μg/ml tet for 96 h) was isolated as described in Materials and Methods. mRNA levels for the indicated sphingolipid genes were determined by qRT-PCR and are displayed as a heat map incorporated into the sphingolipid metabolic pathway. The mean mRNA expression values in ASAH1KD cells range from low (red) to high (green) compared with the mean level of mRNA expression for each gene in H295R WT cells. 3-keto-Sa, 3-Ketosphinganine; Sa, sphinganine; −KSR, 3-keto-Sa reductase; Sa1P, sphinganine-1-phosphate; Ppap2a and Sgpp2, Sa1P phosphatase; SGPL1, S1P lyase; EP, ethanolamine phosphate; C16:0/1-al, hexadecanal; DHCer, dihydroCer; DES, DHCer desaturase; CERK, Cer kinase; C1P, Cer-1-phosphate; SMS1-2, SM synthase; nSMase, neutral SMase; acid (ASAH1), neutral (ASAH2), and alkaline (ACER3) ceramidase. B and C, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 72 h) cells were treated with 0.4 mm Bt2cAMP for 18 h. Total RNA was isolated, and the mRNA levels of CerS (B) or Elovl (C) genes were quantified by qRT-PCR and normalized to the mRNA expression of β-actin. Data are graphed as mean ± sem of three separate experiments, each done in triplicate. Asterisks indicate statistically significant differences from untreated WT cells (P < 0.05). D, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 120 h) cell lysates were separated by SDS-PAGE, and the protein expression of ASAH1, ASAH2, ACER3, CerS2, CerS3, CerS6, SK1, SK2, SPTLC1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was quantified by Western blotting.
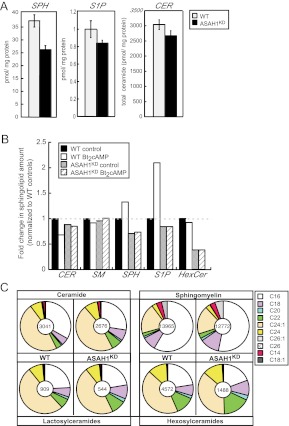
These changes in sphingolipid gene expression prompted us to quantify the intracellular amounts of various sphingolipid species in ASAH1KD cells. Liquid chromatography, electrospray ionization, tandem mass spectrometry (LC-ESI-MS/MS) analysis in WT and ASAH1KD cells revealed that total cellular SPH and S1P amounts were decreased by 30 and 16%, respectively, when ASAH1 was suppressed (Fig. 5A). Significantly, suppression of ASAH1 expression prevented the acute Bt2cAMP-stimulated sphingolipid turnover that was observed in WT cells (Fig. 5B) (30). ASAH1KD altered the relative levels of distinct Cer and SM subspecies (Fig. 5C). The amounts of saturated long-chain (C18–24) Cer and unsaturated long-chain (C18:1)Cer were proportionally increased, whereas C16-Cer and unsaturated very long-chain (C24:1 and C26:1) Cer were decreased in ASAH1KD cells compared with WT (Fig. 5C). Similarly, long-chain (C18–22), very long-chain (C26), and unsaturated long-chain (C18:1)SM species were proportionally increased, whereas C16-SM was decreased in ASAH1KD cells (Fig. 5C). Intriguingly, the amounts of lactosylCer (LacCer) and hexosylCer (HexCer) were decreased by 40 and 70%, respectively, in response to ASAH1KD, with decreases in specific subspecies of LacCer and HexCer mirroring the pattern of acyl-chain composition decreases observed for SM subspecies (Fig. 5C).
Fig. 5.
ASAH1KD leads to complex changes in steady-state sphingolipid content. A, The intracellular amounts of SPH, S1P, and Cer (CER) were quantified in H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cells by LC-ESI-MS/MS as described in Materials and Methods. B, LC-ESI-MS/MS quantification analysis of the intracellular amounts of Cer, SM, SPH, S1P, and HexCer in H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cells treated with 0.4 mm Bt2cAMP for 30 min. Data are graphed as fold change of sphingolipid content and normalized to untreated controls. C, The intracellular content of Cer, SM, LacCer, and HexCer subspecies was quantified in H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cells by LC-ESI-MS/MS. The relative distribution of different subspecies is represented with the chain length and number of double bonds depicted in the legend by x:y, respectively. Pie charts represent the relative changes in sphingolipid subspecies abundance between H295R WT and ASAH1KDcells. The number at the center of each chart refers to the total amount of each sphingolipid specie quantified.
Depletion of ASAH1 alters the expression of various nuclear receptor genes
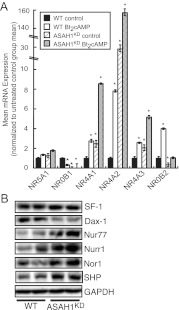
As discussed earlier, several nuclear receptors regulate the transcription of steroidogenic genes (3). Quantitative RT-PCR (qRT-PCR) analysis of genes encoding various nuclear receptors revealed that suppression of ASAH1 expression significantly increased the mRNA levels of NR4A1 and NR4A2 by 2.4- and 30.5-fold, respectively (Fig. 6A). NR4A1 encodes growth factor-inducible immediate early gene nur/77-like receptor (NGFI-B)/Nur77, which regulates the transcription of CYP21A2 (50) and StAR (19), whereas NR4A2 (encodes Nurr1) activates CYP11B2 gene expression in response to angiotensin II (16). The transcription of NR4A3, which encodes the neuron-derived orphan receptor (Nor)1 and regulates CYP11B2 and 3β-HSD reporter gene activity (51), as well as CYP21A2 gene transcription (17, 51), was induced by 2.1-fold in ASAH1KD cells (Fig. 6A). Notably, ASAH1 suppression potentiated Bt2cAMP-induced NR4A1, NR4A2, and NR4A3 mRNA expression. In contrast to the induction observed for NR4A family members, ASAH1KD cells exhibited an 89 and 66% decrease in NR0B1 (encodes DAX-1) and NR0B2 [small heterodimer partner (SHP)] mRNA transcripts, respectively (Fig. 6A). Consistent with the changes in mRNA expression, Nur77, Nurr1, and Nor1 protein levels were increased in ASAH1KD cells, whereas DAX-1 protein expression was decreased under the same conditions (Fig. 6B). Intriguingly, even though SHP mRNA expression was reduced in ASAH1KD cells (Fig. 6A), the protein expression of this nuclear receptor was increased in ASAH1KD cells (Fig. 6B). Finally, although SPH is a ligand for SF-1 (28), reduced levels of this sphingolipid in ASAH1KD cells (Fig. 5A) did not affect the mRNA or protein expression of this receptor (Fig. 6, A and B). In mouse Y-1 cells, NR0B1 and NR4A2 mRNA expression in response to ASAH1KD mirrored their respective pattern of expression in ASAH1KD cells (Supplemental Fig. 2D). In ASAH1 siRNA-transfected Y-1 cells, NR0B1 mRNA transcripts were decreased by 95%, whereas NR4A2 mRNA expression was induced by 2.7-fold compared with scrambled siRNA-transfected controls (Supplemental Fig. 2D).
Fig. 6.
ASAH1KD cells display altered expression of multiple steroidogenesis-associated nuclear receptor genes. A, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 72 h) cells were treated with 0.4 mm Bt2cAMP for 18 h. Total RNA was isolated as described in Materials and Methods, and the mRNA levels of NR5A1, NR4A1, NR4A2, NR4A3, NR0B1, and NR0B2 were quantified by qRT-PCR and normalized to the mRNA expression of β-actin. Data are graphed as mean ± sem of three separate experiments, each done in triplicate. Asterisks indicate statistically significant differences from untreated WT cells (P < 0.05). B, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 120 h) cell lysates were separated by SDS-PAGE, and the protein expression of SF-1 (encoded by NR5A1), DAX-1 (NR0B1), Nur77 (NR4A1), Nurr1 (NR4A2), Nor1 (NR4A3), SHP (NR0B2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was quantified by Western blotting.
ASAH1 regulates intracellular cAMP levels and ACTH responsiveness
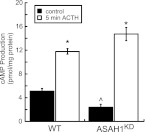
Because down-regulation of ASAH1 altered the transcription of multiple cAMP-regulated genes (Figs. 2 and 6), we hypothesized that ASAH1 suppression increased cAMP amounts. To test this hypothesis, we quantified the amount of intracellular cAMP in WT and ASAH1KD cells under basal and ACTH-stimulated conditions. Unexpectedly, basal intracellular cAMP levels were decreased by 46% in ASAH1KD cells compared with WT (Fig. 7). However, ASAH1KD cells displayed a significantly higher increase in the magnitude (6.1-fold) of intracellular cAMP produced in response to ACTH when compared with WT cells (2.3-fold).
