Abstract
The mammalian circadian clock lying in the suprachiasmatic nucleus (SCN) controls daily rhythms and synchronizes the organism to its environment. In all organisms studied, circadian timekeeping is cell-autonomous, and rhythmicity is thought to be generated by a feedback loop involving clock proteins that inhibit transcription of their own genes. In the present study, we examined how these cellular properties are organized within the SCN tissue to produce rhythmicity and photic entrainment. The results show that the SCN has two compartments regulating Period genesPer1, Per2, and Per3 mRNA expression differentially. One compartment shows endogenous rhythmicity in Per1, Per2, and Per3mRNA expression. The other compartment does not have rhythmic mRNA expression but has gated light-induced Per1 andPer2 and high levels of endogenous nonrhythmicPer3 mRNA expression. These results reveal the occurrence of differential regulation of clock genes in two distinct SCN regions and suggest a potential mechanism for producing functional differences in distinct SCN subregions.
Keywords: suprachiasmatic nuclei, circadian rhythms, clock gene, Per1, Per2, Per3 Bma1, calbindinD28K, vasopressin, Fos, light pulse, oscillator, pacemaker
Circadian regulation of physiology and behavior has been observed in organisms from cyanobacteria to mammals. The discovery of mammalian homologs of nonmammalian clock genes has focused attention on the conservation of circadian mechanisms in evolution (Dunlap, 1999). Molecular models of circadian oscillators in mammals are based on findings in Drosophila andNeurospora (for review, see Young 1998; Dunlap 1999). In these models, rhythmicity is produced by a cell-based negative transcriptional feedback loop, in which expression of the putative clock genes is suppressed by their own protein products, and entrainment results from their modification by light-induced signals. Although many clock genes are conserved, there are distinct species differences, as noted in the reviews cited above. Mice have threePeriod genes (mPer1, mPer2, andmPer3), whereas Drosophila has one (dPer). Although the expression of dPer is rhythmic, dPer is not acutely responsive to light, as aremPer1 and mPer2 mRNAs. These data suggest that certain features of entrainment and oscillation may be unique to mammals.
A model explaining the mammalian circadian system will require understanding the circadian clock in the suprachiasmatic nucleus (SCN) at both the cellular and tissue levels. There is substantial evidence that SCN cells are autonomous oscillators. Individual dispersed SCN cells exhibit a circadian rhythm of electrical activity (Welsh et al., 1995; Liu et al., 1997; Herzog et al., 1998), rhythmically secrete vasopressin (VP; Murakami et al., 1991; Watanabe et al., 1993), and restore rhythmicity in locomotor activity to SCN-ablated hamsters (Ralph et al., 1990; Silver et al., 1990). These results demonstrate the cell-autonomous nature of the SCN oscillators but cannot account for entrainment. Mechanisms of entrainment in the mammalian SCN have been addressed only at the tissue level. Photic cues are the most important entraining signals, and it is well established that the greatest density of retinal fibers travels to an area of the SCN termed the “ventrolateral” or “core” region and are more sparse in the area called the “dorsomedial” or “shell” region (Leak and Moore, 1996; Miller et al., 1996; Moore, 1996). These reports indicate that SCN cells are neither functionally nor regionally homogeneous.
The present study explores how cell-autonomous oscillators are organized within SCN tissue to produce rhythmicity and photic entrainment. We examine the localization of light-induced and endogenously rhythmic expression of Per1, Per2, andPer3 mRNAs using calbindinD28K(CalB), FOS, and VP mRNA or protein as markers. The results show that the regulation of clock genes is not uniform among SCN cells. Expression of rhythmic and light-induced Per mRNA is regionally specific. Endogenous rhythmicity in Per1 andPer2 mRNA expression was primarily restricted to the VP region of the SCN, whereas light-induced Per1 andPer2 mRNA expression occurred primarily in the CalB region, where rhythmic Per1 and Per2 mRNA expression was not detectable. Interestingly, Per3 mRNA was strongly expressed in the CalB region but was not rhythmic in this area. In the VP region, Per3 mRNA expression showed low-amplitude rhythmicity. These data suggest that two compartments, one rhythmic and the other nonrhythmic, constitute the SCN pacemaker and form the basis of functional differences within the SCN.
Parts of this paper were presented previously at the seventh meeting of the Society for Research in Biological Rhythms, 2000 (Amelia Island, FL).
MATERIALS AND METHODS
Animals and housing. Adult male hamsters (Mesocricetus auratus) were given food and water ad libitum. The animal colony room was kept on a 12 hr light/dark cycle (LD), with light intensity of 600 lux. The testing rooms were equipped with a white noise generator (91 dB sound pressure level) to mask environmental noise. For animals housed in constant darkness (DD), a dim red light (<1 lux; Delta 1, Dallas, TX) allowed for maintenance.
For studies of zeitgeber time (ZT), animals were maintained in LD for at least 2 weeks before being killed. For studies of circadian time (CT), animals were housed in LD and then placed in DD for at least 1 week before being killed. In this case, hamsters transferred to DD were placed in cages equipped with running wheels (diameter, 16 cm), and locomotor activity was monitored continuously using a computer-based data acquisition system (Dataquest; Data Sciences, St Paul, MN).
All handling of animals was done in accordance with the Institutional Animal Care and Use Committee guidelines of Columbia University.
Free-floating digoxigenin in situhybridization. Hamster Per1, Per2, andPer3 cDNA fragments were PCR-amplified using the following oligonucleotides: 5′-CGAGATGTGTTTCGGGGTG-3′ and 5′-AGAGTGGTCAAAGGGCTGC-3′ for Per1, 5′-TGCCGTGTCAGCGTTGGAA-3′ and 5′-CGCTGGATGATGTCTGGCT-3′ forPer2, and 5′-GAAGAAGCCAAGCAGAGCC-3′ and 5′-GGGAGAGCAGACAACAGAG-3′ for Per3. Hamster CalB, VP, andBmal1 cDNA fragments were amplified by PCR using the following oligonucleotides: 5′-CTGGAAGGAAAGGAGCTG-3′ and 5′-GTATCCGTTGCCATCCTG-3′ for CalB, 5′-AGTGTCTCCCCTGCGGCCC-3′ and 5′-CAGCTGCGTGGCGTTGCTC-3′ for VP, and 5′-GCAACCGCAAGAGGAAAGG-3′ and 5′-AACAGGTGGAGGCGAAGTC-3′ for Bmal1. These PCR products were then cloned into the pGEM-T Easy vector (Promega, Madison, WI) and sequenced to verify their identity. Cloned partial hamsterPer1 and Per3 show 93 and 75% homology to mousePer1 and Per3, respectively. HamsterPer2 was 90% homologous to rat Per2. HamsterBmal1, CalB, and VP were 96, 94, and 90% homologous to ratBmal1, CalB, and VP, respectively. Antisense and sense cRNA probes (digoxigenin-labeled) were generated using the MEGAscriptin vitro transcription kit (Ambion, Austin, TX). For in situ hybridization using digoxigenin (DIG) cRNA probes in free-floating tissue, we examined 50–100 serial sections (20–30 μm) through the hypothalamic region, including the entire extent of the SCN, for each animal reported here. In situhybridization was performed as described previously (Hamada et al., 1999), except for the use of free-floating sections in the present study. Sections were photographed on Fuji 35 mm film, and color prints were made. For quantification of optical density, images of brain sections were captured using a CCD video camera (Sony XC77) attached to a light microscope (BH-2; Olympus Optical, Tokyo, Japan). mRNA expression was quantified by measuring stain density using the NIH Image program version 1.61.
