Abstract
In the title compound, C20H14Cl2F3NO3, the trifluromethyl group is disordered over two sets of sites in a 0.784 (10):0.216 (10) ratio. The quinoline ring system is essentially planar with a maximum deviation of 0.058 (2) Å for the N atom and forms dihedral angles of 89.23 (11) and 8.13 (17)°, respectively with the mean planes of the benzene ring and the carboxylate group. In the crystal, pairs of weak C—H⋯O and C—H⋯F hydrogen bonds link molecules into centrosymmetric dimers. The crystal structure is further stabilized by weak π–π [centroid–centroid distance = 3.624 (2) Å] interactions.
Related literature
For background to the properties and uses of quinoline derivatives, see: Kaur et al. (2010 ▶); Eswaran et al. (2010 ▶); Chou et al. (2010 ▶); Chen et al. (2004 ▶); Shingalapur et al. (2009 ▶). For a related structure, see: Fun et al. (2011 ▶). For standard bond-length data, see: Allen et al. (1987 ▶).
Experimental
Crystal data
C20H14Cl2F3NO3
M r = 444.22
Triclinic,
a = 8.090 (2) Å
b = 9.547 (3) Å
c = 14.047 (4) Å
α = 77.299 (6)°
β = 76.198 (5)°
γ = 67.488 (4)°
V = 963.3 (5) Å3
Z = 2
Mo Kα radiation
μ = 0.39 mm−1
T = 296 K
0.43 × 0.18 × 0.07 mm
Data collection
Bruker APEX DUO CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.852, T max = 0.972
13916 measured reflections
5071 independent reflections
2759 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.056
wR(F 2) = 0.179
S = 1.04
5071 reflections
288 parameters
7 restraints
H-atom parameters constrained
Δρmax = 0.35 e Å−3
Δρmin = −0.34 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812001249/lh5400sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812001249/lh5400Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812001249/lh5400Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16A⋯O3i | 0.93 | 2.52 | 3.292 (5) | 141 |
| C18—H18A⋯F2i | 0.97 | 2.50 | 3.355 (7) | 147 |
Symmetry code: (i) 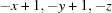 .
.
Acknowledgments
HKF and CWO thank Universiti Sains Malaysia (USM) for the Research University Grant (1001/PFIZIK/811160). CWO also thanks the Malaysian Government and USM for the award of the post of research assistant under the Research University Grant (1001/PFIZIK/811151). AMI is thankful to the Department of Atomic Energy, Board for Research in Nuclear Sciences, Government of India for the Young Scientist award.
supplementary crystallographic information
Comment
The quinoline moiety is of great importance to chemists as well as biologists as it is one of the key building elements for many naturally occurring compounds. Members of this family have wide range of applications as pharmaceuticals as antimalarial (Kaur et al., 2010), anti-tuberculosis (Eswaran et al., 2010), antitumor (Chou et al., 2010), anticancer (Chen et al., 2004) and antiviral (Shingalapur et al., 2009) agents. Some of the present day drugs such as chloroquine, mefloquine, tafenoquine and primaquine contain quinoline as the basic unit in their structures. In view of the biological importance, we have synthesized the title compound to study its crystal structure.
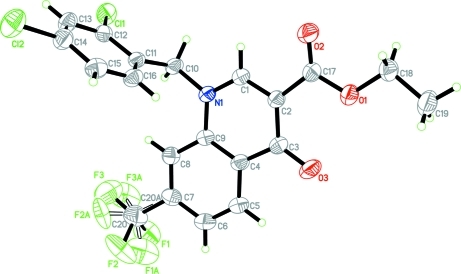
In the molecular structure (Fig. 1), the trifluromethyl group is disordered over two sets of sites in a ratio of 0.784 (10):0.216 (10). The quinoline ring (N1/C1–C9) is essentially planar with a maximum deviation of 0.058 (2) Å at atom N1. The quinoline ring makes dihedral angles of 89.23 (11) and 8.13 (17)°, respectively with the chloro-substituted benzene ring (C11–C16) and the carboxylate group (O1/O2/C17–C19). The bond lengths (Allen et al., 1987) and angles are within normal ranges and are comparable to a related structure (Fun et al., 2011).