Fig. 7.
ASAH1 regulates intracellular cAMP levels and ACTH responsiveness. H295R WT or ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cells were treated with 50 nm ACTH for 5 min. Intracellular cAMP levels were quantified by ELISA as described in Materials and Methods. cAMP amounts are normalized to total cellular protein content, and data graphed represent the mean ± sem of three experiments, each done in triplicate. Asterisks and carats indicate statistically significant differences from untreated controls within each group or from untreated WT controls, respectively (P < 0.05).
ASAH1KD affects histone H3 acetylation levels at target promoters
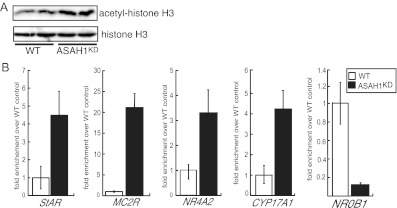
Because ASAH1 suppression altered the transcriptional rate of multiple genes, we postulated that ASAH1 may alter nuclear patterns of histone acetylation. Western blot analysis of histone H3 acetylation in WT and ASAH1KD cells revealed that suppression of ASAH1 resulted in a 1.7-fold increase in total acetyl-histone H3 levels (Fig. 8A). Therefore, we carried out chromatin immunoprecipitation (ChIP) analysis in WT and ASAH1KD cells to quantify the relative amounts of acetylated histone H3 at the proximal promoter region of a subset of steroidogenic genes that exhibited altered mRNA expression in response to ASAH1KD (Figs. 2, A and B, and 6A). Consistent with increased mRNA expression, acetyl-histone H3 levels at the proximal promoter region of StAR, MC2R, NR4A2, and CYP17A1 genes were increased by 4.2-, 21.1-, 3.2-, and 4.1-fold, respectively, in ASAH1KD cells compared with WT (Fig. 8B). Further, an 8.6-fold decrease in histone H3 acetylation at the DAX-1 proximal promoter was observed in ASAH1KD cells (Fig. 8B).
Fig. 8.
Suppression of ASAH1 affects histone H3 acetylation levels. A, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cell lysates were isolated and separated by SDS-PAGE. Acetyl-histone H3 and total histone H3 levels were determined by Western blotting. B, H295R WT and ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cells were crossed linked with 1% formaldehyde and the sheared chromatin immunoprecipitated with an antiacetyl histone H3 antibody. Acetyl-histone H3 levels at the proximal promoter regions of StAR, MC2R, CYP17A1, NR4A2, and DAX-1 genes were assessed by quantitative reverse chain reaction (qPCR). Purified DNA was quantified by real-time PCR and normalized to the ΔΔ cycle threshold values of input DNA. Data are expressed as fold change over untreated WT controls.
Discussion
In the zonae fasciculata and reticularis of the human adrenal cortex, steroid hormone biosynthesis requires the coordinate action of steroid hydroxylases and dehydrogenases (2, 52, 53), lipoprotein receptors (46, 54), and cholesterol-binding proteins (6, 55–58). The transcription of most of these genes is regulated by ACTH, which upon binding to MC2R activates a cAMP signaling cascade. Sphingolipids, such as Cer, SPH, and S1P, regulate steroidogenesis by acting as secondary modulators of steroidogenic gene transcription (31, 28, 59–61) and/or steroidogenic regulatory pathways (62–70). We have previously demonstrated that dynamic sphingolipid metabolism is a component of ACTH/cAMP-dependent steroidogenesis (30, 32).
A hallmark of adrenocortical steroid hormone biosynthesis is the induction of steroidogenic gene transcription (Fig. 2A) in response to chronic ACTH stimulation. Our studies demonstrate that ASAH1 suppression mimics ACTH/cAMP-dependent CYP11A1, CYP17A1, CYP11B1/2, and CYP21A2 gene expression and potentiates the effect of Bt2cAMP on CYP11A1 and CYP21A2 mRNA levels (Fig. 2B). Moreover, depletion of ASAH1 resulted in the up-regulation of StAR, TSPO, HSL, and SR-BI transcription (Fig. 2C), all of which encode proteins that are required for the cellular uptake of lipoproteins, the deesterification of cholesteryl esters, and the transport of free cholesterol into the inner mitochondria membrane (71–74). The effect of ASAH1 suppression on the transcription of these steroidogenic genes is consistent with our previously published data establishing SPH as an antagonist of SF-1 and suppressor of CYP17A1 transcription (28). Because SF-1 regulates the transcription of CYP17A1, CYP11A1, StAR, and CYP11B1 (3, 11), lower SPH intracellular levels (Fig. 5A) as a result of ASAH1KD positively correlates with increased basal mRNA expression of these genes (Fig. 2B). Altered steroidogenic gene expression in ASAH1KD cells can also be explained by higher mRNA and protein levels of the nuclear receptors Nur77 and Nor1 (Fig. 6, A and B), both of which regulate CYP21A2 transcription (17, 50). Nur77 also modulates StAR gene expression (18). Further, the expression of the transcription factor DAX-1 is significantly down-regulated in ASAH1KD cells (Fig. 6, A and B). DAX-1 represses the transactivation potential of many nuclear receptors (75), including SF-1 (15, 76), and its expression negatively regulates steroid hormone production at multiple levels (75, 77–79). Further, the absence of DAX-1 potentiates the ability of the adrenal to respond to ACTH in vivo (14). Therefore, repression of this nuclear receptor may contribute to increased steroidogenic gene expression in ASAH1KD cells. The fact that not all SF-1-regulated genes are basally up-regulated in ASAH1KD cells (e.g. 3β-HSD) suggests that additional factors are required for inducing the expression of those genes. Consistent with our mRNA and protein expression data (Fig. 2), we show that ASAH1KD cells secrete significantly higher levels of cortisol and DHEA than WT cells, both basally and in response to Bt2cAMP stimulation (Fig. 3). ASAH1KD also increased the magnitude cAMP production in response to ACTH (Fig. 7). This finding positively correlates with higher mRNA and protein expression of MC2R (Fig. 2, C and D), the gene that encodes the cognate receptor for ACTH. Significantly, quantification of steroidogenic gene transcription and steroid hormone production in mouse Y-1 adrenocortical cells yielded similar results as in H295R cells (Supplemental Fig. 2). Conversely, no change in steroidogenic gene transcription was observed in a different steroidogenic cell type, gonadal MA-10 Leydig cells, after transient ASAH1KD (data not shown), suggesting that ASAH1 plays distinct roles in adrenocortical and gonadal tissues. Consistent with this premise, Cer stimulates progesterone production from MA-10 cells (80) but inhibits the secretion of aldosterone by bovine adrenal glomerulosa cells (69). Of note, we have found that ASAH1 is expressed in the nucleus of Y1 and H295R cells but in the cytoplasmic compartment of MA-10 cells (Lucki, N. C., and M. B. Sewer, unpublished observations). Studies are ongoing to define the functional significance of subcellular localization of ASAH1 in regulating adrenocortical vs. gonadal steroid hormone biosynthesis.
Significantly, we show that ASAH1 suppression alters the amount of acetylated histone H3 at the proximal promoter region of multiple genes, including MC2R, StAR, DAX-1, CYP17A1, and NR4A2 (Fig. 8B). Lysine acetylation of histones is an epigenetic modification that is usually associated with active chromatin regions (81). We found that increased histone H3 acetylation at the proximal promoter regions of MC2R, StAR, CYP17A1, and NR4A2 genes positively correlated with their increased mRNA expression in response to ASAH1KD (Fig. 2B). Moreover, decreased mRNA levels of DAX-1 in ASAH1KD cells (Fig. 6A) positively correlated with lowered amounts of acetylated histone H3 (Fig. 8B).
Sphingolipids are rapidly emerging as important mediators of various nuclear processes (27). Recently, Hait et al. (82) uncovered a novel role for nuclear SK2 and S1P in regulating histone deacetylase (HDAC) activity and histone acetylation. S1P was found to inhibit HDAC1 and HDAC2 enzymatic activity and enhance local histone H3 acetylation and transcription of target genes (82). Along this line of evidence, it would follow that lower ASAH1 expression would result in lower S1P levels and thus decreased histone H3 acetylation. However, our data show that ASAH1 suppression leads to increased cellular amounts of acetylated histone H3 (Fig. 8A) and an increase in the acetylation of histone H3 levels at selected promoters (Fig. 8B). Given that we observed no significant change in S1P levels in ASAH1KD cells (Fig. 5A), this discrepancy between our findings and the work of Hait et al. (82) may reflect cell-specific differences. Of note, intracellular S1P amounts remained unchanged in response to ASAH1 suppression despite an increase in SK1 and SK2 protein levels (Fig. 4D), which is likely due to lower substrate (SPH) amount (Fig. 5A).