Measurements of relative optical density (ROD), assessing the mean gray value per pixel of the measured area, were used to quantify the intensity of the signal. Optical density of staining for Per1, Per2, Per3, and Bmal1 mRNA were assessed in the CalB and VP regions, as defined in alternate sections stained for these peptides. To calculate the ROD, the background intensity of staining was subtracted from the intensity of staining in the SCN region of interest. Background OD for Per1, Per2, Per3, andBmal1 mRNA was measured in the lateral hypothalamic area. Background OD for CalB mRNA was measured in the VP area of the SCN. This was done because that CalB immunoreactivity occurs in the extra-SCN hypothalamic region, whereas the VP region of the SCN is devoid of CalB mRNA and protein (Fig. 1A,B; Silver et al., 1996).
Fig. 1.
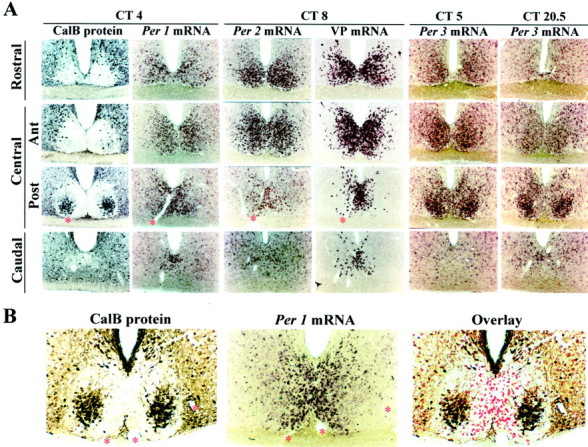
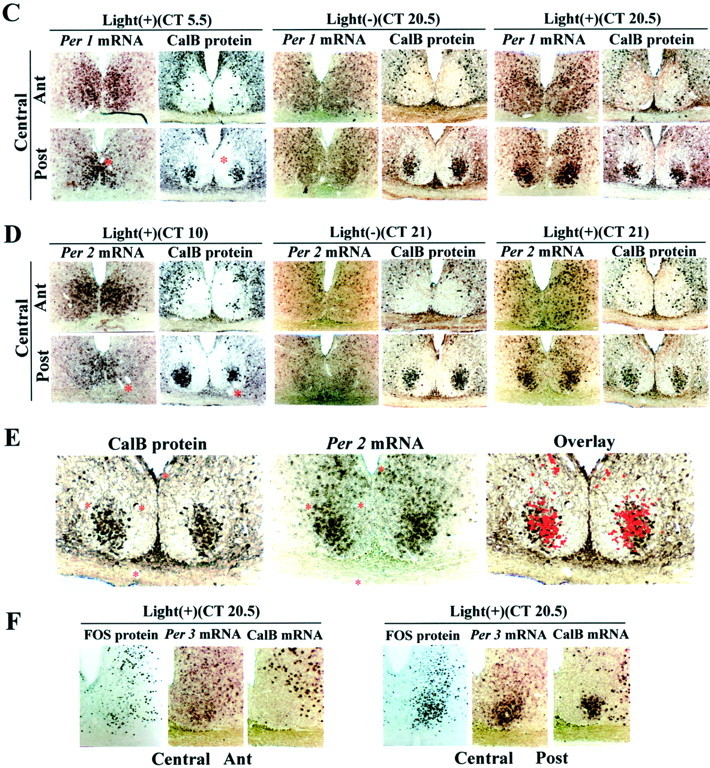
Photomicrographs depicting the localization of endogenously rhythmic (A, B) and light-induced (C–F) expression of Per1, Per2,and Per3 mRNA with respect to other known markers of the SCN, namely, CalB, VP, and FOS. Asterisks denote adjacent sections. A, The columns show coronal sections of each SCN quadrant from rostral to caudal from the same animal. At circadian times when Per1 andPer2 mRNA expression peaks (CT4, CT8, respectively), both signals are localized to the VP region of the SCN, and there is little expression (background level) in the CalB region of the central SCN, posterior aspect. This is highlighted in higher-power photomicrographs (B) in which the image of thePer1mRNA at ZT4 is captured in Adobe Photoshop, converted to a red signal, and superimposed in an overlay on the image of the adjacent section immunoreacted for CalB. The last two columns of A showPer3 mRNA at two circadian times, CT5 andCT20.5. Comparison of expression in each SCN quadrant at these times reveals weak Per3 rhythmic expression outside the CalB region. In contrast to Per1,Per2, and VP mRNA, Per3 mRNA is strongly expressed in the CalB region at both CT5 and CT20.5 but is not rhythmic. C, D, Comparison of responses in the presence and absence of light [Light(+) vsLight(−)] indicates that photic input induces Per1 (C) andPer2 (D) mRNA expression in the CalB region. The light-induced response can be seen during subjective night (CT20.5, Per1; CT21,Per2) but not during subjective day (CT5.5, Per1; CT10,Per2). Note that light-induced Per1 andPer2 mRNA and rhythmic Per1 andPer2 mRNA are expressed in different SCN compartments.E, Data shown in C and Dhighlighted in a high-power photomicrograph and overlay in which the image of the Per2 mRNA is captured in Adobe Photoshop, converted to a red signal, and superimposed in an overlay on the image of the adjacent section immunoreacted for CalB protein. The time of killing is given at the top of eachcolumn. Light(+), Presentation of a light pulse (600 lux for 30 min);light(−), no light pulse. Eachcolumn shows sections from two central SCN quadrants [anterior (Ant) and posterior (Post)] of the same animal. F,Photomicrographs showing the localization of light-induced FOS,Per3, and CalB mRNA in the central anterior and posterior aspects of the SCN. It is clear that light induces FOS in the CalB region of the SCN. Comparison of Per3 atCT5 in A and at CT20.5 in F indicates that light does not affect the expression of eitherPer3 or CalB mRNA.
