Abstract
Studies on cell division traditionally focus on the mechanisms of chromosome segregation and cytokinesis, yet we know comparatively little about how organelles segregate. Analysis of organelle partitioning in asymmetrically dividing cells has provided insights into the mechanisms through which cells control organelle distribution. Interestingly, these studies have revealed that segregation mechanisms frequently link organelle distribution to organelle growth and formation. Furthermore, in many cases, cells use organelles, such as the endoplasmic reticulum and P granules, as vectors for the segregation of information. Together, these emerging data suggest that the coordination between organelle growth, division, and segregation plays an important role in the control of cell fate inheritance, cellular aging, and rejuvenation, i.e., the resetting of age in immortal lineages.
Introduction
Compared with chromosome segregation, which needs to be very precise to ensure that each cell inherits only one copy of each duplicated chromosome, organelle segregation appears at first to require less control. In this context, it is only necessary that each daughter inherits some material, sufficient for seeding the further expansion or de novo assembly of the considered organelle. We define organelles here in its broadest sense that is not restricted to membrane-delimited organelles but includes other larger bodies of the cell. Although organelle segregation seems like a much less structured process than chromosome segregation, the flexibility of this process underlies its contribution to the generation of asymmetry during cell division. But because of both their plasticity and the fact that they were originally less studied, we still understand little about how organelle distribution and segregation are controlled.
The process of asymmetric cell division takes place in both prokaryotes and eukaryotes, where it plays a central role in the generation of cellular diversity. Whereas asymmetric division contributes to the generation of distinct cellular lineages and the renewal of stem cells in metazoans (Gönczy, 2008; Neumüller and Knoblich, 2009), it promotes phenotypic diversity and counterbalances aging in unicellular organisms such as yeast and bacteria (Gershon and Gershon, 2000; Ksiazek, 2010). Cells that divide asymmetrically frequently partition organelles in a specialized manner. Studies in these cells have revealed that they use a variety of mechanisms to generate asymmetry (Knoblich, 2008, 2010). Therefore, they are particularly interesting models to study the mechanisms that govern organelle segregation. In asymmetrically dividing cells, the segregation of organelles seems to follow three general types of scenarios. First, asymmetrically dividing cells frequently assemble nonessential organelles, which then segregate to only one daughter. Generally, these organelles contribute to fate determination or aging. The P granules of nematode Caenorhabditis elegans embryos and aggresomes are prime examples. Second, some organelles might divide in a seemingly symmetrical manner between daughters but yet contribute to the asymmetrical segregation of cellular components, for example when the organelle fragments inherited by the daughters are not functionally equivalent. We will describe and discuss particularly the cases of the ER, the nuclear envelope during closed mitoses, and centrosomes. Finally, the segregation of other organelles, such as the vacuole, depends on specific transport mechanisms. These have already been extensively reviewed (Weisman, 2003, 2006; Ostrowicz et al., 2008), and we will therefore not discuss them here.
Organelles that segregate asymmetrically: The case of P granules and aggresomes
P granules are massive round organelles composed of protein and RNA and segregate specifically to the precursors of the germ cells, where they specify germinal identity (Strome, 2005). Like the nucleus, these granules selectively exclude molecules >40 kD (Updike et al., 2011), reinforcing the notion that they are bona fide organelles. P granules are neither transported nor anchored to any structure, and the mechanism underlying their unique segregation pattern has remained mysterious until recently. Studies performed by the Hyman laboratory demonstrated that the partition of P granules to the posterior end of the one-cell embryo and hence their subsequent segregation to the corresponding daughter cell depend on their assembly dynamics (Brangwynne et al., 2009). Their assembly is driven by the condensation of P granule components into granules. This process is efficient in the posterior half of the one-cell embryo, whereas P granules disassemble when located in the anterior of the embryo. P granule condensation into large and massive structures considerably slows down the diffusion kinetics of these particles and of their constituents, which become near stationary in the posterior end of the cell. On the anterior of the embryo, the disassembling granules release their material into smaller particles, which are free to rapidly disperse throughout the cell. Hence, these components become available to increase the size of the granules in the posterior half of the cell. Consequently, P granule constituents accumulate in the posterior of the embryo, whereas their concentration drops at its anterior. Thus, the dynamics of granules components are very reminiscent of the process of evaporation/condensation leading to the separation of chemicals during distillation.
