Abstract
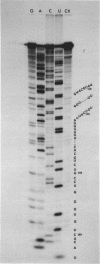
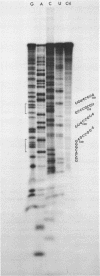
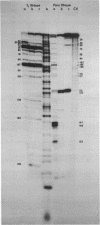
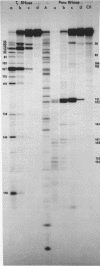
We have re-examined the nucleotide sequence of Vicia faba (broad bean) 5.8S rRNA using partial chemical degradation and a new approach to high temperature (65-80 degrees C) sequencing gels. The results indicate that the secondary structure was not completely disrupted in previous studies (Tanaka, Y., Dyer, T.A. and Brownlee, G.G. (1980) Nucleic Acid Res. 8, 1259-1272) and explain ambiguities between the nucleotide sequence and T1 ribonuclease digests. Despite this revision, estimates in the secondary structure suggest that this 5.8S rRNA differs from previously examined examples in two respects, more open conformations in both the "GC-rich" and "AU-rich" stems. The secondary structure was probed under a variety of ionic conditions using limited pancreatic and T1 ribonuclease digestion and rapid gel sequencing techniques. These studies and theoretical considerations generally supported the "burp gun" model previously proposed for all 5.8S rRNAs and were inconsistent with the recently suggested "cloverleaf" configuration. More importantly, they were also consistent with more open stem structures in this higher plant.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambartsumyan N. S., Mazo A. M. Elimination of the secondary structure effect in gell sequencing of nucleic acids. FEBS Lett. 1980 Jun 2;114(2):265–268. doi: 10.1016/0014-5793(80)81130-6. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S rRNA sequences and their precursors. Nucleic Acids Res. 1980 Jan 11;8(1):r31–r47. doi: 10.1093/nar/8.1.197-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. C., Randerath K. Rapid print-readout technique for sequencing of RNA's containing modified nucleotides. Nucleic Acids Res. 1979 Aug 10;6(11):3443–3458. doi: 10.1093/nar/6.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 25S-rRNA and the location of a 5.8S-rRNA binding site. Nucleic Acids Res. 1981 Mar 11;9(5):1111–1121. doi: 10.1093/nar/9.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Maden B. E. Chemical modification studies and the secondary structure of HeLa cell 5.8S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4521–4534. doi: 10.1093/nar/8.19.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp G., Gross H. J. Rapid RNA sequencing: nucleases from Staphylococcus aureus and Neurospora crassa discriminate between uridine and cytidine. Nucleic Acids Res. 1979 Aug 10;6(11):3481–3490. doi: 10.1093/nar/6.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma G. A., Marshall A. G. Laser Raman evidence for new cloverleaf secondary structures for eukaryotic 5.8S RNA and prokaryotic 5S RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4901–4905. doi: 10.1073/pnas.75.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N., Matheson A. T. Nucleotide sequence of Thermus aquaticus ribosomal 5 S ribonucleic acid. Sequence homologies in thermophilic organisms. J Biol Chem. 1977 Jun 25;252(12):4256–4261. [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Structural analyses of mammalian ribosomal ribonucleic acid and its precursors. Nucleotide sequence of ribosomal 5.8 S ribonucleic acid. J Biol Chem. 1975 Nov 25;250(22):8591–8597. [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A., Brimacombe R. Application of a rapid gel method to the sequencing of fragments of 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1978 Jan;5(1):241–256. doi: 10.1093/nar/5.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Sitz T. O., Kuo S. C., Nazar R. N. Multimer forms of eukaryotic 5.8S ribosomal RNA. Biochemistry. 1978 Dec 26;17(26):5811–5815. doi: 10.1021/bi00619a031. [DOI] [PubMed] [Google Scholar]
- Speirs J., Birnstiel M. Arrangement of the 5-8 S RNA cistrons in the genome of Xenopus laevis. J Mol Biol. 1974 Aug 5;87(2):237–256. doi: 10.1016/0022-2836(74)90146-6. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Dyer T. A., Brownlee G. G. An improved direct RNA sequence method; its application to Vicia faba 5.8S ribosomal RNA. Nucleic Acids Res. 1980 Mar 25;8(6):1259–1272. doi: 10.1093/nar/8.6.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wildeman A. G., Nazar R. N. Studies on the secondary structure of 5.8 S rRNA from a thermophile, Thermomyces lanuginosus. J Biol Chem. 1981 Jun 10;256(11):5675–5682. [PubMed] [Google Scholar]
- Woledge J., Corry M. J., Payne P. I. Ribosomal RNA homologies in flowering plants: comparison of the nucleotide sequences in 5.8-S rRNA from broad bean, dwarf bean, tomato, sunflower and rye. Biochim Biophys Acta. 1974 May 31;349(3):339–350. [PubMed] [Google Scholar]