ASAH1KD had various effects on the transcription of sphingolipid genes (Fig. 4). However, the changes observed in mRNA expression cannot fully explain the changes seen in the steady-state pool of sphingolipids (Fig. 5). Total Cer and SM levels were unchanged in ASAH1KD cells, whereas the total amounts of LacCer and HexCer were significantly lower compared with WT cells (Fig. 5B). As predicted, a reduction in SPH levels in ASAH1KD cells was observed (Fig. 5A). However, this decrease was inconsistent with the lack of a significant change in total Cer levels. Given that ASAH1 catalyzes Cer hydrolysis and Cer degradation is the primary source of cellular SPH (36), we anticipated that Cer would accumulate in these cells. Meroni et al. (59) previously reported that inhibition of ceramidase activity by (1S,2R)-d-erythro-2-(N-myristoylamino)-1-phenyl-1-propanol, an inhibitor of ACER3 (83), resulted in Cer accumulation and led to a decrease in testosterone secretion from rat Leydig cells. Although our cell system differs from the one used by Meroni et al. (59), the quantitative differences in Cer amounts when comparing these two studies support the notion that each ceramidase isoform has unique cellular functions. This premise is supported by the differences in subcellular localization (84–89) and substrate specificity (86, 90–93) of each ceramidase isoform. As mentioned above, we have preliminary evidence indicating that each ceramidase isoform exhibits distinct subcellular expression patterns in H295R cells (Lucki, N. C., and M. B. Sewer, unpublished observations). Our findings suggest that alternate mechanisms are employed by the cell to maintain total Cer levels at a constant level despite low ASAH1 expression.
Our data indicate that ASAH1KD had varied effect of sphingolipid homeostasis. Depletion of ASAH1 caused a shift in the acyl-chain lengths of Cer, SM, HexCer, and LacCer (Fig. 5B). We observed an accumulation in saturated long-chain (C18–24) Cer and SM species, a decrease in unsaturated C24:1- and C26:1-Cer, and an overall reduction in C16-sphingolipids. A decrease in unsaturated very long-chain Cer species may be partially due to decreased mRNA levels of Elovl2 (Fig. 4C), which catalyzes the elongation of polyunsaturated fatty acyl-CoA of up to 24 carbons (48), and lower CerS2 protein levels (Fig. 4D), which uses very long-chain acyl-CoA as substrates (47). It is equally probable that the protein levels and/or activity of Elovl, as well as the enzymatic activity of different CerS enzymes, are increased in ASAH1KD cells and may contribute to the acyl-chain composition of sphingolipids in these cells. Interestingly, the levels of C16-Cer species are decreased in ASAH1KD cells despite an increase in CerS6 protein levels (Fig. 4D). Also, although ACER3 was recently reported to hydrolyze unsaturated long-chain C18:1-Cer (90), we observed higher levels of this particular Cer subspecies in ASAH1KD cells, which was inversely correlated to the levels of ACER3 mRNA expression in these cells (Fig. 4A). The underlying mechanism for these changes in the sphingolipidome is unknown, and further studies are necessary to elucidate the specific roles of ASAH1 in regulating sphingolipid acyl-chain composition. However, it is notable that similar unpredicted changes in sphingolipid content were recently observed by Mullen et al. (94) in response to CerS2 knockdown. Park et al. (95) reported that the lipid backbone composition of Cer subspecies and fatty acyl-CoA from mouse embryonic stem cells and embryoid bodies correlated fairly well with CerS and Elovl genomic analysis. Thus, the changes in sphingolipid amounts resulting from sphingolipid gene knockdown are multifaceted and sometimes cannot be readily explained by changes in gene expression. Given that recently lipidomic analysis has revealed that Cer with varying fatty acyl-chain length may have distinct cellular functions (96, 97), it is tempting to speculate that some of the physiological changes observed in response to ASAH1 suppression are partially due to a shift in the acyl-chain length of sphingolipid species.
Because ceramidases regulate the intracellular levels of two proliferation-associated molecules, they have key implications in cell fate. ASAH1 is overexpressed in various cancers (41–44), whereas ASAH1 knockout leads to embryonic lethality in mice (40). Further, ACER3 and ASAH2 were both reported to modulate cell proliferation either by up-regulating cyclin-dependent kinase inhibitor p21CIP1/WAF1 expression (90) or inducing cell cycle arrest at G0/G1 and retinoblastoma protein dephosphorylation (98), respectively. Accordingly, our data suggest that ASAH1 controls the rate of cell proliferation in H295R cells (Fig. 3B), at least in part, by decreasing the expression of multiple proliferation protein markers, including β-catenin, PCNA, and cyclin B2 (Fig. 3C). Because ASAH1 suppression had no effect on cell viability (data not shown), we excluded apoptosis as a reason for reduced proliferation. We have recently shown that the phytoestrogen genistein induces MCF-7 cell growth in an ASAH1-dependent manner, and suppression of ASAH1 expression by siRNA prevented genistein-induced cyclin B2 protein expression (99). The present study is consistent with our previous findings in MCF-7 cells and further supports a role for ASAH1 in controlling cell growth, at least in part, through the regulation of proliferation- associated protein expression (Fig. 3C). Additionally, emerging evidence suggest that LacCer levels positively correlate with various phenotypic changes, including cell proliferation and migration (100–102). Therefore, lower LacCer amounts in response to ASAH1KD may also contribute to the decrease in proliferation of ASAH1KD cells.
In summary, we show here that ASAH1KD leads to global changes in various classes of genes, including steroidogenic, nuclear receptors, and sphingolipid- metabolizing enzymes. ASAH1KD cells secrete significantly higher levels of cortisol and DHEA than WT cells, which positively correlate with higher mRNA expression of CYP11A1, CYP11B1/2, CYP17A1, CYP21A2, StAR, TSPO, HSL, SR-BI, and MC2R genes. These changes were largely the result of increased acetylation of histone H3 at the proximal promoters of these genes. ASAH1KD cells were also more sensitive to ACTH and display 6-fold higher intracellular cAMP accumulation upon hormone stimulation than WT cells, likely due to induced MC2R expression (Fig. 2D). Intriguingly, ASAH1 silencing resulted in the induction of the NR4A family and the suppression of DAX-1 (Fig. 6), indicating a novel role for this ceramidase in modulating nuclear receptor expression. Finally, we demonstrate that down-regulation of ASAH1 causes a shift in sphingolipid metabolism toward Cer, SM, LacCer, and HexCer subspecies with long saturated acyl-chain lengths. We conclude that ASAH1 is a global mediator of steroidogenic gene expression and adrenocortical steroidogenesis.
Materials and Methods
Reagents
Bt2cAMP was obtained from Sigma (St. Louis, MO). Blasticidin, tet, and zeocin were purchased from Invitrogen (Carlsbad, CA). Human ACTH-(1-39) was purchased from American Peptide Co. (Sunnyvale, CA).
Cell culture
H295R adrenocortical cells (103, 104) were generously donated by William E. Rainey (Medical College of Georgia, Augusta, GA) and cultured in DMEM/F12 medium (Mediatech, Inc., Manassas, VA) supplemented with 10% Nu-Serum I (BD Biosciences, Palo Alto, CA), 1% ITS Plus (BD Biosciences), and antibiotics. H295R ASAH1KD cells were cultured in the same medium as the parental cell line with the addition of 1 μg/ml blasticidin and 50 μg/ml zeocin.
Generation of the H295R ASAH1KD stable cell line
H295R cells expressing tet-inducible ASAH1 shRNA were generated using the BLOCK-iT Inducible H1 RNAi Entry Vector kit (Invitrogen) following the manufacturer's instructions. First, the TetR expression plasmid (pcDNA6-TR; Invitrogen) was stably transfected into H295R cells using GeneJuice (EMD Biosciences, San Diego, CA). Each clone was selected using 1 μg/ml blasticidin, and TetR expression was determined by Western blotting using an anti-TetR antibody (Sigma). The clone that expressed the highest TetR protein (H295R-TetR cells) was transfected with the pENTR/H1/TO-ASAH1shRNA vector. To construct an inducible vector for ASAH1 shRNA, the following sequences were cloned into pENTR/H1/TO: 5′-ACC GCA CCA ATG CTA AAG GTT ATA GTG AAC GAA TTC ACT ATA ACC TTT AGC ATT GGT G-3′ and 5′-AAA ACA CCA ATG CTA AAG GTT ATA GTG AAT TCG TTC ACT ATA ACC TTT AGC ATT GGT GC-3′, corresponding to positions 221–246 of the ASAH1 mRNA sequence (NM_004315). H295R-TetR cells were stably transfected with the constructed pENTR/H1/TO-ASAH1shRNA expression vector or the control vector using GeneJuice (EMD Biosciences), and cell clones were selected using 50 μg/ml zeocin. Clones were treated with 5 μg/ml tet for 96 h, and suppression of ASAH1 protein levels in each clone was confirmed by Western blotting using an anti-ASAH1 antibody (HPA005468; Sigma).