Immunohistochemistry. Hamsters were heavily anesthetized (pentobarbital, 200 mg/kg), and perfused intracardially with 150 ml of 0.9% saline followed by 300–400 ml of 4% paraformaldehyde in 0.1m phosphate buffer, pH 7.3. Brains were post-fixed for 18–24 hr at 4°C and cryoprotected in 20% sucrose in 0.1m phosphate buffer overnight. For immunocytochemistry, sections (20–30 μm) were processed using the avidin–biotin–immunoperoxidase method (Silver et al., 1996; LeSauter et al., 1999a). The primary antibodies used were rabbit polyclonal FOS (1:5000; Calbiochem, Cambridge, MA) and mouse monoclonal CalB (1:20,000; Sigma, St. Louis, MO.).
Delineation of distinct SCN regions. We used immunoreactivity or in situ hybridization for CalB, VP, and light-induced FOS to demarcate distinct regions of the SCN. To examine the localization of Per1, Per2, Per3, CalB, and VP mRNA, alternate free-floating sections were processed for DIG in situ hybridization and immunochemistry.
Various terms have been used previously to delineate distinct SCN regions. We use the descriptive terms “CalB region” and “VP region” to demarcate nonoverlapping regions of the hamster SCN (Fig.1A), with explicit recognition that SCN regions delineated by these markers contain other cell types. We concur with the characterization of the rodent SCN as heterogeneous on the basis of the peptidergic content of its neuronal populations and their projections. Further analysis will be required to determine how the SCN regions characterized by their clock gene expression in the present study correspond to those previously defined as ventrolateral and dorsomedial or core and shell in hamster, mouse, and rat (Miller et al., 1996; Moore, 1996; Moore and Silver, 1998; Leak et al., 1999).
RESULTS
Rhythmic expression of Per1 and Per2 mRNA occurs outside the calbindinD28K region
By examining alternate sections by in situhybridization and immunochemistry, we characterized the regional distribution of Per1, Per2, and VP mRNA and CalB protein in coronal sections from the rostral through the central (anterior and posterior portions) and caudal quadrants of the SCN. Peak expression ofPer1 mRNA in whole SCN occurs around CT4 and ZT4, whereasPer2 mRNA in whole SCN peaks at CT8–10 and ZT10–12 in the hamster (Maywood et al., 1999; Horikawa et al., 2000, Moriya et al., 2000; Yokota et al. 2000). Figure1A shows photomicrographs of brain sections harvested at CT4 and stained for CalB taken through the extent of the SCN. As previously reported (Silver et al., 1996), CalB is highly restricted to the central posterior SCN. Photomicrographs of Per1 mRNA at CT4 and CalB protein in adjacent sections indicate that Per1 mRNA is expressed throughout most of the SCN but not in the CalB-positive region. This point is highlighted in an enlarged view showing an overlay of Per1 mRNA and CalB taken from adjacent sections at ZT4 (Fig. 1B). There is little or no detectable overlap in the distribution of cells expressing Per mRNA and CalB protein (Fig. 1B). Photomicrographs from animals killed at ZT4 indicate that the expression and distribution ofPer1 mRNA is similar under both LD and DD. The localization of Per2 mRNA does not differ from that of Per1mRNA under LD (data not shown) and DD (Fig. 1A), although the expression level of Per2 mRNA is much higher.
Expression of VP mRNA was examined at CT8, when it peaks (Jin et al., 1999). Some VP mRNA can be seen in each quadrant of the SCN, but its distribution does not overlap with that of CalB protein (Fig.1A). Comparison of mRNA expression forPer1 and Per2 in sections adjacent to those showing VP mRNA at two circadian times indicates that rhythmicPer1 (Fig. 1, A vs C) andPer2 (Fig. 1, A vs D) mRNAs are expressed in the VP region.
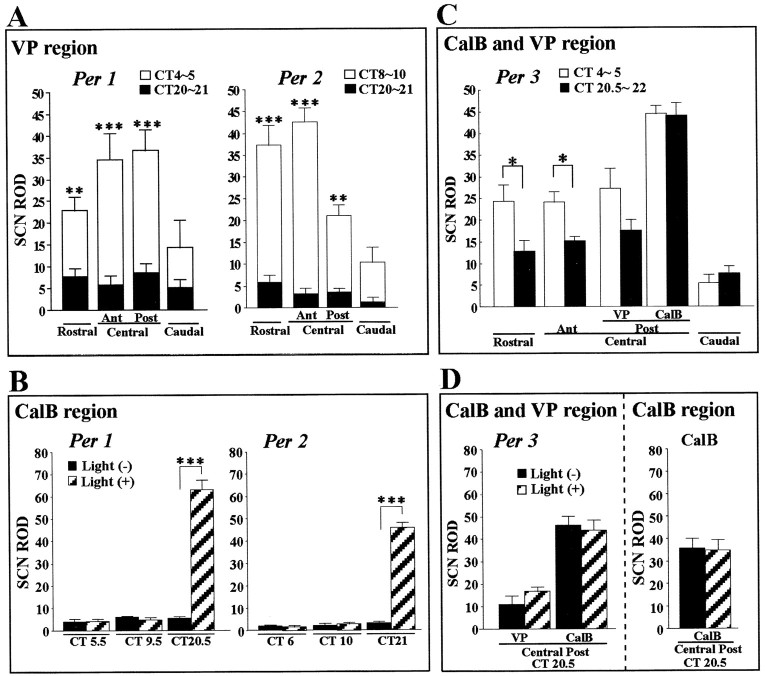
Quantitative analysis (Fig.2A) of the ROD in each quadrant of the SCN outside the CalB area shows marked differences inPer1 and Per2 mRNA expression during subjective day and night (Fig. 2A). In contrast, within the CalB area, expression of Per1 and Per2 mRNA is at low, background levels evident at all circadian times (Fig.2B).
Fig. 2.
Quantification of the expression ofPer genes in distinct SCN regions for each quadrant in the presence (+) and absence (−) of light. Values shown are mean ± SEM; n = 3∼4 hamsters per time point. *p < 0.05; **p < 0.01; ***p < 0.001, Student's t test).A, In the VP area, ROD measurements ofPer1 and Per2 mRNA at times of peak and trough expression reveal rhythmicity in each SCN quadrant.B, In the CalB region, comparison of circadian day and night for Per1 and Per2 mRNA in the absence of light indicates no detectable change in either mRNA. On the other hand, marked light-induced Per1 andPer2 mRNA occurs in the CalB region of the SCN.C, ROD measurements for Per3 mRNA in each SCN region indicate differences between subjective day and night in the VP region of the rostral and central SCN. Note that Per3mRNA is strongly expressed but is not rhythmic in the CalB region of the central posterior SCN in both subjective day and night.D, Expression of Per3 and CalB mRNA is not affected by a light pulse given during the subjective night.Ant, Anterior; Post, posterior.