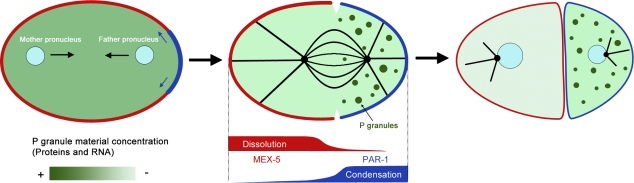
Instead of heat and cold, the asymmetric distribution of the granules is driven by the presence of polarity factors promoting their dissociation or condensation at opposite ends of the one-cell oocyte (Fig. 1). Among these polarity factors, the kinase PAR-1 and the RNA-binding protein MEX-5 are prime candidates to act in the control of P granule condensation and dissociation, respectively (Guo and Kemphues, 1995; Schubert et al., 2000). During P granule partition, the procondensation factor PAR-1 localizes to the posterior cortex of the embryo, whereas the dissociation promoter MEX-5 localizes to the cytoplasm of the anterior half of the cell. The importance of these two proteins in granule dynamics is underlined by the observation that P granules disassemble throughout the cell in par-1 mutant embryos, whereas they assemble and accumulate throughout the cell when MEX-5 is depleted (Brangwynne et al., 2009). However, it is unclear at this point whether both PAR-1 and MEX-5 act directly on P granule components. Indeed, PAR-1 is also known to control MEX-5 distribution (Tenlen et al., 2008; Daniels et al., 2010). Therefore, PAR-1 may act by promoting the condensation of granule components through their direct phosphorylation at the posterior end of the oocyte and by mediating the confinement of MEX-5 and P-granule disassembly to the anterior end of the cell. Alternatively, it may control P granule partition solely through this last process. It may also regulate the function of pptr-1, a regulatory subunit of PP2A recently shown to be required for P granule formation (Gallo et al., 2010). Regardless, it is attractive to think that the simple mechanism of dissociation/condensation might very generally drive the partition of cytoplasmic material into specialized organelles such as Cajal bodies, P bodies, and stress granules and control their spatial distribution. Together, these studies indicate that at least one mechanism controlling the distribution and the symmetric or asymmetric segregation of an organelle is to spatially control the dynamics of its assembly and disassembly. However, alternative pathways appear to coexist and ensure that asymmetry is achieved with high fidelity. In the case of P granules, such an alternative pathway is provided after division by autophagy, which eliminates missegregated granules in the somatic cells (Zhang et al., 2009; Zhao et al., 2009). However, this mechanism assumes that division is already asymmetric enough to allow the emergence of a somatic lineage. Therefore, autophagy appears to enhance asymmetry rather than generate it in the first place.
Figure 1.
P granule formation in the C. elegans one-cell embryo. Formation of this organelle is proposed to occur through a dissolution/aggregation mechanism. After fertilization, P granule components (both RNAs and proteins) are distributed uniformly throughout the cytoplasm. Upon specification of the anterior–posterior axis, the posterior polarity protein PAR-1 (blue) promotes their aggregation. As a consequence, P granules assemble specifically in the posterior of the embryo. Once aggregated, P granule components diffuse more slowly and therefore remain preferentially in the posterior compartment of the embryo. On the anterior of the embryo, MEX-5 (red) promotes the dissolution of P granules. Once the different components are in solution in the anterior, they diffuse more rapidly and can replenish the posterior pool. Cleavage results in the inheritance of P granules only in the posterior daughter cell (the P1 cell).
Aggresomes are a second type of organelle that segregate asymmetrically at mitosis (Macara and Mili, 2008). The aggresome is formed of ubiquitinated and aggregating misfolded proteins and is characterized by the accumulation of the proteasome on its surface (Fig. 2; Johnston et al., 1998; Wigley et al., 1999; Garcia-Mata et al., 2002). Whereas the aggresome is constitutively present in some cell types such as HEK293 and HeLa cells (Wigley et al., 1999), in other cells, its formation is induced by the expression of proteins that fold inefficiently (such as mutant or poly-Q/N–rich proteins) or through inhibition of the ubiquitin-dependent degradation pathway (Bence et al., 2001; Chiti and Dobson, 2006; Gidalevitz et al., 2006; Link et al., 2006). Therefore, the aggresome is thought to accumulate misfolded and aggregating proteins that the cell is not able to properly degrade, particularly amyloid structures. It generally localizes as a single entity to the vicinity of the centrosome and therefore segregates with only one of the two spindle poles at mitosis (Wigley et al., 1999). Its asymmetric segregation is thought to help clear one of the two daughter cells, generally the self-renewing stem cell, from damaged and potentially damaging proteins. Current models suggest that aggresome formation results from the transport of smaller aggregates to the centrosome and their accumulation around it (Fig. 2 A, top). In favor of such a scenario, cells lacking microtubules or the microtubule-dependent motor protein dynein fail to assemble an aggresome but instead display smaller aggregates throughout the cell (Johnston et al., 1998, 2002; García-Mata et al., 1999). This phenotype is very reminiscent of that of cells lacking chaperones, such as Hsp104 in yeast. Thus, beyond their function in disaggregating these chaperones, Hsp104 might also function in the transport of the aggregating proteins, perhaps by acting as an adaptor between them and transport motors such as dynein (Fig. 2 A, bottom). However, it is unclear whether microtubules and motor activity are required for transport and delivery of aggresome constituents or rather for the localization of a condensation activity required for aggresome assembly.