DNA microarray
H295R WT or tet-treated (5 μg/ml for 72 h) ASAH1KD cells were subcultured into 100-mm dishes and treated with 0.4 mm Bt2cAMP for 18 h. Total RNA was isolated using the QIAGEN RNeasy kit (QIAGEN, Valencia, CA), and gene expression profiling was done by Phalanx Biotech Group, Inc. (Palo Alto, CA) using the Human Whole Genome OneArray DNA Microarray (HOA_004).
RNA isolation and qRT-PCR
H295R WT or ASAH1KD cells were subcultured into 12-well plates and shRNA cells treated with 5 μg/ml tet for 48–72 h. In some experiments, cells were stimulated with 0.4 mm Bt2cAMP for 18 h before RNA extraction. Total RNA was isolated using Isol-RNA Lysis reagent (5 Prime, Inc., Gaithersburg, MD) and amplified using a One-Step SYBR Green RT-PCR kit (Thermo Scientific, Inc., Waltham, MA) and the primer sets listed in Table 1. Gene expression was normalized to β-actin mRNA content and calculated using the ΔΔ cycle threshold method.
Table 1.
Primer sets used in RT-PCR
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| β-Actin | ACGGCTCCGGCATGTGCAAG | TGACGATGCCGTGCTGCATG |
| ASAH1 | GCACAAGTTATGAAGGAAGCCAAG | TCCAATGATTCCTTTCTGTCTCG |
| ASAH2 | GCATCAACACAGGAGAGTC | GGAGGCAGAGGCATAGAG |
| ACER3 | ATCCGCCTGGTCTTCATC | CTCCTTATTGCTGGTCTTCC |
| SK1 | CTGGCAGCTTCCTTGAACCAT | TGTGCAGAGACAGCAGGTTCA |
| SK2 | CCAGTGTTGGAGAGCTGAAGGT | GTCCATTCATCTGCTGGTCCTC |
| SPTLC1 | AGTCCCTTTCTCCAGCCTTTC | TTCCACCGTGACCACAACC |
| SPTLC2 | CAAGAAGAAATACAAGGCATAC | CCATCATAACATCCACATCC |
| LASS1 | CCTTCTACTTCTTCTTCAATGC | TCGGCTGTGTCATACTCC |
| LASS2 | GTCATCCTGCCCTTCTGG | CACTGCTGGCATCTTCTACC |
| LASS3 | TCATACATCTTCCTCAACCTACAG | GCCTCTTCTTCTTCCTCTTCC |
| LASS5 | ATGGTGGCTCCTCAATGG | AGGTGGTCACATCTTCTTCC |
| LASS6 | GCTGACGAGGTTCTGTGAG | AGTTGTGAGTGGCTGATAGG |
| ELOVL1 | GTCTACAACTTCTCACTGGTGGC | AAGTGCCTCAGGGCTGTTGGAA |
| ELOVL2 | TCCACTTGGGAAGGAGGCTACA | CCAGGAACTCTACTGATTTGGAG |
| ELOVL4 | CCGAGAACCTTTTCAGATGCGTC | AATCCACACTCTGGCAAATATAG |
| ELOVL5 | ACGTCTACCACCATGCCTCGAT | TGGAAGGGACTGACGACAAACC |
| ELOVL6 | CCATCCAATGGATGCAGGAAAAC | CCAGAGCACTAATGGCTTCCTC |
| ELOVL7 | CCTACTATGGACTTTCTGCATTGG | GAACTGGCTTATGTGGATGGCG |
| CYP17A1 | CTCTTGCTGCTTCACCTA | TCAAGGAGATGACATTGGTT |
| CYP11A1 | CGTGGAGTCGGTTTATGTC | CTCTGGTAATACTGGTGATAGG |
| CYP11B1/2a | ACGGCGACAACTGTATCC | AGAGCGTCATCAGCAAGG |
| CYP21A2 | TGTGGAACTGGTGGAAGC | GGTGGAGCCTGTAGATGG |
| 3β-HSD | CCAGTAGCATAGAGGTAGCC | TCAGATTCCACCCGTTAGC |
| NR5A1 | GGAGTTTGTCTGCCTCAAGTTCA | CGTCTTTCACCAGGATGTGGTT |
| NR5A2 | TACCGACAAGTGGTACATGGAA | CGGCTTGTGATGCTATTATGGA |
| NR4A1 | GGACAACGCTTCATGCCAGCAT | CCTTGTTAGCCAGGCAGATGTAC |
| NR4A2 | AAACTGCCCAGTGGACAAGCGT | GCTCTTCGGTTTCGAGGGCAAA |
| NR4A3 | ACTGCCCAGTAGACAAGAGACG | GTTTGGAAGGCAGACGACCTCT |
| NR0B1 | CCAAATGCTGGAGTCTGAACATC | CCCACTGGAGTCCCTGAATGTA |
| NR0B2 | TGCCTGAAAGGGACCATCCTCT | GTTCCAGGACTTCACACAGCAC |
| StAR | GCTCTCTACTCGGTTCTC | GCTGACTCTCCTTCTTCC |
| TSPO | GCAGATTCCGTGATTACAGTG | TCCTCCTCGTCGTCATCG |
| HSL | CACTACAAACGCAACGAGAC | CCAGAGACGATAGCACTTCC |
| MC2R | GTGGTGCTTACGGTCATCTGGA | AGGCACAGGATGAAGACCAGCA |
| PDE4B | TAGTCAGCCTCCTGTCTCCAGA | GAAGCCATCTCACTGACAGACC |
| SGPP2 | ACTCCTCATCGTCCTCAC | CAGAATCCTATGGTCACTCC |
| nSMase 1 | GTGCTCAACGCCTATGTG | GTCTGCCTTCTTGGATGTG |
| nSMase 2 | ACTGCTCCTCTGACGACAAG | CCACGGCTTCTCCTCACC |
| SGMS2 | ACTCTACCTGTGCCTGGAATG | AATAAGTCAGTGTCAGCGTAACC |
| aSMase 1 | TTGTAGCCAGGTATGAGAAC | GCCGATGTAGGTAGTTGC |
| CERK | ACCACTGACATCATCGTTACTG | GCACCTCGCTGAACATACC |
| DEGS1 | GTTTGGAATGTTTGCTAATC | GAGAGGCTGAAGAATAACC |
The primer set used to assess the mRNA expression of CYP11B1/2 cannot distinguish between CYP11B1 and CYP11B2 isoforms.
Western blotting
H295R WT or ASAH1KD (5 μg/ml tet for 0–120 h) cells were harvested into radioimmunoprecipitation assay buffer [1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1× protease inhibitor cocktail set I (EMD Biosciences)]. Cells were then lysed by sonication (one 2-sec burst) followed by incubation on ice for 30 min. Lysates were centrifuged at 12,000 rpm for 15 min at 4 C and the supernatant collected for analysis by SDS-PAGE. Aliquots of each sample (30 μg of protein) were resolved on 8% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes (Millipore, Temecula, CA). Blots were probed with antibodies listed in Table 2, and expression was detected using an ECF Western blotting reagent kit (GE Healthcare, Piscataway, NJ) and visualized by scanning the blots on a VersaDoc 4000 imager (Bio-Rad, Hercules, CA). Protein amounts were determined using the BCA Protein Assay (Pierce, Rockford, IL).
Table 2.