Light-induced and rhythmic Per1 andPer2 mRNA are expressed in different SCN compartments
The hamster SCN receives photic input primarily in the CalB region. This is based on both tract-tracing (Moore and Silver, 1998) electron microscopic studies (Bryant et al., 2000) and FOS induction after a light pulse (Hastings et al., 1996; Silver et al., 1996). The present results show that rhythmic Per1 and Per2mRNAs are not detectable in this region (Fig. 1A,B). In the mouse, however, Per1 and Per2 are reported to have an important role in light-induced phase shifts (Shigeyoshi et al., 1997; Akiyama et al., 1999; Wakamatsu et al., 2001), andPer1 is induced by light in the ventromedial SCN (Shigeyoshi et al., 1997). We therefore examined the regional distribution of light-induced Per1 andPer2 mRNAs. Hamsters were exposed to a 30 min light pulse at suitable circadian times and killed 1.5–2 hr later. Control animals were not exposed to light but were otherwise treated identically. Photomicrographs through the two central quadrants of the SCN reveal that a light pulse during the subjective day did not inducePer1 or Per2 mRNA expression (Fig.1C,D). Control animals not exposed to a light pulse are shown in Figure 1A. In contrast, during late subjective night, there was strong light induction of Per1and Per2 mRNA, highly concentrated in the CalB region. Further evidence of the overlap in their distribution is shown in adjacent sections of Per2 mRNA and CalB protein at high power (Fig. 1E). This is different from the area that expresses endogenously rhythmic Per1 and Per2mRNAs. Quantification of the results shows significant light-inducedPer1 and Per2 mRNA during subjective night but not during subjective day in the CalB region (Fig.2B).
Per3 mRNA expression is high but is neither rhythmic nor affected by a light pulse in the calbindinD28Kregion
The pattern of Per3 mRNA expression differs substantially from that of Per1, Per2, and VP mRNA (Figs.1A, 2C,D). Endogenous Per3 mRNA is expressed in both the VP and CalB regions (Fig.1A). There is circadian expression of Per3mRNA in the rostral and central VP region of the SCN (CT4–5 vs CT20.5–22). The amplitude of the Per3 mRNA rhythm is less than that of Per1 and Per2 mRNA in this area. In contrast, there is no detectable circadian expression ofPer3 mRNA within the CalB region, where rhythmic Per1, Per2, and VP mRNA expression is also not detectable (Fig.1A,B). Per3 mRNA expression in the CalB region is constant and high (Fig. 2C). Notably, this is the region where light induces Per1 and Per2 mRNA (Fig. 1C–E). Per3 mRNA, however, is not detectably induced by light during subjective night in any part of the central SCN (Figs. 1F, 2D). Quantification confirms the weak but significant (p < 0.05, Student's t test) rhythmic expression of Per3 mRNA in the VP region (Fig.2D).
Bmal1 mRNA expression occurs outside the calbindinD28K region
To further define the regulatory mechanisms that underlie the differential regulation of the Per genes in the two SCN regions, we used the expression of the clock gene Bmal1 as a marker to study its relation to light-induced FOS protein expression with the following rationale: Most light-induced expression of FOS is concentrated in the CalB area rather than in the VP region of the SCN (Fig. 1F; Hastings et al., 1996; Silver et al., 1996). BMAL1 and CLOCK are important regulators of rhythmic VP andPer1 gene expression in the SCN (Jin et al., 1999). To determine whether there is differential expression of the regulatory gene Bmal1 in distinct SCN regions, we compared Bmal1, Per3, and CalB mRNA expression in each SCN quadrant. Results indicate that at its peak, the Bmal1 mRNA signal is strong in the VP region in the rostral and central anterior SCN, but it is very weakly expressed in the central posterior CalB region (Fig.3). In contrast, Per3 mRNA is very highly expressed in the CalB region at CT16. Light-induced FOS also occurs here (Fig. 1F).
Fig. 3.

Expression of Bmal1 andPer3 mRNA occur in different regions. Photomicrographs show the localization of Bmal1 mRNA with respect toPer3 and CalB mRNA in each SCN quadrant of the SCN. (ForCT16, each row shows serial sections from the same animal.) Expression of Bmal1 mRNA is stronger outside the CalB region of the SCN, whereas Per3 mRNA expression is high within this region. Comparison between CT4 and CT16 shows thatBmal1 mRNA is expressed rhythmically outside the CalB region. Ant, Anterior; Post, posterior.
DISCUSSION
The identification and analysis of clock genes have been very successful in demonstrating how a cellular oscillator can be built both mechanistically (Young, 1998; Dunlap, 1999; King and Takahashi, 2000) and formally (Goldbeter, 1995). In mammals, the mechanism of cellular oscillation is likely to be similar to the Drosophila model, although some components of the oscillator are different (Shearman et al., 2000b). The Drosophila model, however, does not speak to the mechanism of photic entrainment in mammals. The present analysis uses the information derived from molecular events to understand SCN organization at the tissue level. The results indicate that understanding mechanisms of entrainment in mammals will require knowledge of SCN circuitry. The present studies of cellular organization of the SCN reveal novel aspects of the mammalian circadian clock organization that set the parameters for further studies of mechanisms at the level of the cell, the tissue, and the organism as a whole.