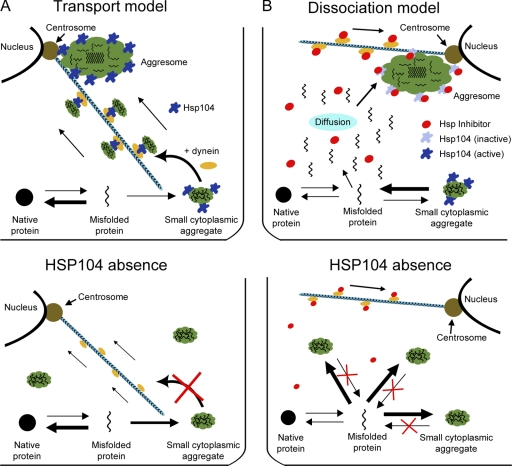
Figure 2.
Two models for the formation of aggresomes. (A, top) In the transport model, small cytoplasmic aggregates are formed throughout the cell and accumulate the chaperone protein Hsp104. The association with Hsp104 is required for the loading of the small aggregates onto microtubules and their transport to the centrosome in a dynein-dependent manner. At this location, the small aggregates merge with the aggresome. In this model, Hsp104 is active everywhere. (B, top) In the dissociation/condensation model, Hsp104 activity is high throughout the cytoplasm and on the small aggregates formed, whereas it is low on the aggresome. Therefore, the small aggregates rapidly release their material for incorporation in the aggresome. In this model, microtubules may mediate the transport of an Hsp104 inhibitor to the centrosome to allow aggresome condensation at this place. (A and B, bottom illustrations) Predictions for the effects of Hsp104 inhibiting in each model. Inhibition of Hsp104 leads to formation of smaller aggregates throughout the cytoplasm. In the transport model (A, bottom), this is because of the fact that they are no longer transported to the centrosome. In the dissociation model (B, bottom), condensation occurs throughout the cytoplasm.
Clues about the mechanism of aggresome formation were provided by studies from the Frydman laboratory; these investigations indicate that in both yeast and mammalian cells, two distinct compartments can accumulate misfolded proteins (Kaganovich et al., 2008). One of these compartments, called JUNQ, forms as an indentation of the nuclear envelope. It is enriched in disaggregases and proteasome complexes and is the destination of ubiquitinated substrates. Fluorescence loss in photobleaching experiments shows that proteins enriched in JUNQ exchange with the cytoplasm, consistent with JUNQ-promoting protein disaggregation. The second compartment, called IPOD, accumulates terminally aggregated proteins, such as yeast prion proteins or Huntingtin-Q103, and any unfolded proteins when the proteasome is inactivated or saturated. As opposed to proteins in JUNQ, IPOD proteins do not exchange with the cytoplasm, indicating that IPOD is the final destination for protein aggregates that cannot be disassembled and that are thought to be cytotoxic (Kaganovich et al., 2008). Importantly, both IPOD and JUNQ localize exclusively to the yeast mother cell and segregate asymmetrically during mitosis. The mechanisms of IPOD and JUNQ formation and localization are unclear, but, at least for the IPOD compartment, it seems to depend on controlled dissolution/condensation (Fig. 2 B). Indeed, IPOD fails to form properly when the chaperone Hsp104 is inhibited, and, instead, smaller aggregates form throughout the cell (Fig. 2 B, bottom). Hsp104, which is related to ClpB in prokaryotes, is a protein of the AAA+ ATPase family that functions as a molecular chaperone able to mediate protein disaggregation (Doyle and Wickner, 2009). Therefore, dissolution of the smaller aggregates by Hsp104 appears to be required for the accumulation of these misfolded proteins in IPOD, where condensation is favored at least through inhibition of dissolution. Accordingly, the lack of recovery upon photobleaching suggests that dissolution is very inefficient in this organelle. The localization of Hsp104 to the aggresome might be, in that regard, misleading: it may accumulate on the aggresome not so much because it acts at that site but because it is inactive and trapped there (Fig. 2 B). Therefore, it is tempting to speculate that controlled dissolution/condensation may be the prime mechanism for the asymmetric partition of IPOD to the yeast mother cell.