List of antibodies used for Western blotting
| Target protein | Catalog no. vendor |
|---|---|
| ASAH1 | HPA005468, Sigma-Aldrich (St. Louis, MO) |
| ASAH2 | PRS4743, Sigma-Aldrich |
| ACER3 | sc-18822, Santa Cruz Biotechnology, Inc. |
| GAPDH | sc-25778, Santa Cruz Biotechnology, Inc. |
| P450c17 | sc-66849, Santa Cruz Biotechnology, Inc. |
| P450scc | |
| P450c11Ba | sc-28205, Santa Cruz Biotechnology, Inc. |
| 3β-HSD | sc-28206, Santa Cruz Biotechnology, Inc. |
| MC2R | sc-13107, Santa Cruz Biotechnology, Inc. |
| StAR | sc-25806, Santa Cruz Biotechnology, Inc. |
| HSL | sc-25843, Santa Cruz Biotechnology, Inc. |
| CerS2 | ab57136, Abcam (Cambridge, MA) |
| CerS3 | ab28637, Abcam |
| CerS6 | ab56582, Abcam |
| SK1 | sc-48825, Santa Cruz Biotechnology, Inc. |
| SK2 | ab37977, Abcam |
| SPTLC1 | sc-136076, Santa Cruz Biotechnology, Inc. |
| SF-1 | 07-618, Millipore |
| DAX-1 | ab60144, Abcam |
| SHP | sc-301069, Santa Cruz Biotechnology, Inc. |
| Nur77 | SAB2101650, Sigma-Aldrich |
| Nurr1 | N6538, Sigma-Aldrich |
| Nor1 | sc-100906, Santa Cruz Biotechnology, Inc. |
| β-Catenin | 05-613, Millipore |
| Cyclin B2 | K0189-3, MBL Labs (Woods Hole, MA) |
| PCNA | sc-7907, Santa Cruz Biotechnology, Inc. |
The antibody used to assess the protein levels of P450c11B cannot distinguish between P450c11B1 and P450c11B2 isoforms.
Analysis of sphingolipid amounts
H295R WT or ASAH1KD cells (pretreated with 5 μg/ml tet for 96 h) were subcultured into 100-mm dishes, and sphingolipid amounts in whole cells were analyzed by LC-ESI-MS/MS as described previously (105, 106).
Cortisol and DHEA enzyme-linked immune assays (EIA)
H295R WT or ASAH1KD cells (pretreated with 5 μg/ml tet for 72 h) were subcultured into 12-well plates and treated with 0.4 mm Bt2cAMP for 48 h before media collection. Cortisol and DHEA released into the media was determined in triplicate against cortisol or DHEA standards made up in DMEM/F12 medium using a 96-well plate enzyme-linked immune DHEA or cortisol assay (Assay Designs, Inc., Ann Arbor, MI). Steroid hormone amounts were normalized to the total cellular protein content, as determined using the BCA protein assay (Pierce).
Quantification of intracellular cAMP
H295R WT or ASAH1KD cells (pretreated with 5 μg/ml tet for 96 h) were cultured into 12-well plates and treated with 50 nm ACTH for 5 min. Cells were lysed in 0.1 m HCl for 20 min at room temperature, collected by scraping, and centrifuged at 4000 rpm for 10 min. The supernatant was collected, acetylated, and used in a Direct Cyclic AMP Correlate-EIA (Assay Designs, Inc.) following the manufacturer's instructions. Data were normalized to the total protein amount of each sample and are expressed as picomoles per milligram of total protein.
Chromatin immunoprecipitation
H295R WT or ASAH1KD (pretreated with 5 μg/ml tet for 96 h) cells were subcultured into 150-mm dishes, and ChIP assays were performed as described previously (107). The purified chromatin solutions were precleared and immunoprecipitated using 3 μg of antiacetyl-histone H3 (06-599; Millipore) or 3 μg rabbit anti-IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies and 30 μl of protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.). Quantitative PCR was carried out using 20% of output, 5% input (diluted 1:4), the ABsolute quantitative reverse chain reaction SYBR Green Fluorescein Mix (Thermo Scientific, Inc.), and the primer sets listed in Table 3.
Table 3.
Primer sets used in ChIP studies
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| StAR | GCAGTGTGAGGCAATCGCTCT | TGTTTCCTGGCAAATGTGGCA |
| MC2R | TTGCCCAGAAAGTTCCTGCTT | TTCTCCTGCTTGTGGTTAAGG |
| CYP17A1 | GGCTGGGCTCCAGGAGAATCTTTCTTCCAC | CGGCAGGCAAGATAGACAGCAGTGGAGTAG |
| DAX-1 | TCCTGCTTTTAAAGAGCACCCGCCCCT | CGGCGCCCGTAGCCCAGTTCT |
| NR4A2 | GATCAGCTTGGACTCCCTTGAAAGT | CATTCCAGAAATTGGCATCTTTTGA |
Cell proliferation
For quantitative proliferative assays, H295R WT, H295R-TetR, and ASAH1KD cells were seeded in 96-well plates (5 × 103 cells/well). H295R-TetR and ASAH1KD cells were treated for 96 h with 5 μg/ml tet. After treatment, the cultures were incubated for an additional 6 h in the presence of 5-bromo-2-deoxyuridine (BrdU) (10 μm). Cell proliferation was assayed by BrdU incorporation measurements with an ELISA kit (Roche Applied Science, Indianapolis, IN).
Statistical analysis
One-way ANOVA, Tukey-Kramer multiple comparison, and unpaired Student's t tests were performed using GraphPad InStat software (GraphPad Software, Inc., San Diego, CA). Significant differences from a compared value were defined as P < 0.05 and denoted by asterisks or carats. Hierarchical clustering analysis of DNA microarray studies was done by performing observation and variable tree computation using complete linkage clustering and correlation distance matrix with robust center scale normalization.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health (NIH) Grants DK084178 (to M.B.S.), GM069338 (to A.H.M.), and the American Heart Association (AHA) Predoctoral Fellowship 10PRE3230019 (to N.C.L.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- ACER
- Alkaline ceramidase
- ASAH1
- acid ceramidase
- ASAH1KD
- ASAH1 knockdown
- aSMase1
- acid sphingomyelinase 1
- BrdU
- 5-bromo-2-deoxyuridine
- Bt2
- dibutyryl
- Cer
- ceramide
- CerS
- Cer synthase
- ChIP
- chromatin immunoprecipitation
- CoA
- coenzyme A
- CYP
- cytochrome P450 monooxygenase
- DAX-1
- dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1
- DHEA
- dehydroepiandrosterone
- EIA
- enzyme-linked immune assay
- Elovl
- fatty acyl-CoA elongase
- HDAC
- histone deacetylase
- HexCer
- hexosylCer
- 3β-HSD
- 3β- hydroxysteroid dehydrogenase
- HSL
- hormone-sensitive lipase
- LacCer
- lactosylCer
- LC-ESI-MS/MS
- liquid chromatography, electrospray ionization, tandem mass spectrometry
- MC2R
- melanocortin-2 receptor
- Nor
- neuron-derived orphan receptor
- NR
- nuclear receptor
- PCNA
- proliferating cell nuclear antigen
- qRT-PCR
- quantitative RT-PCR
- SF-1
- steroidogenic factor 1
- SHP
- small heterodimer partner
- shRNA
- short hairpin RNA
- siRNA
- small interfering RNA
- SK
- SPH kinase
- SM
- sphingomyelin
- S1P
- SPH-1-phosphate
- SPH
- sphingosine
- SPTLC
- serine palmitoyltransferase long-chain base subunit
- SR-BI
- scavenger receptor type B class I
- StAR
- steroidogenic acute regulatory protein
- tet
- tetracycline
- TetR
- tet repressor
- TSPO
- 18-kDa translocator protein
- WT
- wild type.