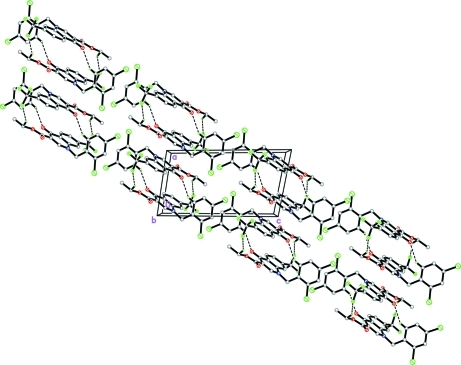
In the crystal (Fig. 2), intermolecular C16—H16A···O3i and C18—H18A···F2i hydrogen bonds (Table 1) link molecules to form dimers. The crystal is further stabilized by weak π–π interactions between the quinoline (N1/C1–C9) and the benzene ring (C4–C9) [centroid-to-centroid (-x, 1 - y, -z); distance = 3.624 (2) Å].
Experimental
A mixture of ethyl 4-hydroxy-7-(trifluoromethyl)quinoline-3-carboxylate (0.10 g, 0.00035 mol), potassium carbonate (0.053 g, 0.00038 mol) and 1-(bromomethyl)-2,4-dichlorobenzene (0.091 g, 0.00038 mol) in dimethylformamide (5 ml) was stirred at 353K for 3 h. After completion of the reaction, the reaction mixture was poured into ice-cold water. The solid product obtained was filtered, washed with water and recrystallized using ethanol. Yield: 0.150 g, 96.77%. M. p.: 428–429 K.
Refinement
All H atoms were positioned geometrically and refined using a riding model with Uiso(H) = 1.2 or 1.5Ueq(C) (C—H = 0.93, 0.96 or 0.97 Å). A rotating group model was applied to the methyl group. In the final refinement, the outliners (-4 - 6 1), (3 2 0), (5 0 6), (5 1 8) were omitted. A bond-distance restraint was applied to C20A–C7. The same Uij parameters were used for atom pairs F3A/F1A and C20A/C7.
Figures
Fig. 1.
The molecular structure of the title compound, showing 30% probability displacement ellipsoids. Both disorder components are shown.
Fig. 2.
The crystal packing of the title compound. The dashed lines represent the hydrogen bonds. Only the major disordered component is shown.
Crystal data
| C20H14Cl2F3NO3 | Z = 2 |
| Mr = 444.22 | F(000) = 452 |
| Triclinic, P1 | Dx = 1.531 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.090 (2) Å | Cell parameters from 3077 reflections |
| b = 9.547 (3) Å | θ = 2.3–24.2° |
| c = 14.047 (4) Å | µ = 0.39 mm−1 |
| α = 77.299 (6)° | T = 296 K |
| β = 76.198 (5)° | Block, colourless |
| γ = 67.488 (4)° | 0.43 × 0.18 × 0.07 mm |
| V = 963.3 (5) Å3 |
Data collection
| Bruker APEX DUO CCD area-detector diffractometer | 5071 independent reflections |
| Radiation source: fine-focus sealed tube | 2759 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| φ and ω scans | θmax = 29.1°, θmin = 1.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −10→11 |
| Tmin = 0.852, Tmax = 0.972 | k = −13→13 |
| 13916 measured reflections | l = −19→19 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.056 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.179 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0711P)2 + 0.3522P] where P = (Fo2 + 2Fc2)/3 |