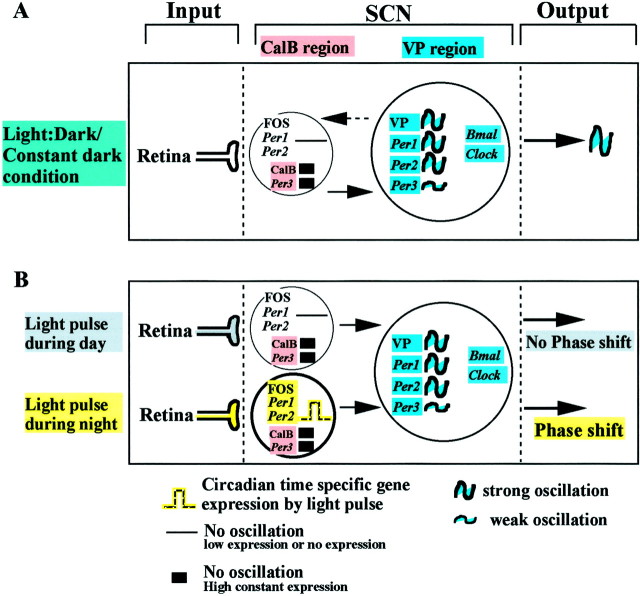
First, cells in one subregion of the hamster SCN do not oscillate with respect to Per production. The relationship between nonoscillating and oscillating cells is shown schematically in Figure4. The commonly used tripartite organization of the SCN, with an input, clock, and output, is retained. The key feature is the segregation of the SCN clock into two distinct elements, shown as a small nonrhythmic region and a larger rhythmic region expressing the clock genes. It is noteworthy that cells in the nonrhythmic CalB region are easily missed, because they form a small proportion of the SCN population. In the rat, each SCN has ∼8000–10,000 cells (van den Pol, 1980); assuming that the hamster SCN has a comparable number of cells, the CalB subregion constitutes no more than 10% of the nucleus (Silver et al., 1996). In the hamster the CalB region is restricted to the posterior aspect of the central region of the nucleus (Fig. 1). In other species, cells with comparable properties may be more dispersed among other SCN neurons and difficult to localize.
Fig. 4.
Model of rhythmicity and photic entrainment at the tissue level, comprising two functionally different SCN compartments. As is widely accepted, the circadian system has three primary components: input, SCN, and output. A,Model of rhythmicity. In both Light:darkand Constant dark conditions, there are two distinct regions of the SCN showing the same pattern of Per gene expression. In the hamster SCN, these regions can readily be delineated by cells containing CalB and VP respectively, as indicated in Materials and Methods. These two compartments constitute the SCN pacemaker. In the CalB region, rhythmic expression of Per1 andPer2 mRNA are not detectable (SCN,left). Another characteristic of this area is that CalB and Per3 mRNA are highly expressed but not rhythmic. Endogenous rhythmic expression of clock genes (Per1, Per2, Per3, and Bmal1 mRNA) and VP mRNA occurs in a large number of SCN cells restricted to the VP region (SCN, right). B. Model of photic entrainment. Gated light induced Per1 and Per2 mRNA and FOS expression. A light pulse during the day (top) has no effect on the expression of Per1 and Per2mRNA or on FOS. A light pulse during the night (bottom) induces Per1 and Per2 mRNA and FOS expression in the CalB region. Importantly, some genes in this region are not induced by light pulse during the night (Per3, CalB). See Discussion for the proposed mechanism of entrainment.Ant, Anterior; Post, posterior.
As shown in the model, clock gene oscillation is a property of some but not all SCN cells. One SCN region, marked by VP mRNA in this study, shows rhythmic expression of Per1, Per2, and Per3mRNA. It has been reported that spontaneous expression of Jun-B and FOS occur in the dorsomedial region of hamster SCN (Guido et al., 1999). The model also shows that the other SCN region, marked by CalB cells, has time-gated light-induced Per1 and Per2 mRNA and FOS protein and constant high expression of Per3 mRNA. The key concept emerging from the present results is this separation of the nonrhythmic and rhythmic compartments of the SCN expressing clock genes, along with the demonstration that responses of nonrhythmic cells are themselves gated by the circadian system (Fig.4A,B). Photic information from the retina reaches the CalB region of the SCN directly (Bryant et al., 2000). This SCN compartment is not rhythmic in Per mRNA (Fig. 2), CalB mRNA (data not shown), or CalB protein expression (LeSauter et al., 1999a). At appropriate times of day, in the presence of a light pulse, photic input induces Per1 and Per2 mRNA (Fig.4B). These results are consistent with many reports in hamsters of light-induced responses concentrated in the CalB region, including induction of FOS (Silver et al., 1996; Fig. 3D),vgf (Wisor and Takahashi, 1997), and egr-3(Morris et al., 1998).
An important question is how (and whether) phase-setting information, reflected in high expression of light-induced Per1 andPer2, travels from the CalB compartment to other SCN regions. One possibility is that the VP region itself receives direct retinal input. Tract-tracing studies, which label retinofugal pathways, indicate that retinal fibers occupy a large region of the SCN, although they are most dense in the CalB (i.e., ventrolateral or core) region (Pickard and Silverman 1981; Johnson et al.; 1988; Youngstrom et al., 1991; Miller et al., 1996; Silver et al., 1996). Synaptic connections made by retinal fibers in the hamster have been documented only for CalB (Bryant et al., 2000) and gastrin-related peptide, which is located in the CalB region (Aioun et al., 1998). It seems unlikely that photic input directly resets rhythmic Per1 andPer2 expression in the VP region, given the sparse expression of these mRNAs after a light pulse (Fig. 1C,D). The relatively sparse induction of FOS in this region after photic input also argues against this possibility (Kornhauser et al., 1990;Rea, 1992; but see Guido et al., 1999). The alternative possibility is that Per1 and Per2 mRNA expression in the VP region are produced indirectly, via information relayed from the CalB region. It has been shown in anatomical studies of rats that information travels selectively within the SCN, from the retinorecipient to the nonretinorecipient regions (Moore 1996; Leak et al., 1999), even though available evidence on intra-SCN circuitry suggests that SCN neurons make hundreds of intranuclear synapses and appositions in rats and hamsters (Guldner 1976; van den Pol and Gorcs, 1986; Daikoku et al., 1992; Romijn et al., 1997; Jacomy et al., 1999; LeSauter et al., 1999b). In support of this hypothesis, in SCN cell culture, vasoactive intestinal polypeptide (VIP) induces phase shifts of VP secretion in a light-like phase shift (Watanabe et al., 2000). Importantly, gastrin-related peptide (Moore and Silver, 1998) strongly induces a light pulse-type phase shift in hamster locomotor activity (Piggins et al., 1995).
The absence of rhythmic expression of clock genes in the CalB region begs the question of why lesions of the CalB region, which spare other compartments of the SCN, result in loss of circadian rhythms in hamsters (LeSauter and Silver, 1999). One possibility is that there is a population of pacemaker cells, yet to be identified, lying near but not in the CalB region, with rhythmic Per expression. An alternative possibility is that in the absence of the cell group with gated expression of clock genes, oscillators in extra-SCN brain areas drift out of synchrony. One puzzling feature of SCN organization is the fact that efferents from the VP region and CalB regions both reach common target areas in the hamster brain (LeSauter et al., 1999b). Although the function of these overlapping pathways remains to be examined directly, such results indicate how phase information might travel.