Studies in yeast and mammalian cells have suggested that protein aggregates undergo directed motion. Whereas in mammals transport seems to depend on the microtubules (Johnston et al., 2002), studies have suggested that motion might depend on actin in yeast (Liu et al., 2010). Upon exposure to acute and prolonged stresses such as heat shocks or equivalent proteotoxic stresses, particles decorated by Hsp104 are formed throughout the cell. Whereas most of these aggregates are rapidly dissolved (Zhou et al., 2011), some of them remain stable long enough upon return to physiological conditions to allow the monitoring of their dynamics (Liu et al., 2010). According to Liu et al. (2010), the population of such aggregates that were originally located in the bud translocates back to the mother cell in an actin-dependent manner. The movement of these inclusion bodies and their colocalization with actin cables suggest that they are anchored on the cables that are growing from the bud tip and follow their retrograde flow toward the mother cells (Evangelista et al., 2003). This mechanism would suggest the existence of a sweeping process that pushes back inclusion bodies from the mother into the bud. However, this model is challenged by closer analysis of particle movement (Zhou et al., 2011). Indeed, these studies indicate that the vast majority of the particles undergo random movements rather than directed retrograde transport. All taken in consideration, it remains unclear whether the movement of these aggregates is of any relevance under normal physiological conditions. Whereas it is generally accepted that at least one aggregate forms in yeast mother cells as they age, daughter cells are generally born free of such a structure (Erjavec et al., 2007). Thus, the relevant question might not be to understand how aggregates are transported from the bud into the mother but how and why aggregates are being retained in aging mother cells. The idea that retention of aggregates in the mother cell contributes to the rejuvenation of the daughter cell has been recently challenged. Indeed, expression of the meiosis-specific transcription factor Ndt80 during vegetative growth causes old yeast mother cells to reset their age (Unal et al., 2011). Remarkably, these rejuvenation events were nearly as efficient as those undergone by yeast daughter cells as they emerge from their mothers, yet they did not involve clearance from aggregates. Altogether, the available data suggest that the asymmetric segregation of aggresomes in yeast and animal cells depends mainly on a dissolution/condensation process that determines where stable endogenous aggresomes form and remain. Yet, it remains possible that under stress situations, transport mechanisms also contribute to the faithful partition of aggregates.
Segregation of the ER: Emergence and function of organelle polarity
In contrast to P granules and aggresomes, many organelles (such as the ER and mitochondria) are essential for the viability of every single cell. In these cases, it is crucial that both daughters inherit each a fraction of the organelle. As a demonstration for this point, in response to acute stress, yeast cells inhibit ER inheritance by the bud (Babour et al., 2010). In these extreme cases of asymmetric division, the daughter produced without an ER is unviable, as predicted. However, even in normal divisions, the organelle fragments that are inherited by each daughter can be nonequivalent. Again, the case of the budding yeast is illuminating. After cell division, mother and daughter cells show several physiological differences. Provided that it is haploid, the mother cell expresses a specific endonuclease, called HO, which promotes recombination at the mating-type locus and thereby the switch of the sexual identity of the cell between the two possible mating types (Haber, 1998). Asymmetry of the HO endonuclease is a result of the expression of a transcriptional repressor, Ash1, in the bud only. This asymmetry is achieved through the transport of the ASH1 mRNA to the bud (Darzacq et al., 2003; Cosma, 2004). In turn, transport and anchorage in the bud depend, at least in part, on the ER protein She3 (Jansen et al., 1996; Estrada et al., 2003; Buvelot Frei et al., 2006; Schmid et al., 2006; Aronov et al., 2007). She3 recruits the myosin V Myo4 to ER tubules and thereby mediates their migration into the bud, as well as that of polar mRNAs, along actin cables. Therefore, before cell division, the bud ER is homogenously labeled with She3, whereas She3 is absent from the surface of the ER in the mother cell. Loss of She3 function leads to delayed ER segregation and impairs inheritance of polar mRNA by the bud. Thus, the yeast ER is asymmetric during mitosis, at least with respect to its She3 content and its decoration with polar mRNAs, and this asymmetry is required for proper segregation of the ER between mother and bud. Together, ER asymmetry and myosin-dependent movement of bud-specific ER into the bud drive not only the asymmetric partition of ER-associated factor but, in the first place, ensure that both mother and bud inherit ER.