References
- 1. Ghayee HK, Auchus RJ. 2007. Basic concepts and recent developments in human steroid hormone biosynthesis. Rev Endocr Metab Disord 8:289–300 [DOI] [PubMed] [Google Scholar]
- 2. Arlt W, Stewart PM. 2005. Adrenal corticosteroid biosynthesis, metabolism, and action. Endocrinol Metab Clin North Am 34:293–313, viii [DOI] [PubMed] [Google Scholar]
- 3. Sewer MB, Dammer EB, Jagarlapudi S. 2007. Transcriptional regulation of adrenocortical steroidogenic gene expression. Drug Metab Rev 39:371–388 [DOI] [PubMed] [Google Scholar]
- 4. Sewer MB, Waterman MR. 2003. ACTH modulation of transcription factors responsible for steroid hydroxylase gene expression in the adrenal cortex. Microsc Res Tech 61:300–307 [DOI] [PubMed] [Google Scholar]
- 5. Miller WL, Auchus RJ. 2011. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev 32:81–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stocco DM, Wang X, Jo Y, Manna PR. 2005. Multiple signaling pathways regulating steroidogenesis and steroidogenic acute regulatory protein expression: more complicated than we thought. Mol Endocrinol 19:2647–2659 [DOI] [PubMed] [Google Scholar]
- 7. Stocco DM, Clark BJ. 1996. Regulation of the acute production of steroids in steroidogenic cells. Endocr Rev 17:221–244 [DOI] [PubMed] [Google Scholar]
- 8. Chen WY, Juan LJ, Chung BC. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol Cell Biol 25:10442–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hammer GD, Krylova I, Zhang Y, Darimont BD, Simpson K, Weigel NL, Ingraham HA. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol Cell 3:521–526 [DOI] [PubMed] [Google Scholar]
- 10. Jacob AL, Lund J, Martinez P, Hedin L. 2001. Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J Biol Chem 276:37659–37664 [DOI] [PubMed] [Google Scholar]
- 11. Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X, Wong M, Bakke M, Zhao L, Frigeri C, Hanley NA, Stallings N, Schimmer BP. 2002. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res 57:19–36 [DOI] [PubMed] [Google Scholar]
- 12. Gummow BM, Winnay JN, Hammer GD. 2003. Convergence of Wnt signaling and steroidogenic factor-1 (SF-1) on transcription of the rat inhibin α gene. J Biol Chem 278:26572–26579 [DOI] [PubMed] [Google Scholar]
- 13. Jordan BK, Shen JH, Olaso R, Ingraham HA, Vilain E. 2003. Wnt4 overexpression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/β-catenin synergy. Proc Natl Acad Sci USA 100:10866–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Babu PS, Bavers DL, Beuschlein F, Shah S, Jeffs B, Jameson JL, Hammer GD. 2002. Interaction between Dax-1 and steroidogenic factor-1 in vivo: increased adrenal responsiveness to ACTH in the absence of Dax-1. Endocrinology 143:665–673 [DOI] [PubMed] [Google Scholar]
- 15. Ito M, Yu R, Jameson JL. 1997. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol 17:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bassett MH, Suzuki T, Sasano H, White PC, Rainey WE. 2004. The orphan nuclear receptors NURR1 and NGFIB regulate adrenal aldosterone production. Mol Endocrinol 18:279–290 [DOI] [PubMed] [Google Scholar]
- 17. Fernandez PM, Brunel F, Jimenez MA, Saez JM, Cereghini S, Zakin MM. 2000. Nuclear receptors Nor1 and NGFI-B/Nur77 play similar, albeit distinct, roles in the hypothalamo-pituitary-adrenal axis. Endocrinology 141:2392–2400 [DOI] [PubMed] [Google Scholar]
- 18. Havelock JC, Smith AL, Seely JB, Dooley CA, Rodgers RJ, Rainey WE, Carr BR. 2005. The NGFI-B family of transcription factors regulates expression of 3β-hydroxysteroid dehydrogenase type 2 in the human ovary. Mol Hum Reprod 11:79–85 [DOI] [PubMed] [Google Scholar]
- 19. Martin LJ, Boucher N, Brousseau C, Tremblay JJ. 2008. The orphan nuclear receptor NUR77 regulates hormone-induced StAR transcription in Leydig cells through cooperation with Ca2+/calmodulin-dependent protein kinase I. Mol Endocrinol 22:2021–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lucki NC, Sewer MB. 2008. Multiple roles for sphingolipids in steroid hormone biosynthesis. Subcell Biochem 49:387–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saddoughi SA, Song P, Ogretmen B. 2008. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem 49:413–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hannun YA, Obeid LM. 2008. Principles of bioactive lipid signaling: lessons from sphingolipids. Nat Rev Mol Cell Biol 9:139–150 [DOI] [PubMed] [Google Scholar]
- 23. Hla T, Lee MJ, Ancellin N, Liu CH, Thangada S, Thompson BD, Kluk M. 1999. Sphingosine-1-phosphate: extracellular mediator or intracellular second messenger? Biochem Pharmacol 58:201–207 [DOI] [PubMed] [Google Scholar]
- 24. Huwiler A, Kolter T, Pfeilschifter J, Sandhoff K. 2000. Physiology and pathophysiology of sphingolipid metabolism and signaling. Biochim Biophys Acta 1485:63–99 [DOI] [PubMed] [Google Scholar]
- 25. Kihara A, Mitsutake S, Mizutani Y, Igarashi Y. 2007. Metabolism and biological functions of two phosphorylated sphingolipids, sphingosine 1-phosphate and ceramide 1-phosphate. Prog Lipid Res 46:126–144 [DOI] [PubMed] [Google Scholar]
- 26. Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. 2008. Sphingolipids in macroautophagy. Methods Mol Biol 445:159–173 [DOI] [PubMed] [Google Scholar]
- 27. Ledeen RW, Wu G. 2008. Nuclear sphingolipids: metabolism and signaling. J Lipid Res 49:1176–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Urs AN, Dammer E, Sewer MB. 2006. Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology 147:5249–5258 [DOI] [PubMed] [Google Scholar]
- 29. Lucki NC, Sewer MB. 2010. The interplay between bioactive sphingolipids and steroid hormones. Steroids 75:390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ozbay T, Merrill AH, Jr, Sewer MB. 2004. ACTH regulates steroidogenic gene expression and cortisol biosynthesis in the human adrenal cortex via sphingolipid metabolism. Endocr Res 30:787–794 [DOI] [PubMed] [Google Scholar]
- 31. Ozbay T, Rowan A, Leon A, Patel P, Sewer MB. 2006. Cyclic adenosine 5′-monophosphate-dependent sphingosine-1-phosphate biosynthesis induces human CYP17 gene transcription by activating cleavage of sterol regulating element binding protein 1. Endocrinology 147:1427–1437 [DOI] [PubMed] [Google Scholar]
- 32. Lucki N, Sewer MB. 2009. The cAMP-responsive element binding protein (CREB) regulates the expression of acid ceramidase (ASAH1) in H295R human adrenocortical cells. Biochim Biophys Acta 1791:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mao C, Obeid LM. 2008. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta 1781:424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shtraizent N, Eliyahu E, Park JH, He X, Shalgi R, Schuchman EH. 2008. Autoproteolytic cleavage and activation of human acid ceramidase. J Biol Chem 283:11253–11259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernardo K, Hurwitz R, Zenk T, Desnick RJ, Ferlinz K, Schuchman EH, Sandhoff K. 1995. Purification, characterization, and biosynthesis of human acid ceramidase. J Biol Chem 270:11098–11102 [DOI] [PubMed] [Google Scholar]
- 36. Wang E, Norred WP, Bacon CW, Riley RT, Merrill AH., Jr 1991. Inhibition of sphingolipid biosynthesis by fumonisins. Implications for diseases associated with Fusarium moniliforme. J Biol Chem 266:14486–14490 [PubMed] [Google Scholar]
- 37. Cuvillier O, Pirianov G, Kleuser B, Vanek PG, Coso OA, Gutkind S, Spiegel S. 1996. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 381:800–803 [DOI] [PubMed] [Google Scholar]
- 38. Kolesnick R. 2002. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J Clin Invest 110:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maceyka M, Payne SG, Milstien S, Spiegel S. 2002. Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim Biophys Acta 1585:193–201 [DOI] [PubMed] [Google Scholar]