Another feature suggested by our data, that the regulation of clock genes is not uniform among SCN cells, is counter to current models of circadian regulation of Per genes (Dunlap 1999; Shearman et al., 2000b). In rat and mouse, the Per1 promoter contains both an E-box (CACGTG) and a cAMP response element (CRE; Hida et al., 2000; Yamaguchi et al., 2000). VP also has an E-box and CRE sequence in its promoter, and circadian expression of VP is thought to be controlled by BMAL1 and CLOCK protein binding to the E-box (Jin et al., 1999). Bmal1 has an important role in activating E-box-dependent rhythmic Per1 and VP mRNA expression. Although the underlying mechanism is not yet known, rhythmicity in FOS and in CRE-mediated gene expression (using CRE-β-galactosidase transgenic mice) has also been reported (Obrietan et al., 1999;Schwartz et al., 2000). The present studies show that the VP area is rich in Bmal1 mRNA (Fig. 3), suggesting that the circadian expression of Per1, Per2, Per3, and VP mRNA in this region occurs through the activation of E-boxes. In contrast, in the CalB region, where the expression of BMAL1 is low,Per1 and Per2 mRNA and FOS protein are strongly light-induced. This suggests that Per1 and Per2mRNA regulation in this region may be achieved through a light response element such as the CRE. In further support of the notion that light induction involves the CRE, expression of phosphorylated CRE binding protein, a positive regulator of the c-fos gene, also occurs in the region where light induces FOS protein (Schurov et al., 1999). Last, Per3 mRNA is strongly expressed in the CalB region but is not induced by light, suggesting that Per3 regulation in this region is achieved by a distinct transcriptional system from that of Per1 and Per2. These results suggest that cells expressing rhythmic Per1, Per2, Clock, andBmal1 mRNA in the VP region of the SCN are candidates for principal oscillator cells.
Many previous reports indicate that the SCN may have functionally distinct regions. In the rat and hamster, a substantial population of VP-containing cells lies in the dorsomedial SCN, whereas VIP is located mainly in the ventrolateral SCN (Card and Moore, 1984; Inouye and Shibata, 1994; Miller et al., 1996; Moore, 1996; Moore and Silver, 1998). The ventrolateral region of the rat SCN has a dense region of CalB expression (Arvanitogiannis et al., 2000), although these are not as densely packed as in the hamster. The VP content of the SCN shows circadian rhythmicity in both LD and DD and is not affected by a light pulse (Inouye and Shibata, 1994). In contrast, the VIP content of the SCN has diurnal variation in LD but not in DD in vivo. Light-induced fos family genes are concentrated in the ventrolateral region, whereas circadian rhythms of fos occur primarily in the dorsomedial subdivision of the rat and hamster SCN (Chambille et al., 1993; Guido et al., 1999; Schwartz et al., 2000). In support of the notion that functional retinal input is highly localized to part of the SCN, electrical stimulation of the optic nerve evokes fast positive and late large negative waves in the ventrolateral but not the dorsolateral SCN in a horizontal slice preparation of the rat SCN (Shibata et al., 1984). These reports suggest that the SCN has two functionally distinct regions, wherein one receives light information and the other does not. Importantly, daily rhythms driven by the LD cycle, such as SCN VIP content, do not involve an E-box-dependent negative feedback loop. This is consistent with the present results showing that there is no detectable rhythm in expression ofPer1 and Per2 mRNA in the CalB region. Furthermore, in this region, Per1 and Per2 mRNA expression are the same in both LD and DD (Figs.1A,B, 4A). In support of this idea, individual SCN neurons in homozygous Clock mutant mice are arrhythmic in electrical activity, paralleling the effects on locomotor activity in these animals (Herzog et al., 1998) and suggesting a role for E-box-dependent mechanisms in SCN pacemaker function. The occurrence of regional differences in E-box-dependent negative feedback loops of clock genes and their products, shown in the present study, seems to play an important role in regional differences within the SCN.
Our results also explain some of the behavioral phenotypes of various circadian mutants. Per2 mutant mice, characterized by a deletion mutation in the PAS domain of the Per2 gene, show arrhythmic responses in constant darkness (Zheng et al., 1999). This is consistent with our results showing that rhythmicity in Per2occurs in SCN pacemakers (located in the VP region). Our results suggest that regulation of light-induced clock genes is distinct from that of endogenously occurring clock gene expression in the VP region. In this context, it is interesting that Clock mutant mice (Vitaterna et al., 1994) and Mop3 (also known asBmal1) knock-out mice (Bunger et al., 2000) entrain but show disrupted circadian rhythms of locomotor activity in DD. As might be predicted from the present results, Clock mutant mice express light-induced Per1 and Per2 mRNA (Shearman and Weaver, 1999). Bmal1 and Clock are both important for the regulation of rhythmicity (King and Takahashi, 2000). On the other hand, very low expression of CLOCK protein (van Esseveldt et al., 2000) and Bmal1 mRNA (Fig. 4) in the CalB region bring into question the role of these genes in light-inducedPer1 and Per2 mRNA expression. Finally, mice with targeted disruption of the mPer3 gene had normal activity rhythms, with a slightly shorter free-running period than wild-type animals (Shearman et al., 2000a), consistent with our finding of lowPer3 mRNA expression in the rhythmic compartment.
Footnotes
This research was supported by National Institutes of Health Grant NS37919 (R.S.) and a grant from the Japan Society for the Promotion of Science (T.H.). We thank Drs. Paul Hardin, Lance Kriegsfeld, and William J. Schwartz for comments on drafts of this manuscript and Honor Kirwan for technical assistance.
Correspondence should be addressed to Dr. Rae Silver, Columbia University, Mail Code 5501, 1190 Amsterdam Avenue, Room 406, Schermerhorn Hall, New York, NY 10027. Email: QR@columbia.edu.
Dr. Venuti's present address: Department of Cell Biology and Anatomy, Louisiana State University Medical Center, 1901 Perdido Street, New Orleans, LA 70112.