Remarkably, several additional mRNAs cofractionate with the ASH1 mRNA, and most of these mRNAs encode transmembrane proteins (Takizawa et al., 2000; Shepard et al., 2003), at least two of which are ER resident proteins involved in lipid biosynthesis (Erg2 and Lcb1). These observations are consistent with these mRNAs being translated on the surface of the ER. Furthermore, they also suggest that cotransport of mRNAs and ER could serve two reciprocally beneficial functions. First, the specialized ER in the bud provides an anchor for the retention of polar mRNAs in this compartment. Second, at least a subfraction of these mRNAs probably contributes to de novo synthesis and expansion of the ER in the bud. Therefore, some ER constituents might be synthesized mainly in the bud, whereas the preexisting molecules remain in the mother, as it has been proposed for the multidrug transporters present on the plasma membrane (Eldakak et al., 2010). However, this would require that proteins synthesized at the ER surface in the bud remain in the bud. Supporting this idea, a previous study has indicated that a diffusion barrier assembles very early after bud emergence in the ER membrane at the bud neck (Luedeke et al., 2005). Although this barrier has no effect on the exchange between mother and bud for luminal proteins of the ER, it strongly impedes the migration of ER membrane proteins through the bud neck and hence partially insulates the ER membranes of mother and bud from each other. Hence, ER asymmetry and segregation might correlate with the formation of a young ER in the bud. In any case, this insulation suggests that the membranes of mother and bud ER are distinct. Clarifying the nature and function of their dissimilarities might provide profound insights about a novel role of the ER in cellular physiology and the mechanisms of ER segregation.
Diffusion barriers and age segregation: The yeast nucleus
Although we generally consider the nucleus from the point of view of its chromosomal content, its structure and its envelope make it a bona fide organelle as well. Furthermore, in cells that undergo closed mitosis, division of the nucleus comprises a certain level of asymmetry. For example, nonchromosomal DNA is generally segregated asymmetrically between yeast mother and daughter cells (Murray and Szostak, 1983), and retention of nonchromosomal DNA circles in the mother cell is one of the driving mechanisms of replicative aging (Sinclair and Guarente, 1997; Sinclair et al., 1998). Furthermore, whereas closed mitosis is not observed in animal cells, at least some animal stem cells undergo semiclosed mitoses, such as in Drosophila melanogaster (Katsani et al., 2008).
Indeed, the best-characterized aging factors in yeast are the extrachromosomal ribosomal DNA (rDNA) circles, which are thought to contribute to aging by titrating cellular components, such as the replication machinery, as they accumulate. Extrachromosomal rDNA circles pop out from the chromosome as a result of homologous recombination between adjacent copies of the rDNA repeats in the nucleolar locus during repair of double strand breaks. These circular DNA molecules, which are ∼9 kb in length, contain a replication origin and replicate during S phase (Bell et al., 1977; Sinclair and Guarente, 1997). However, they do not segregate symmetrically at mitosis but remain confined in the mother cell at a retention frequency >98% per circle and per mitosis (Sinclair and Guarente, 1997; Gillespie et al., 2004). Remarkably, most replicative plasmids that do not contain a segregation sequence, such as a centromere or the segregation sequence of the 2-µm plasmid, are also retained in the mother cell and promote aging (Murray and Szostak, 1983; Falcón and Aris, 2003). However, the strength of the bias varies. For some plasmids, the retention frequency can be up to 96–97% (unpublished data), whereas for others, it is lower (Gehlen et al., 2011). Recent studies have provided different insights into how nuclear asymmetry might be achieved (Fig. 3; Shcheprova et al., 2008; Gehlen et al., 2011).
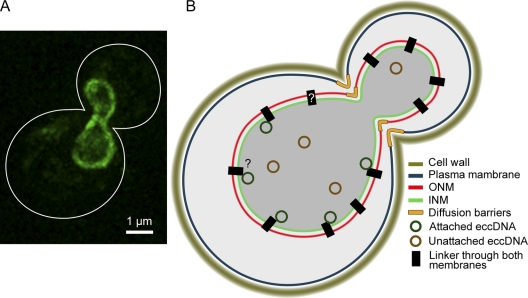
Figure 3.
A nuclear diffusion barrier and nuclear morphology promote the retention of DNA circles in the yeast mother cell. (A) The morphology of the nucleus in early anaphase reveals by the expression of Nsg1-GFP a component of the outer nuclear envelope. The cell outline is represented by a white line. (B) A schematic representation of a yeast nucleus at the same stage as the cell shown in A. Both the morphology of the nucleus and anchorage to the envelope contribute to the high level of retention observed for most extrachromosomal circles (eccDNA). INM, inner nuclear membrane; ONM, outer nuclear membrane.
A modeling approach indicated that the diffusion properties of circular plasmids in the nucleoplasm and the geometry of the nucleus alone can easily explain the retention of plasmids in the mother up to a frequency of 85% (Gehlen et al., 2011). Addition of several hypotheses may explain up to 90% retention by geometry, although the actual validity of these premises is not yet demonstrated. In any case, reaching a retention frequency >90% certainly depends on additional mechanisms. These mechanisms are crucial, as modeling has established that rejuvenation of the daughter cell relies on high retention frequencies (Gillespie et al., 2004; Shcheprova et al., 2008). Indeed, by the retention frequency of 85% predicted by the geometry of the nucleus, it takes only four copies of the plasmid to reach a probability of 50% that the bud inherits one of them. In contrast, by a retention frequency of 96–98%, it takes 16–35 copies to reach this probability. Therefore, such additional mechanisms are likely to be crucial for the proper control of cellular longevity and for the rejuvenation of the daughters.