| 5071 reflections | (Δ/σ)max < 0.001 |
| 288 parameters | Δρmax = 0.35 e Å−3 |
| 7 restraints | Δρmin = −0.34 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cl1 | −0.33323 (10) | 0.73631 (10) | 0.39029 (6) | 0.0780 (3) | |
| Cl2 | 0.24014 (16) | 0.66624 (13) | 0.54781 (7) | 0.1052 (4) | |
| C20 | 0.1448 (11) | 0.0460 (8) | 0.2381 (6) | 0.0900 (15) | 0.784 (10) |
| F1 | 0.0710 (8) | −0.0345 (4) | 0.2048 (3) | 0.1144 (17) | 0.784 (10) |
| F2 | 0.2991 (6) | −0.0610 (5) | 0.2577 (5) | 0.137 (2) | 0.784 (10) |
| F3 | 0.0375 (13) | 0.0915 (6) | 0.3154 (4) | 0.197 (5) | 0.784 (10) |
| C20A | 0.134 (3) | 0.055 (2) | 0.2391 (13) | 0.0638 (7) | 0.216 (10) |
| F1A | 0.207 (3) | −0.0777 (17) | 0.2083 (14) | 0.149 (7) | 0.216 (10) |
| F2A | 0.204 (2) | 0.042 (2) | 0.3172 (9) | 0.103 (6) | 0.216 (10) |
| F3A | −0.026 (2) | 0.078 (2) | 0.2781 (19) | 0.149 (7) | 0.216 (10) |
| O1 | 0.3593 (3) | 0.7747 (2) | −0.19081 (13) | 0.0747 (6) | |
| O2 | 0.2188 (3) | 0.9285 (2) | −0.07639 (15) | 0.0818 (6) | |
| O3 | 0.4032 (4) | 0.4757 (3) | −0.14202 (17) | 0.0992 (8) | |
| N1 | 0.0638 (3) | 0.5929 (2) | 0.11707 (14) | 0.0525 (5) | |
| C1 | 0.1139 (3) | 0.6989 (3) | 0.04830 (18) | 0.0538 (6) | |
| H1A | 0.0652 | 0.7997 | 0.0611 | 0.065* | |
| C2 | 0.2309 (3) | 0.6694 (3) | −0.03871 (18) | 0.0519 (6) | |
| C3 | 0.3065 (4) | 0.5147 (3) | −0.0630 (2) | 0.0614 (7) | |
| C4 | 0.2558 (3) | 0.4008 (3) | 0.01455 (19) | 0.0546 (6) | |
| C5 | 0.3257 (4) | 0.2471 (3) | −0.0008 (2) | 0.0668 (7) | |
| H5A | 0.4018 | 0.2206 | −0.0600 | 0.080* | |
| C6 | 0.2850 (4) | 0.1353 (3) | 0.0693 (2) | 0.0726 (8) | |
| H6A | 0.3294 | 0.0344 | 0.0570 | 0.087* | |
| C7 | 0.1763 (4) | 0.1747 (3) | 0.1588 (2) | 0.0638 (7) | |
| C8 | 0.1019 (4) | 0.3237 (3) | 0.1768 (2) | 0.0580 (6) | |
| H8A | 0.0275 | 0.3481 | 0.2369 | 0.070* | |
| C9 | 0.1394 (3) | 0.4388 (3) | 0.10383 (18) | 0.0506 (6) | |
| C10 | −0.0705 (3) | 0.6443 (3) | 0.20509 (18) | 0.0570 (6) | |
| H10A | −0.1461 | 0.5812 | 0.2247 | 0.068* | |
| H10B | −0.1484 | 0.7490 | 0.1876 | 0.068* | |
| C11 | 0.0122 (3) | 0.6370 (3) | 0.29170 (17) | 0.0497 (6) | |
| C12 | −0.0999 (3) | 0.6848 (3) | 0.37876 (18) | 0.0546 (6) | |
| C13 | −0.0312 (4) | 0.6930 (3) | 0.4573 (2) | 0.0692 (8) | |
| H13A | −0.1089 | 0.7262 | 0.5148 | 0.083* | |
| C14 | 0.1529 (4) | 0.6517 (3) | 0.4499 (2) | 0.0680 (8) | |
| C15 | 0.2693 (4) | 0.5999 (3) | 0.3659 (2) | 0.0651 (7) | |
| H15A | 0.3942 | 0.5695 | 0.3619 | 0.078* | |
| C16 | 0.1983 (4) | 0.5938 (3) | 0.2878 (2) | 0.0580 (6) | |