- 40. Eliyahu E, Park JH, Shtraizent N, He X, Schuchman EH. 2007. Acid ceramidase is a novel factor required for early embryo survival. FASEB J 21:1403–1409 [DOI] [PubMed] [Google Scholar]
- 41. Saad AF, Meacham WD, Bai A, Anelli V, Elojeimy S, Mahdy AE, Turner LS, Cheng J, Bielawska A, Bielawski J, Keane TE, Obeid LM, Hannun YA, Norris JS, Liu X. 2007. The functional effects of acid ceramidase overexpression in prostate cancer progression and resistance to chemotherapy. Cancer Biol Ther 6:1455–1460 [DOI] [PubMed] [Google Scholar]
- 42. Mahdy AE, Cheng JC, Li J, Elojeimy S, Meacham WD, Turner LS, Bai A, Gault CR, McPherson AS, Garcia N, Beckham TH, Saad A, Bielawska A, Bielawski J, Hannun YA, Keane TE, Taha MI, Hammouda HM, Norris JS, Liu X. 2009. Acid ceramidase upregulation in prostate cancer cells confers resistance to radiation: AC inhibition, a potential radiosensitizer. Mol Ther 17:430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elojeimy S, Liu X, McKillop JC, El-Zawahry AM, Holman DH, Cheng JY, Meacham WD, Mahdy AE, Saad AF, Turner LS, Cheng J, A Day T, Dong JY, Bielawska A, Hannun YA, Norris JS. 2007. Role of acid ceramidase in resistance to FasL: therapeutic approaches based on acid ceramidase inhibitors and FasL gene therapy. Mol Ther 15:1259–1263 [DOI] [PubMed] [Google Scholar]
- 44. Seelan RS, Qian C, Yokomizo A, Bostwick DG, Smith DI, Liu W. 2000. Human acid ceramidase is overexpressed but not mutated in prostate cancer. Gene Chromosome Canc 29:137–146 [DOI] [PubMed] [Google Scholar]
- 45. Park JH, Schuchman EH. 2006. Acid ceramidase and human disease. Biochim Biophys Acta 1758:2133–2138 [DOI] [PubMed] [Google Scholar]
- 46. Connelly MA. 2009. SR-BI-mediated HDL cholesteryl ester delivery in the adrenal gland. Mol Cell Endocrinol 300:83–88 [DOI] [PubMed] [Google Scholar]
- 47. Levy M, Futerman AH. 2010. Mammalian ceramide synthases. IUBMB Life 62:347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tvrdik P, Westerberg R, Silve S, Asadi A, Jakobsson A, Cannon B, Loison G, Jacobsson A. 2000. Role of a new mammalian gene family in the biosynthesis of very long chain fatty acids and sphingolipids. J Cell Biol 149:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamura K, Makino A, Hullin-Matsuda F, Kobayashi T, Furihata M, Chung S, Ashida S, Miki T, Fujioka T, Shuin T, Nakamura Y, Nakagawa H. 2009. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res 69:8133–8140 [DOI] [PubMed] [Google Scholar]
- 50. Wilson TE, Mouw AR, Weaver CA, Milbrandt J, Parker KL. 1993. The orphan nuclear receptor NGFI-B regulates expression of the gene encoding steroid 21-hydroxylase. Mol Cell Biol 13:861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nogueira EF, Xing Y, Morris CA, Rainey WE. 2009. Role of angiotensin II-induced rapid response genes in the regulation of enzymes needed for aldosterone synthesis. J Mol Endocrinol 42:319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. 2005. Molecular biology of the 3β-hydroxysteroid dehydrogenase/δ5-δ4 isomerase gene family. Endocr Rev 26:525–582 [DOI] [PubMed] [Google Scholar]
- 53. Sewer MB, Nguyen VQ, Huang CJ, Tucker PW, Kagawa N, Waterman MR. 2002. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation among p54(nrb)/NonO, protein-associated splicing factor, and SF-1, a complex that also participates in repression of transcription. Endocrinology 143:1280–1290 [DOI] [PubMed] [Google Scholar]
- 54. Azhar S, Leers-Sucheta S, Reaven E. 2003. Cholesterol uptake in adrenal and gonadal tissues: the SR-BI and ‘selective’ pathway connection. Front Biosci 8:s998–s1029 [DOI] [PubMed] [Google Scholar]
- 55. Liu J, Li H, Papadopoulos V. 2003. PAP7, a PBR/PKA-RIα-associated protein: a new element in the relay of the hormonal induction of steroidogenesis. J Steroid Biochem Mol Biol 85:576–586 [DOI] [PubMed] [Google Scholar]
- 56. Lacapère JJ, Papadopoulos V. 2003. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids 68:569–585 [DOI] [PubMed] [Google Scholar]
- 57. Stocco DM. 2001. StAR protein and the regulation of steroid hormone biosynthesis. Annu Rev Physiol 63:193–213 [DOI] [PubMed] [Google Scholar]
- 58. Baker BY, Lin L, Kim CJ, Raza J, Smith CP, Miller WL, Achermann JC. 2006. Nonclassic congenital lipoid adrenal hyperplasia: a new disorder of the steroidogenic acute regulatory protein with very late presentation and normal male genitalia. J Clin Endocrinol Metab 91:4781–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meroni SB, Pellizzari EH, Cánepa DF, Cigorraga SB. 2000. Possible involvement of ceramide in the regulation of rat Leydig cell function. J Steroid Biochem Mol Biol 75:307–313 [DOI] [PubMed] [Google Scholar]
- 60. Morales V, Santana P, Díaz R, Tabraue C, Gallardo G, López Blanco F, Hernández I, Fanjul LF, Ruiz de Galarreta CM. 2003. Intratesticular delivery of tumor necrosis factor-α and ceramide directly abrogates steroidogenic acute regulatory protein expression and Leydig cell steroidogenesis in aadult rats. Endocrinology 144:4763–4772 [DOI] [PubMed] [Google Scholar]
- 61. Santana P, Llanes L, Hernandez I, Gallardo G, Quintana J, Gonzalez J, Estevez F, Ruiz de Galarreta C, Fanjul LF. 1995. Ceramide mediates tumor necrosis factor effects on P450-aromatase activity in cultured granulosa cells. Endocrinology 136:2345–2348 [DOI] [PubMed] [Google Scholar]
- 62. Pörn MI, Tenhunen J, Slotte JP. 1991. Increased steroid hormone secretion in mouse Leydig tumor cells after induction of cholesterol translocation by sphingomyelin degradation. Biochim Biophys Acta 1093:7–12 [DOI] [PubMed] [Google Scholar]
- 63. Rábano M, Peña A, Brizuela L, Marino A, Macarulla JM, Trueba M, Gómez-Muñoz A. 2003. Sphingosine-1-phosphate stimulates cortisol secretion. FEBS Lett 535:101–105 [DOI] [PubMed] [Google Scholar]
- 64. Li QL, Ni J, Bian SL, Yao LC, Zhu H, Zhang W. 2001. Inhibition of steroidogenesis and induction of apoptosis in rat luteal cells by cell-permeable ceramide in vitro. Sheng Li Xue Bao 53:142–146 [PubMed] [Google Scholar]
- 65. McClellan DR, Bourdelat-Parks B, Salata K, Francis GL. 1997. Sphingomyelinase affects hormone production in Jeg-3 choriocarcinoma cells. Cell Endocrinol Metab 9:19–24 [Google Scholar]
- 66. Degnan BM, Bourdelat-Parks B, Daniel A, Salata K, Francis GL. 1996. Sphingomyelinase inhibits in vitro Leydig cell function. Ann Clin Lab Sci 26:234–242 [PubMed] [Google Scholar]
- 67. Santana P, Llanes L, Hernandez I, Gonzalez-Robayna I, Tabraue C, Gonzalez-Reyes J, Quintana J, Estevez F, Ruiz de Galarreta CM, Fanjul LF. 1996. Interleukin-1β stimulates sphingomyelin hydrolysis in cultured granulosa cells: evidence for a regulatory role of ceramide on progesterone and prostaglandin biosynthesis. Endocrinology 137:2480–2489 [DOI] [PubMed] [Google Scholar]
- 68. Brizuela L, Rábano M, Gangoiti P, Narbona N, Macarulla JM, Trueba M, Gómez-Muñoz A. 2007. Sphingosine-1-phosphate stimulates aldosterone secretion through a mechanism involving the PI3K/PKB and MEK/ERK 1/2 pathways. J Lipid Res 48:2264–2274 [DOI] [PubMed] [Google Scholar]
- 69. Brizuela L, Rábano M, Peña A, Gangoiti P, Macarulla JM, Trueba M, Gómez-Muñoz A. 2006. Sphingosine-1-phosphate: a novel stimulator of aldosterone secretion. J Lipid Res 47:1238–1249 [DOI] [PubMed] [Google Scholar]
- 70. Budnik LT, Jähner D, Mukhopadhyay AK. 1999. Inhibitory effects of TNFα on mouse tumor Leydig cells: possible role of ceramide in the mechanism of action. Mol Cell Endocrinol 150:39–46 [DOI] [PubMed] [Google Scholar]
- 71. Kraemer FB, Shen WJ, Harada K, Patel S, Osuga J, Ishibashi S, Azhar S. 2004. Hormone-sensitive lipase is required for high-density lipoprotein cholesteryl ester-supported adrenal steroidogenesis. Mol Endocrinol 18:549–557 [DOI] [PubMed] [Google Scholar]
- 72. Hauet T, Liu J, Li H, Gazouli M, Culty M, Papadopoulos V. 2002. PBR, StAR, and PKA: partners in cholesterol transport in steroidogenic cells. Endocr Res 28:395–401 [DOI] [PubMed] [Google Scholar]