REFERENCES
- 1.Aioun J, Chambille I, Peytevin J, Martinet L. Neurons containing gastrin-releasing peptide and vasoactive intestinal polypeptide are involved in the reception of the photic signal in the suprachiasmatic nucleus of the Syrian hamster: an immunocytochemical ultrastructural study. Cell Tissue Res. 1998;291:239–253. doi: 10.1007/s004410050994. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, Maetani M, Watanabe S, Tei H, Sakaki Y, Shibata S. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–11121. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvanitogiannis A, Robinson B, Beaule C, Amir S. Calbindin-D28k immunoreactivity in the suprachiasmatic nucleus and the circadian response to constant light in the rat. Neuroscience. 2000;99:397–401. doi: 10.1016/s0306-4522(00)00327-4. [DOI] [PubMed] [Google Scholar]
- 4.Bryant DN, LeSauter J, Silver R, Romero MT. Retinal innervation of calbindin-D28K cells in the hamster suprachiasmatic nucleus: ultrastructural characterization. J Biol Rhythms. 2000;15:103–111. doi: 10.1177/074873040001500204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Card JP, Moore RY. The suprachiasmatic nucleus of the golden hamster: immunohistochemical analysis of cell and fiber distribution. Neuroscience. 1984;13:415–431. doi: 10.1016/0306-4522(84)90240-9. [DOI] [PubMed] [Google Scholar]
- 7.Chambille I, Doyle S, Serviere J. Photic induction and circadian expression of Fos-like protein. Immunohistochemical study in the retina and suprachiasmatic nuclei of hamster. Brain Res. 1993;612:138–150. doi: 10.1016/0006-8993(93)91654-b. [DOI] [PubMed] [Google Scholar]
- 8.Daikoku S, Hisano S, Kagotani Y. Neuronal associations in the rat suprachiasmatic nucleus demonstrated by immunoelectron microscopy. J Comp Neurol. 1992;325:559–571. doi: 10.1002/cne.903250408. [DOI] [PubMed] [Google Scholar]
- 9.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 10.Guido ME, Goguen D, De Guido L, Robertson HA, Rusak B. Circadian and photic regulation of immediate-early gene expression in the hamster suprachiasmatic nucleus. Neuroscience. 1999;90:555–571. doi: 10.1016/s0306-4522(98)00467-9. [DOI] [PubMed] [Google Scholar]
- 11.Goldbeter A. A model for circadian oscillations in the Drosophila period protein (PER). Proc R Soc Lond B Biol Sci. 1995;261:319–324. doi: 10.1098/rspb.1995.0153. [DOI] [PubMed] [Google Scholar]
- 12.Guldner F-H. Synaptology of the rat suprachiasmatic nucleus. Cell Tissue Res. 1976;165:509–544. doi: 10.1007/BF00224478. [DOI] [PubMed] [Google Scholar]
- 13.Hamada T, Ootomi M, Horikawa K, Niki T, Wakamatu H, Ishida N. The expression of the melatonin synthesis enzyme: arylalkylamine N-acetyltransferase in the suprachiasmatic nucleus of rat brain. Biochem Biophys Res Commun. 1999;258:772–777. doi: 10.1006/bbrc.1999.0668. [DOI] [PubMed] [Google Scholar]
- 14.Hastings MH, Best JD, Ebling FJ, Maywood ES, McNulty S, Schurov I, Selvage D, Sloper P, Smith KL. Entrainment of the circadian clock. Prog Brain Res. 1996;111:147–174. doi: 10.1016/s0079-6123(08)60406-9. [DOI] [PubMed] [Google Scholar]
- 15.Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 16.Hida A, Koike N, Hirose M, Hattori M, Sakaki Y, Tei H. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 17.Horikawa K, Yokota S, Fuji K, Akiyama M, Moriya T, Okamura H, Shibata S. Nonphotic entrainment by 5-HT1A/7 receptor agonists accompanied by reduced Per1 and Per2 mRNA levels in the suprachiasmatic nuclei. J Neurosci. 2000;20:5867–5873. doi: 10.1523/JNEUROSCI.20-15-05867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inouye ST, Shibata S. Neurochemical organization of circadian rhythm in the suprachiasmatic nucleus. Neurosci Res. 1994;20:109–130. doi: 10.1016/0168-0102(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 19.Jacomy H, Burlet A, Bosler O. Vasoactive intestinal peptide neurons as synaptic targets for vasopressin neurons in the suprachiasmatic nucleus. Double-label immunocytochemical demonstration in the rat. Neuroscience. 1999;88:859–870. doi: 10.1016/s0306-4522(98)00259-0. [DOI] [PubMed] [Google Scholar]
- 20.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RF, Morin LP, Moore RY. Retinohypothalamic projections in the hamster and rat demonstrated using cholera toxin. Brain Res. 1988;462:301–312. doi: 10.1016/0006-8993(88)90558-6. [DOI] [PubMed] [Google Scholar]
- 22.King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–742. doi: 10.1146/annurev.neuro.23.1.713. [DOI] [PubMed] [Google Scholar]
- 23.Kornhauser JM, Nelson DE, Mayo KE, Takahashi JS. Photic and circadian regulation of c-fos gene expression in the hamster suprachiasmatic nucleus. Neuron. 1990;5:127–134. doi: 10.1016/0896-6273(90)90303-w. [DOI] [PubMed] [Google Scholar]
- 24.Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- 25.Leak RK, Card JP, Moore RY. Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res. 1999;819:23–32. doi: 10.1016/s0006-8993(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 26.LeSauter J, Silver R. Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J Neurosci. 1999;19:5574–5585. doi: 10.1523/JNEUROSCI.19-13-05574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeSauter J, Stevens P, Jansen H, Lehman MN, Silver R. Calbindin expression in the hamster SCN is influenced by circadian genotype and by photic conditions. NeuroReport. 1999a;10:3159–3163. doi: 10.1097/00001756-199910190-00007. [DOI] [PubMed] [Google Scholar]
- 28.LeSauter J, Leak RK, Silver R, Moore RY. Hamster suprachiasmatic nucleus: chemoarchitecture and topography of projections. Soc Neurosci Abstr. 1999b;29:552.2. [Google Scholar]
- 29.Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 30.Maywood ES, Mrosovsky N, Field MD, Hastings MH. Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc Natl Acad Sci USA. 1999;96:15211–15216. doi: 10.1073/pnas.96.26.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JD, Morin LP, Schwartz WJ, Moore RY. New insights into the mammalian circadian clock. Sleep. 1996;8:641–667. doi: 10.1093/sleep/19.8.641. [DOI] [PubMed] [Google Scholar]
- 32.Moore RY. Entrainment pathways and the functional organization of the circadian system. Prog Brain Res. 1996;111:103–119. doi: 10.1016/s0079-6123(08)60403-3. [DOI] [PubMed] [Google Scholar]
- 33.Moore RY, Silver R. Suprachiasmatic nucleus organization. Chronobiol Int. 1998;15:475–487. doi: 10.3109/07420529808998703. [DOI] [PubMed] [Google Scholar]