| H16A | 0.2769 | 0.5598 | 0.2307 | 0.070* | |
| C17 | 0.2670 (3) | 0.8046 (3) | −0.10207 (19) | 0.0566 (6) | |
| C18 | 0.3971 (5) | 0.9011 (3) | −0.2582 (2) | 0.0729 (8) | |
| H18A | 0.4611 | 0.9439 | −0.2290 | 0.087* | |
| H18B | 0.2848 | 0.9811 | −0.2730 | 0.087* | |
| C19 | 0.5111 (6) | 0.8397 (4) | −0.3499 (2) | 0.1007 (13) | |
| H19A | 0.5278 | 0.9226 | −0.3993 | 0.151* | |
| H19B | 0.4521 | 0.7878 | −0.3740 | 0.151* | |
| H19C | 0.6271 | 0.7691 | −0.3356 | 0.151* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0572 (4) | 0.0910 (6) | 0.0667 (5) | −0.0187 (4) | 0.0125 (3) | −0.0118 (4) |
| Cl2 | 0.1179 (8) | 0.1260 (8) | 0.0906 (7) | −0.0464 (7) | −0.0256 (6) | −0.0378 (6) |
| C20 | 0.091 (4) | 0.062 (3) | 0.116 (4) | −0.028 (3) | −0.016 (3) | −0.010 (3) |
| F1 | 0.120 (4) | 0.097 (2) | 0.156 (3) | −0.073 (3) | −0.038 (2) | 0.003 (2) |
| F2 | 0.099 (3) | 0.099 (3) | 0.207 (5) | −0.048 (2) | −0.072 (3) | 0.069 (3) |
| F3 | 0.325 (11) | 0.088 (3) | 0.129 (4) | −0.094 (5) | 0.092 (6) | −0.020 (3) |
| C20A | 0.0626 (17) | 0.0525 (15) | 0.0778 (19) | −0.0208 (13) | −0.0151 (14) | −0.0079 (13) |
| F1A | 0.096 (9) | 0.116 (10) | 0.226 (15) | −0.066 (7) | 0.006 (8) | 0.010 (9) |
| F2A | 0.083 (9) | 0.145 (15) | 0.076 (8) | −0.057 (10) | −0.038 (7) | 0.053 (8) |
| F3A | 0.096 (9) | 0.116 (10) | 0.226 (15) | −0.066 (7) | 0.006 (8) | 0.010 (9) |
| O1 | 0.0975 (15) | 0.0655 (12) | 0.0540 (11) | −0.0334 (11) | 0.0120 (10) | −0.0122 (9) |
| O2 | 0.1094 (17) | 0.0645 (13) | 0.0696 (13) | −0.0377 (12) | 0.0106 (12) | −0.0200 (10) |
| O3 | 0.1143 (19) | 0.0721 (14) | 0.0803 (15) | −0.0225 (13) | 0.0394 (13) | −0.0285 (12) |
| N1 | 0.0536 (12) | 0.0498 (11) | 0.0481 (11) | −0.0138 (9) | 0.0006 (9) | −0.0124 (9) |
| C1 | 0.0558 (15) | 0.0509 (14) | 0.0526 (14) | −0.0154 (11) | −0.0049 (11) | −0.0139 (11) |
| C2 | 0.0520 (14) | 0.0529 (14) | 0.0486 (13) | −0.0178 (11) | −0.0027 (11) | −0.0103 (11) |
| C3 | 0.0539 (15) | 0.0601 (16) | 0.0569 (15) | −0.0089 (12) | 0.0025 (12) | −0.0153 (12) |
| C4 | 0.0477 (13) | 0.0523 (14) | 0.0582 (15) | −0.0091 (11) | −0.0061 (11) | −0.0153 (11) |
| C5 | 0.0615 (17) | 0.0576 (16) | 0.0692 (18) | −0.0067 (13) | −0.0025 (13) | −0.0198 (14) |
| C6 | 0.0751 (19) | 0.0515 (16) | 0.087 (2) | −0.0132 (14) | −0.0137 (16) | −0.0176 (15) |
| C7 | 0.0626 (17) | 0.0525 (15) | 0.0778 (19) | −0.0208 (13) | −0.0151 (14) | −0.0079 (13) |
| C8 | 0.0548 (15) | 0.0585 (16) | 0.0601 (15) | −0.0212 (12) | −0.0063 (12) | −0.0087 (12) |
| C9 | 0.0476 (13) | 0.0481 (13) | 0.0553 (14) | −0.0139 (11) | −0.0070 (11) | −0.0131 (11) |