A second study based on a photobleaching approach established the existence of diffusion barriers in the outer membrane of the envelope of the anaphase nucleus, similar to the barrier present in the cortical ER (Shcheprova et al., 2008). This barrier cannot be explained simply by the geometry of the nuclear envelope, which is pinched at the bud neck, because no diffusion barrier is observed in the inner membrane of the envelope (Shcheprova et al., 2008; unpublished data). Remarkably, all mutations that affect the strength of the diffusion barrier in the outer membrane of the nuclear envelope affect the retention of the plasmid in the yeast mother cell (Shcheprova et al., 2008; Gehlen et al., 2011; Lindstrom et al., 2011), and the level of plasmid retention correlates well with the strength of the diffusion barrier (Shcheprova et al., 2008). Thus, it has been proposed that this diffusion barrier is required for plasmid retention. Obviously, it remains to be determined how a diffusion barrier in the outer membrane could affect the distribution of DNA circles in the nucleus. One hypothesis raised by Gehlen et al. (2011) is that mutants affecting the barrier may affect the duration of anaphase. Indeed, a longer anaphase would leave more time to plasmids to diffuse into the bud. However, there is no evidence so far that barrier mutants do indeed affect anaphase progression.
One possibility to explain how the barrier in the outer nuclear membrane retains plasmids in the mother cell could be that plasmids are attached to some structure at the nuclear envelope, as already postulated by Murray and Szostak (1983), and, more specifically, a structure spanning both inner and outer membranes. Against this hypothesis, some plasmids are retained in the mother cell without being continuously localized at the nuclear periphery (Gehlen et al., 2011). However, such plasmids are not among the best-retained plasmids. In contrast, both rDNA units and well-retained plasmids do localize to the nuclear periphery (Dvorkin et al., 1991; Oakes et al., 1998; Scott-Drew et al., 2002; Shcheprova et al., 2008). Although the hypothesis has been made that attachment sites at the nuclear periphery might be nuclear pores (Shcheprova et al., 2008; Chadrin et al., 2010), recent data have reopened the question (Khmelinskii et al., 2010, 2011). In any case, together, these data suggest that there are probably several mechanisms working together to ensure a high retention of nonchromosomal DNA circles in the mother cell. These mechanisms appear to involve the morphology, the dynamics, and the compartmentalization of the nuclear envelope. The convergence of several parallel mechanisms might be necessary to achieve a high retention frequency.
Interestingly, extrachromosomal DNA are not unique to yeast, as they have been observed in plants and Drosophila as well as mammalian cells (Cohen and Segal, 2009). For example, such DNA fragments are present in many mammalian cancer cells and contain amplified oncogenes or resistance genes that contribute to their overproliferation potential (Hahn, 1993). Their high copy numbers suggest that these too are segregated asymmetrically (Leach and Jackson-Cook, 2004). Interestingly, these DNA fragments are eliminated from the nucleus by yet unknown mechanisms and formed so-called double-minute–type micronuclei that are surrounded by a nuclear envelope membrane (Shimizu, 2011; unpublished data). These extrachromosomal DNA circles have also been shown to accumulate both in vivo and in vitro in aging mammalian cells, as it is the case in yeast cells (Kunisada et al., 1985). Therefore, both in yeast and mammalian cells, extrachromosomal DNA fragments are segregated differently than the rest of the genome, suggesting that there are mechanisms able to recognize them and sort them out of the rest of the genome. It will be interesting to determine how such sorting is achieved and whether it involves interaction with membranes.