- 73. Shen WJ, Patel S, Natu V, Hong R, Wang J, Azhar S, Kraemer FB. 2003. Interaction of hormone-sensitive lipase with steroidogenic acute regulatory protein: facilitation of cholesterol transfer in adrenal. J Biol Chem 278:43870–43876 [DOI] [PubMed] [Google Scholar]
- 74. Miller WL. 2007. Mechanism of StAR's regulation of mitochondrial cholesterol import. Mol Cell Endocrinol 265–266:46–50 [DOI] [PubMed] [Google Scholar]
- 75. Niakan KK, McCabe ER. 2005. DAX1 origin, function, and novel role. Mol Genet Metab 86:70–83 [DOI] [PubMed] [Google Scholar]
- 76. Nachtigal MW, Hirokawa Y, Enyeart-VanHouten DL, Flanagan JN, Hammer GD, Ingraham HA. 1998. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell 93:445–454 [DOI] [PubMed] [Google Scholar]
- 77. Ehrlund A, Anthonisen EH, Gustafsson N, Venteclef N, Robertson Remen K, Damdimopoulos AE, Galeeva A, Pelto-Huikko M, Lalli E, Steffensen KR, Gustafsson JA, Treuter E. 2009. E3 ubiquitin ligase RNF31 cooperates with DAX-1 in transcriptional repression of steroidogenesis. Mol Cell Biol 29:2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shimizu T, Sudo N, Yamashita H, Murayama C, Miyazaki H, Miyamoto A. 2009. Histone H3 acetylation of StAR and decrease in DAX-1 is involved in the luteinization of bovine granulosa cells during in vitro culture. Mol Cell Biochem 328:41–47 [DOI] [PubMed] [Google Scholar]
- 79. Lalli E, Melner MH, Stocco DM, Sassone-Corsi P. 1998. DAX-1 blocks steroid production at multiple levels. Endocrinology 139:4237–4243 [DOI] [PubMed] [Google Scholar]
- 80. Kwun C, Patel A, Pletcher S, Lyons B, Abdelrahim M, Nicholson D, Morris E, Salata K, Francis GL. 1999. Ceramide increases steroid hormone production in MA-10 Leydig cells. Steroids 64:499–509 [DOI] [PubMed] [Google Scholar]
- 81. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 82. Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. 2009. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325:1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bielawska A, Greenberg MS, Perry D, Jayadev S, Shayman JA, McKay C, Hannun YA. 1996. (1S,2R)-D-erythro-2-(N-myristoylamino)-1-phenyl-1-propanol as an inhibitor of ceramidase. J Biol Chem 271:12646–12654 [DOI] [PubMed] [Google Scholar]
- 84. Mao C, Xu R, Szulc ZM, Bielawska A, Galadari SH, Obeid LM. 2001. Cloning and characterization of a novel human alkaline ceramidase. A mammalian enzyme that hydrolyzes phytoceramide. J Biol Chem 276:26577–26588 [DOI] [PubMed] [Google Scholar]
- 85. Xu R, Jin J, Hu W, Sun W, Bielawski J, Szulc Z, Taha T, Obeid LM, Mao C. 2006. Golgi alkaline ceramidase regulates cell proliferation and survival by controlling levels of sphingosine and S1P. FASEB J 20:1813–1825 [DOI] [PubMed] [Google Scholar]
- 86. Sun W, Xu R, Hu W, Jin J, Crellin HA, Bielawski J, Szulc ZM, Thiers BH, Obeid LM, Mao C. 2008. Upregulation of the human alkaline ceramidase 1 and acid ceramidase mediates calcium-induced differentiation of epidermal keratinocytes. J Invest Dermatol 128:389–397 [DOI] [PubMed] [Google Scholar]
- 87. Hwang YH, Tani M, Nakagawa T, Okino N, Ito M. 2005. Subcellular localization of human neutral ceramidase expressed in HEK293 cells. Biochem Biophys Res Commun 331:37–42 [DOI] [PubMed] [Google Scholar]
- 88. Romiti E, Meacci E, Tani M, Nuti F, Farnararo M, Ito M, Bruni P. 2000. Neutral/alkaline and acid ceramidase activities are actively released by murine endothelial cells. Biochem Biophys Res Commun 275:746–751 [DOI] [PubMed] [Google Scholar]
- 89. Ferlinz K, Kopal G, Bernardo K, Linke T, Bar J, Breiden B, Neumann U, Lang F, Schuchman EH, Sandhoff K. 2001. Human acid ceramidase: processing, glycosylation, and lysosomal targeting. J Biol Chem 276:35352–35360 [DOI] [PubMed] [Google Scholar]
- 90. Hu W, Xu R, Sun W, Szulc ZM, Bielawski J, Obeid LM, Mao C. 2010. Alkaline ceramidase 3 (ACER3) hydrolyzes unsaturated long-chain ceramides, and its down-regulation inhibits both cell proliferation and apoptosis. J Biol Chem 285:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. El Bawab S, Roddy P, Qian T, Bielawska A, Lemasters JJ, Hannun YA. 2000. Molecular cloning and characterization of a human mitochondrial ceramidase. J Biol Chem 275:21508–21513 [DOI] [PubMed] [Google Scholar]
- 92. Sun W, Jin J, Xu R, Hu W, Szulc ZM, Bielawski J, Obeid LM, Mao C. 2010. Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2). J Biol Chem 285:8995–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Momoi T, Ben-Yoseph Y, Nadler HL. 1982. Substrate-specificities of acid and alkaline ceramidases in fibroblasts from patients with Farber disease and controls. Biochem J 205:419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mullen TD, Spassieva S, Jenkins RW, Kitatani K, Bielawski J, Hannun YA, Obeid LM. 2011. Selective knockdown of ceramide synthases reveals complex inter-regulation of sphingolipid metabolism. J Lipid Res 52:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Park H, Haynes CA, Nairn AV, Kulik M, Dalton S, Moremen K, Merrill AH., Jr 2010. Transcript profiling and lipidomic analysis of ceramide subspecies in mouse embryonic stem cells and embryoid bodies. J Lipid Res 51:480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mesicek J, Lee H, Feldman T, Jiang X, Skobeleva A, Berdyshev EV, Haimovitz-Friedman A, Fuks Z, Kolesnick R. 2010. Ceramide synthases 2, 5, and 6 confer distinct roles in radiation-induced apoptosis in HeLa cells. Cell Signal 22:1300–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ben-David O, Futerman AH. 2010. The role of the ceramide acyl chain length in neurodegeneration: involvement of ceramide synthases. Neuromolecular Med 12:341–350 [DOI] [PubMed] [Google Scholar]
- 98. Wu BX, Zeidan YH, Hannun YA. 2009. Downregulation of neutral ceramidase by gemcitabine: implications for cell cycle regulation. Biochim Biophys Acta 1791:730–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lucki NC, Sewer MB. 2011. Genistein stimulates MCF-7 breast cancer cell growth by inducing acid ceramidase (ASAH1) gene expression. J Biol Chem 286:19399–19409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chatterjee S. 1991. Lactosylceramide stimulates aortic smooth muscle cell proliferation. Biochem Biophys Res Commun 181:554–561 [DOI] [PubMed] [Google Scholar]
- 101. Mu H, Wang X, Wang H, Lin P, Yao Q, Chen C. 2009. Lactosylceramide promotes cell migration and proliferation through activation of ERK1/2 in human aortic smooth muscle cells. Am J Physiol Heart Circ Physiol 297:H400–H408 [DOI] [PubMed] [Google Scholar]
- 102. Bhunia AK, Han H, Snowden A, Chatterjee S. 1996. Lactosylceramide stimulates Ras-GTP loading, kinases (MEK, Raf), p44 mitogen-activated protein kinase, and c-fos expression in human aortic smooth muscle cells. J Biol Chem 271:10660–10666 [DOI] [PubMed] [Google Scholar]
- 103. Mountjoy KG, Bird IM, Rainey WE, Cone RD. 1994. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol Cell Endocrinol 99:R17–R20 [DOI] [PubMed] [Google Scholar]
- 104. Staels B, Hum DW, Miller WL. 1993. Regulation of steroidogenesis in NCI-H295 cells: a cellular model of the human fetal adrenal. Mol Endocrinol 7:423–433 [DOI] [PubMed] [Google Scholar]
- 105. Sullards MC, Merrill AH., Jr 2001. Analysis of sphingosine 1-phosphate, ceramides, and other bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Sci STKE 2001:pl1. [DOI] [PubMed] [Google Scholar]
- 106. Sullards MC. 2000. Analysis of sphingomyelin, glucosylceramide, ceramide, sphingosine, and sphingosine 1-phosphate by tandem mass spectrometry. Methods Enzymol 312:32–45 [DOI] [PubMed] [Google Scholar]
- 107. Dammer EB, Leon A, Sewer MB. 2007. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol Endocrinol 21:415–438 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.