| C10 | 0.0511 (14) | 0.0554 (14) | 0.0551 (15) | −0.0129 (11) | 0.0065 (11) | −0.0168 (12) |
| C11 | 0.0544 (14) | 0.0411 (12) | 0.0491 (13) | −0.0184 (10) | 0.0034 (10) | −0.0076 (10) |
| C12 | 0.0558 (14) | 0.0498 (14) | 0.0491 (14) | −0.0196 (11) | 0.0069 (11) | −0.0039 (11) |
| C13 | 0.078 (2) | 0.0700 (18) | 0.0518 (16) | −0.0258 (15) | 0.0095 (14) | −0.0156 (13) |
| C14 | 0.081 (2) | 0.0682 (18) | 0.0591 (16) | −0.0310 (16) | −0.0091 (15) | −0.0132 (14) |
| C15 | 0.0621 (17) | 0.0616 (17) | 0.0757 (19) | −0.0261 (14) | −0.0078 (14) | −0.0140 (14) |
| C16 | 0.0562 (15) | 0.0569 (15) | 0.0578 (15) | −0.0208 (12) | 0.0045 (12) | −0.0156 (12) |
| C17 | 0.0541 (15) | 0.0618 (16) | 0.0529 (14) | −0.0210 (12) | −0.0026 (11) | −0.0119 (12) |
| C18 | 0.084 (2) | 0.0724 (19) | 0.0620 (17) | −0.0388 (16) | 0.0061 (15) | −0.0073 (14) |
| C19 | 0.137 (3) | 0.100 (3) | 0.064 (2) | −0.058 (3) | 0.023 (2) | −0.0218 (18) |
Geometric parameters (Å, °)
| Cl1—C12 | 1.735 (3) | C5—H5A | 0.9300 |
| Cl2—C14 | 1.741 (3) | C6—C7 | 1.388 (4) |
| C20—F3 | 1.262 (8) | C6—H6A | 0.9300 |
| C20—F2 | 1.320 (7) | C7—C8 | 1.371 (4) |
| C20—F1 | 1.344 (8) | C8—C9 | 1.401 (4) |
| C20—C7 | 1.522 (8) | C8—H8A | 0.9300 |
| C20A—F3A | 1.235 (16) | C10—C11 | 1.499 (4) |
| C20A—F1A | 1.303 (16) | C10—H10A | 0.9700 |
| C20A—F2A | 1.311 (16) | C10—H10B | 0.9700 |
| C20A—C7 | 1.494 (17) | C11—C12 | 1.387 (3) |
| O1—C17 | 1.320 (3) | C11—C16 | 1.391 (4) |
| O1—C18 | 1.445 (3) | C12—C13 | 1.377 (4) |
| O2—C17 | 1.204 (3) | C13—C14 | 1.371 (4) |
| O3—C3 | 1.236 (3) | C13—H13A | 0.9300 |
| N1—C1 | 1.349 (3) | C14—C15 | 1.376 (4) |
| N1—C9 | 1.396 (3) | C15—C16 | 1.375 (4) |
| N1—C10 | 1.472 (3) | C15—H15A | 0.9300 |
| C1—C2 | 1.365 (3) | C16—H16A | 0.9300 |
| C1—H1A | 0.9300 | C18—C19 | 1.485 (4) |
| C2—C3 | 1.450 (4) | C18—H18A | 0.9700 |
| C2—C17 | 1.484 (4) | C18—H18B | 0.9700 |
| C3—C4 | 1.465 (4) | C19—H19A | 0.9600 |
| C4—C5 | 1.401 (4) | C19—H19B | 0.9600 |
| C4—C9 | 1.402 (3) | C19—H19C | 0.9600 |
| C5—C6 | 1.367 (4) | ||
| F3—C20—F2 | 112.3 (8) | N1—C9—C8 | 121.8 (2) |
| F3—C20—F1 | 105.5 (6) | N1—C9—C4 | 118.2 (2) |
| F2—C20—F1 | 101.2 (6) | C8—C9—C4 | 120.1 (2) |
| F3—C20—C7 | 113.9 (6) | N1—C10—C11 | 113.9 (2) |
| F2—C20—C7 | 111.8 (6) | N1—C10—H10A | 108.8 |
| F1—C20—C7 | 111.2 (6) | C11—C10—H10A | 108.8 |
| F3A—C20A—F1A | 110.4 (17) | N1—C10—H10B | 108.8 |
| F3A—C20A—F2A | 99.9 (16) | C11—C10—H10B | 108.8 |
| F1A—C20A—F2A | 106.3 (18) | H10A—C10—H10B | 107.7 |
| F3A—C20A—C7 | 118.3 (17) | C12—C11—C16 | 117.1 (2) |