A paradigm for age-dependent segregation at mitosis: The centrosome
The organelle for which the coupling between synthesis and segregation is most striking is the centrosome and its centrioles (Yamashita and Fuller, 2008). Upon mitosis, each daughter cell inherits one pole of the spindle and hence one centrosome consisting of one pair of parental centrioles. This pair splits during early mitosis to seed the formation of the two centrosomes at the opposite poles of the mitotic spindle. During this process, a daughter centriole is synthesized at the side of each of the two parental centrioles to reform a centriole pair for each centrosome. Thus, at mitosis, every cell contains a grandmother, a mother, and two associated daughter centrioles. Throughout their maturation, the centrosomes recruit pericentriolar proteins like ninein, such that the centrosome that contains the oldest centriole has more of this material (Strnad and Gönczy, 2008). In many asymmetrically dividing cells, centrosomes do not segregate randomly. In Drosophila male germinal stem cells (GSCs; Fig. 4 A; Yamashita et al., 2007) and mouse brain stem cells (Fig. 4 B; Wang et al., 2009), the old centrosome segregates stereotypically to the daughter that keeps the stem fate. In contrast, in Drosophila neuroblasts, the oldest centriole segregates to the differentiating daughter (Fig. 4 C; Conduit and Raff, 2010; Januschke et al., 2011). Yet, strikingly, in other stem cells, centrosome segregation is random, such as in the female GSC of Drosophila (Stevens et al., 2007). In cells where centrosome inheritance is not random, we know very little about how the segregation of the centrosomes is specified. However, the fact that inheritance is oriented differently in different lineages suggests the existence of complex mechanisms able to regulate this process and indicates that it is important for the cell which of the old or the young centrosome it inherits. Accordingly, affecting the maturation of the centrioles in mouse brain stem cells randomizes the segregation of the centrosomes and impacts their self-renewal capacity (Wang et al., 2009). Similarly, in old Drosophila males, misorientation of the centrosome in the GSC impairs spermatogenesis (Cheng et al., 2008). In neuroblast, defects in anchoring the old centrosome to the cortical side cause spindle positioning as well as asymmetric division problems (Basto et al., 2006; Siller et al., 2006). However, how the age of centrosomes affects cellular physiology remains unclear. Furthermore, the different modes of centrosome segregation displayed by different types of stem cells provide further complexity to this dilemma. Is spindle asymmetry only a mechanism to guide spindle alignment with the polarity axis of the cell, as it is the case in yeast (Kusch et al., 2003)? Do cellular factors involved in cell fate specification associate specifically with one of the centrosomes? The notion that the aggresome associates with one of the two centrosomes and the association of specific mRNAs with centrosomes in several species (Barral and Liakopoulos, 2009) speak in favor of this last possibility.
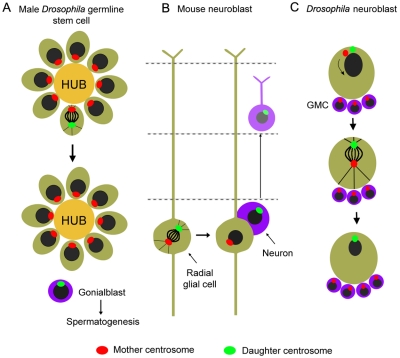
Figure 4.
Asymmetric distribution of centrosomes based on their age. (A and B) The old centrosome, which contains the oldest centriole, segregated preferentially in the daughter cell that maintains the stem cell–like fate in both the male Drosophila germ line (A) and in the mouse neuroblast (B). (C) During neuroblast division, the mother centrosome preferentially segregates to the differentiating daughter. In female Drosophila germ cells, segregation is random (not shown). Therefore, it is impossible at the moment to make general conclusions on the functions and mechanisms of the unequal biases observed. In all panels, the green and the purple cells represent the stem cells and the differentiated somatic cells, respectively. GMC, ganglion mother cell; HUB, region of packed somatic cells required for signaling to the neighboring cells to maintain their germ cell identity.
Conclusion
Over the last few years, a remarkable picture is emerging, in which our understanding of the ill-defined processes underlying organelle segregation becomes unified by the gradual discovery of their tight linkage to processes as profound as aging, rejuvenation, and self-renewal. At this stage, we can already note one recurring theme and raise several major questions. First, the mechanisms of organelle segregation are regularly linked to the mechanisms of organelle biogenesis. This link can be direct, as for P granules, or involve more evolved mechanisms, such as the use of a single multifunctional protein to link both ER tubules and mRNAs together to the actin cytoskeleton. In other cases, although it is clear that synthesis and segregation are connected, how these different processes articulate with each other is not obvious. This is the case for the centrosome, in which we do not understand the mechanisms and functions of their functional differentiation. Clearly, more discoveries are ahead of us.
Second, these findings raised major questions concerning both the molecular mechanisms involved in the processes of organelle biogenesis and their conservation in evolution. Are findings made in yeast, which undergo a closed mitosis, relevant for organisms that undergo open mitoses and vice versa? Are there profound similarities underlying these apparently different processes? If so, we will find them most easily at the molecular level first. Some of them may refer to the use and control of diffusion through both controlled disassembly/condensation mechanisms and diffusion barriers. These mechanisms are simple enough to be widely spread in evolution. More generally, the fact that organelle segregation is so deeply linked to aging and self-renewal, which are probably among the most basic biological processes, suggests that comparative cell biology will be here again a very fruitful approach for future progress.
Acknowledgments
We thank F. Caudron, J. Saarikangas, A. Denoth, S. Baldi, and L. Clay for discussions and critical reading of the manuscript.