- 34.Moriya T, Horikawa K, Akiyama M, Shibata S. Correlative association between N-methyl-d-aspartate receptor-mediated expression of period genes in the suprachiasmatic nucleus and phase shifts in behavior with photic entrainment of clock in hamsters. Mol Pharmacol. 2000;58:1554–1562. doi: 10.1124/mol.58.6.1554. [DOI] [PubMed] [Google Scholar]
- 35.Morris ME, Viswanathan N, Kuhlman S, Davis FC, Weitz CJ. A screen for genes induced in the suprachiasmatic nucleus by light. Science. 1998;279:1544–1547. doi: 10.1126/science.279.5356.1544. [DOI] [PubMed] [Google Scholar]
- 36.Murakami N, Takamure M, Takahashi K, Utunomiya K, Kuroda H, Etoh T. Long-term cultured neurons from rat suprachiasmatic nucleus retain the capacity for circadian oscillation of vasopressin release. Brain Res. 1991;545:347–350. doi: 10.1016/0006-8993(91)91312-o. [DOI] [PubMed] [Google Scholar]
- 37.Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 38.Pickard GE, Silverman AJ. Direct retinal projections to the hypothalamus, piriform cortex, and accessory optic nuclei in the golden hamster as demonstrated by a sensitive anterograde horseradish peroxidase technique. J Comp Neurol. 1981;196:155–172. doi: 10.1002/cne.901960111. [DOI] [PubMed] [Google Scholar]
- 39.Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 41.Rea MA. Different populations of cells in the suprachiasmatic nuclei express c-fos in association with light-induced phase delays and advances of the free-running activity rhythm in hamsters. Brain Res. 1992;579:107–112. doi: 10.1016/0006-8993(92)90748-x. [DOI] [PubMed] [Google Scholar]
- 42.Romijn HJ, Sluiter AA, Pool CW, Wortel J, Buijs RM. Evidence from confocal fluorescence microscopy for a dense, reciprocal innervation between AVP-, somatostatin-, VIP/peptide histidine isoleucine-, GRP-, and peptide histidine isoleucine/GRP-immunoreactive neurons in the rat suprachiasmatic nucleus. Eur J Neurosci. 1997;9:2613–2623. doi: 10.1111/j.1460-9568.1997.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 43.Schurov IL, McNulty S, Best HD, Sloper PJ, Hastings MH. Glutamatergic induction of CREB phosphorylation and Fos expression in primary cultures of the suprachiasmatic hypothalamus in vitro is mediated by co-ordinate activity of NMDA and non-NMDA receptors. J Neuroendocrinol. 1999;11:43–51. doi: 10.1046/j.1365-2826.1999.00289.x. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz WJ, Carpino A, de la Iglesia HO, Baler R, Klein DC, Nakabeppu Y, Aronin N. Differential regulation of fos family genes in the ventrolateral and dorsomedial subdivisions of the rat suprachiasmatic nucleus. Neuroscience. 2000;98:535–547. doi: 10.1016/s0306-4522(00)00140-8. [DOI] [PubMed] [Google Scholar]
- 45.Shearman LP, Weaver DR. Photic induction of Period gene expression is reduced in Clock mutant mice. NeuroReport. 1999;10:613–618. doi: 10.1097/00001756-199902250-00031. [DOI] [PubMed] [Google Scholar]
- 46.Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol. 2000a;20:6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000b;288:1013–1029. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 48.Shibata S, Oomura Y, Kita H, Liou SY, Ueki S. Field potentials in the suprachiasmatic nucleus of rat hypothalamic slice produced by optic nerve stimulation. Brain Res Bull. 1984;12:377–379. doi: 10.1016/0361-9230(84)90108-4. [DOI] [PubMed] [Google Scholar]
- 49.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 50.Silver R, Lehman MN, Gibson M, Gladstone WR, Bittman E. Dispersed cell suspensions of fetal SCN restore circadian rhythmicity in SCN-lesioned adult hamsters. Brain Res. 1990;525:45–58. doi: 10.1016/0006-8993(90)91319-c. [DOI] [PubMed] [Google Scholar]
- 51.Silver R, Romero MT, Besmer HR, Leak R, Nunez JM, LeSauter J. Calbindin-D28K cells in the hamster SCN express light-induced Fos. NeuroReport. 1996;7:1224–1228. doi: 10.1097/00001756-199604260-00026. [DOI] [PubMed] [Google Scholar]
- 52.van den Pol AN. The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J Comp Neurol. 1980;191:661–702. doi: 10.1002/cne.901910410. [DOI] [PubMed] [Google Scholar]
- 53.van den Pol AN, Gorcs T. Synaptic relationships between neurons containing vasopressin, gastrin-releasing peptide, vasoactive intestinal polypeptide, and glutamate decarboxylase immunoreactivity in the suprachiasmatic nucleus: dual ultrastructural immunochemistry with gold-substituted silver peroxidase. J Comp Neurol. 1986;252:507–521. doi: 10.1002/cne.902520407. [DOI] [PubMed] [Google Scholar]
- 54.van Esseveldt KE, Lehman MN, Boer GJ. The suprachiasmatic nucleus and the circadian time-keeping system revisited. Brain Res Rev. 2000;33:34–77. doi: 10.1016/s0165-0173(00)00025-4. [DOI] [PubMed] [Google Scholar]
- 55.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wakamatsu H, Takahashi S, Moriya T, Inouye ST, Okamura H, Akiyama M, Shibata S. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. NeuroReport. 2001;12:127–131. doi: 10.1097/00001756-200101220-00033. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe K, Koibuchi N, Ohtake H, Yamaoka S. Circadian rhythms of vasopressin release in primary cultures of rat suprachiasmatic nucleus. Brain Res. 1993;624:115–120. doi: 10.1016/0006-8993(93)90067-w. [DOI] [PubMed] [Google Scholar]
- 58.Watanabe K, Vanecek J, Yamaoka S. In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res. 2000;877:361–366. doi: 10.1016/s0006-8993(00)02724-4. [DOI] [PubMed] [Google Scholar]
- 59.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 60.Wisor JP, Takahashi JS. Regulation of the vgf gene in the golden hamster suprachiasmatic nucleus by light and by the circadian clock. J Comp Neurol. 1997;378:229–238. [PubMed] [Google Scholar]
- 61.Yamaguchi S, Mitsui S, Miyake S, Yan L, Onishi H, Yagita K, Suzuki M, Shibata S, Kobayashi M, Okamura H. The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr Biol. 2000;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- 62.Yokota S, Horikawa K, Akiyama M, Moriya T, Ebihara S, Komuro G, Ohta T, Shibata S. Inhibitory action of brotizolam on circadian and light-induced Per1 and Per2 expression in the hamster suprachiasmatic nucleus. Br J Pharmacol. 2000;131:1739–1747. doi: 10.1038/sj.bjp.0703735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young MW. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 64.Youngstrom TG, Weiss ML, Nunez AA. Retinofugal projections to the hypothalamus, anterior thalamus and basal forebrain in hamsters. Brain Res Bull. 1991;26:403–411. doi: 10.1016/0361-9230(91)90014-b. [DOI] [PubMed] [Google Scholar]
- 65.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]