| F1A—C20A—C7 | 109.6 (14) | C12—C11—C10 | 119.6 (2) |
| F2A—C20A—C7 | 111.4 (13) | C16—C11—C10 | 123.2 (2) |
| C17—O1—C18 | 116.3 (2) | C13—C12—C11 | 121.8 (3) |
| C1—N1—C9 | 120.0 (2) | C13—C12—Cl1 | 118.4 (2) |
| C1—N1—C10 | 118.3 (2) | C11—C12—Cl1 | 119.8 (2) |
| C9—N1—C10 | 121.7 (2) | C14—C13—C12 | 119.3 (3) |
| N1—C1—C2 | 124.9 (2) | C14—C13—H13A | 120.3 |
| N1—C1—H1A | 117.6 | C12—C13—H13A | 120.3 |
| C2—C1—H1A | 117.6 | C13—C14—C15 | 120.8 (3) |
| C1—C2—C3 | 119.4 (2) | C13—C14—Cl2 | 119.5 (2) |
| C1—C2—C17 | 115.1 (2) | C15—C14—Cl2 | 119.7 (3) |
| C3—C2—C17 | 125.5 (2) | C16—C15—C14 | 119.1 (3) |
| O3—C3—C2 | 125.5 (3) | C16—C15—H15A | 120.4 |
| O3—C3—C4 | 120.0 (2) | C14—C15—H15A | 120.4 |
| C2—C3—C4 | 114.5 (2) | C15—C16—C11 | 121.8 (2) |
| C5—C4—C9 | 118.3 (2) | C15—C16—H16A | 119.1 |
| C5—C4—C3 | 118.9 (2) | C11—C16—H16A | 119.1 |
| C9—C4—C3 | 122.8 (2) | O2—C17—O1 | 122.9 (2) |
| C6—C5—C4 | 121.6 (3) | O2—C17—C2 | 124.4 (2) |
| C6—C5—H5A | 119.2 | O1—C17—C2 | 112.6 (2) |
| C4—C5—H5A | 119.2 | O1—C18—C19 | 107.1 (3) |
| C5—C6—C7 | 119.0 (3) | O1—C18—H18A | 110.3 |
| C5—C6—H6A | 120.5 | C19—C18—H18A | 110.3 |
| C7—C6—H6A | 120.5 | O1—C18—H18B | 110.3 |
| C8—C7—C6 | 121.6 (3) | C19—C18—H18B | 110.3 |
| C8—C7—C20A | 117.5 (8) | H18A—C18—H18B | 108.6 |
| C6—C7—C20A | 120.9 (8) | C18—C19—H19A | 109.5 |
| C8—C7—C20 | 120.7 (4) | C18—C19—H19B | 109.5 |
| C6—C7—C20 | 117.7 (4) | H19A—C19—H19B | 109.5 |
| C7—C8—C9 | 119.3 (3) | C18—C19—H19C | 109.5 |
| C7—C8—H8A | 120.4 | H19A—C19—H19C | 109.5 |
| C9—C8—H8A | 120.4 | H19B—C19—H19C | 109.5 |
| C9—N1—C1—C2 | 3.1 (4) | C20A—C7—C8—C9 | 179.4 (8) |
| C10—N1—C1—C2 | −177.2 (2) | C20—C7—C8—C9 | 177.5 (4) |
| N1—C1—C2—C3 | 1.6 (4) | C1—N1—C9—C8 | 174.9 (2) |
| N1—C1—C2—C17 | −179.3 (2) | C10—N1—C9—C8 | −4.8 (4) |
| C1—C2—C3—O3 | 174.7 (3) | C1—N1—C9—C4 | −5.0 (4) |
| C17—C2—C3—O3 | −4.3 (5) | C10—N1—C9—C4 | 175.2 (2) |
| C1—C2—C3—C4 | −3.8 (4) | C7—C8—C9—N1 | 178.3 (2) |
| C17—C2—C3—C4 | 177.2 (2) | C7—C8—C9—C4 | −1.7 (4) |
| O3—C3—C4—C5 | 3.0 (4) | C5—C4—C9—N1 | −177.3 (2) |
| C2—C3—C4—C5 | −178.4 (3) | C3—C4—C9—N1 | 2.6 (4) |
| O3—C3—C4—C9 | −176.8 (3) | C5—C4—C9—C8 | 2.7 (4) |
| C2—C3—C4—C9 | 1.8 (4) | C3—C4—C9—C8 | −177.4 (2) |
| C9—C4—C5—C6 | −0.8 (4) | C1—N1—C10—C11 | −93.1 (3) |
| C3—C4—C5—C6 | 179.3 (3) | C9—N1—C10—C11 | 86.7 (3) |
| C4—C5—C6—C7 | −2.1 (5) | N1—C10—C11—C12 | 178.9 (2) |
| C5—C6—C7—C8 | 3.2 (5) | N1—C10—C11—C16 | 3.4 (3) |
| C5—C6—C7—C20A | −177.6 (8) | C16—C11—C12—C13 | 1.6 (4) |
| C5—C6—C7—C20 | −175.6 (4) | C10—C11—C12—C13 | −174.2 (2) |
| F3A—C20A—C7—C8 | 49 (2) | C16—C11—C12—Cl1 | −178.55 (18) |
| F1A—C20A—C7—C8 | 176.8 (13) | C10—C11—C12—Cl1 | 5.6 (3) |
| F2A—C20A—C7—C8 | −65.8 (19) | C11—C12—C13—C14 | −0.6 (4) |
| F3A—C20A—C7—C6 | −130.1 (17) | Cl1—C12—C13—C14 | 179.5 (2) |
| F1A—C20A—C7—C6 | −2.5 (18) | C12—C13—C14—C15 | −1.1 (4) |
| F2A—C20A—C7—C6 | 114.9 (15) | C12—C13—C14—Cl2 | 178.5 (2) |
| F3A—C20A—C7—C20 | −159 (16) | C13—C14—C15—C16 | 1.7 (4) |
| F1A—C20A—C7—C20 | −31 (14) | Cl2—C14—C15—C16 | −177.9 (2) |
| F2A—C20A—C7—C20 | 86 (15) | C14—C15—C16—C11 | −0.7 (4) |
| F3—C20—C7—C8 | 4.5 (8) | C12—C11—C16—C15 | −0.9 (4) |
| F2—C20—C7—C8 | −124.1 (6) | C10—C11—C16—C15 | 174.7 (2) |
| F1—C20—C7—C8 | 123.6 (6) | C18—O1—C17—O2 | −1.4 (4) |
| F3—C20—C7—C6 | −176.7 (6) | C18—O1—C17—C2 | 178.7 (2) |
| F2—C20—C7—C6 | 54.7 (8) | C1—C2—C17—O2 | 9.3 (4) |
| F1—C20—C7—C6 | −57.6 (8) | C3—C2—C17—O2 | −171.8 (3) |
| F3—C20—C7—C20A | −24 (14) | C1—C2—C17—O1 | −170.9 (2) |
| F2—C20—C7—C20A | −153 (15) | C3—C2—C17—O1 | 8.1 (4) |
| F1—C20—C7—C20A | 95 (15) | C17—O1—C18—C19 | 176.4 (3) |
| C6—C7—C8—C9 | −1.3 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H16A···O3i | 0.93 | 2.52 | 3.292 (5) | 141 |
| C18—H18A···F2i | 0.97 | 2.50 | 3.355 (7) | 147 |
Symmetry codes: (i) −x+1, −y+1, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5400).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2009). SADABS, APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chen, Y. L., Hung, H. M., Lu, C. M., Li, K. C. & Tzeng, C. C. (2004). Bioorg. Med. Chem. 12, 6539–6546. [DOI] [PubMed]
- Chou, L. C., Tsai, M. T., Hsu, M. H., Wang, S. H., Way, T. D., Huang, C. H., Lin, H. Y., Qian, K., Dong, Y., Lee, K. H., Huang, L. J. & Kuo, S. C. (2010). J. Med. Chem. 53, 8047–8058. [DOI] [PubMed]
- Eswaran, S., Adhikari, A. V., Chowdhury, I. H., Pal, N. K. & Thomas, K. D. (2010). Eur. J. Med. Chem. 45, 3374–3383. [DOI] [PubMed]
- Fun, H.-K., Arshad, S., Garudachari, B., Isloor, A. M. & Satyanarayan, M. N. (2011). Acta Cryst. E67, o3117–o3118. [DOI] [PMC free article] [PubMed]
- Kaur, K., Jain, M., Reddy, R. P. & Jain, R. (2010). Eur. J. Med. Chem. 45, 3245–3264. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shingalapur, R. V., Hosamani, K. M. & Keri, R. S. (2009). Eur. J. Med. Chem. 44, 4244–4248. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812001249/lh5400sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812001249/lh5400Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812001249/lh5400Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report