Y. Barral is supported by Eidgenössische Technische Hochschule Zürich, grants from the Swiss National Foundation, the European Research Council, and the Institut des Hautes Études Scientifiques.
The authors declare no commercial affiliations and no conflicts of interest.
Footnotes
Abbreviations used in this paper:
- GSC
- germinal stem cell
- rDNA
- ribosomal DNA
References
- Aronov S., Gelin-Licht R., Zipor G., Haim L., Safran E., Gerst J.E. 2007. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:3441–3455 10.1128/MCB.01643-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babour A., Bicknell A.A., Tourtellotte J., Niwa M. 2010. A surveillance pathway monitors the fitness of the endoplasmic reticulum to control its inheritance. Cell. 142:256–269 10.1016/j.cell.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y., Liakopoulos D. 2009. Role of spindle asymmetry in cellular dynamics. Int Rev Cell Mol Biol. 278:149–213 10.1016/S1937-6448(09)78004-9 [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. 2006. Flies without centrioles. Cell. 125:1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bell G.I., DeGennaro L.J., Gelfand D.H., Bishop R.J., Valenzuela P., Rutter W.J. 1977. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J. Biol. Chem. 252:8118–8125 [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Buvelot Frei S., Rahl P.B., Nussbaum M., Briggs B.J., Calero M., Janeczko S., Regan A.D., Chen C.Z., Barral Y., Whittaker G.R., Collins R.N. 2006. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol. Cell. Biol. 26:7299–7317 10.1128/MCB.02405-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadrin A., Hess B., San Roman M., Gatti X., Lombard B., Loew D., Barral Y., Palancade B., Doye V. 2010. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 189:795–811 10.1083/jcb.200910043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Türkel N., Hemati N., Fuller M.T., Hunt A.J., Yamashita Y.M. 2008. Centrosome misorientation reduces stem cell division during ageing. Nature. 456:599–604 10.1038/nature07386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. 2006. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75:333–366 10.1146/annurev.biochem.75.101304.123901 [DOI] [PubMed] [Google Scholar]
- Cohen S., Segal D. 2009. Extrachromosomal circular DNA in eukaryotes: Possible involvement in the plasticity of tandem repeats. Cytogenet. Genome Res. 124:327–338 10.1159/000218136 [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Raff J.W. 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20:2187–2192 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Cosma M.P. 2004. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 5:953–957 10.1038/sj.embor.7400251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels B.R., Dobrowsky T.M., Perkins E.M., Sun S.X., Wirtz D. 2010. MEX-5 enrichment in the C. elegans early embryo mediated by differential diffusion. Development. 137:2579–2585 10.1242/dev.051326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Powrie E., Gu W., Singer R.H., Zenklusen D. 2003. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr. Opin. Microbiol. 6:614–620 10.1016/j.mib.2003.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.M., Wickner S. 2009. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem. Sci. 34:40–48 10.1016/j.tibs.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Dvorkin N., Clark M.W., Hamkalo B.A. 1991. Ultrastructural localization of nucleic acid sequences in Saccharomyces cerevisiae nucleoli. Chromosoma. 100:519–523 10.1007/BF00352202 [DOI] [PubMed] [Google Scholar]
- Eldakak A., Rancati G., Rubinstein B., Paul P., Conaway V., Li R. 2010. Asymmetrically inherited multidrug resistance transporters are recessive determinants in cellular replicative ageing. Nat. Cell Biol. 12:799–805 10.1038/ncb2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., Nyström T. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21:2410–2421 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada P., Kim J., Coleman J., Walker L., Dunn B., Takizawa P., Novick P., Ferro-Novick S. 2003. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 163:1255–1266 10.1083/jcb.200304030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Zigmond S., Boone C. 2003. Formins: Signaling effectors for assembly and polarization of actin filaments. J. Cell Sci. 116:2603–2611 10.1242/jcs.00611 [DOI] [PubMed] [Google Scholar]
- Falcón A.A., Aris J.P. 2003. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J. Biol. Chem. 278:41607–41617 10.1074/jbc.M307025200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C.M., Wang J.T., Motegi F., Seydoux G. 2010. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science. 330:1685–1689 10.1126/science.1193697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata R., Bebök Z., Sorscher E.J., Sztul E.S. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239–1254 10.1083/jcb.146.6.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Gao Y.S., Sztul E. 2002. Hassles with taking out the garbage: Aggravating aggresomes. Traffic. 3:388–396 10.1034/j.1600-0854.2002.30602.x [DOI] [PubMed] [Google Scholar]
- Gehlen L.R., Nagai S., Shimada K., Meister P., Taddei A., Gasser S.M. 2011. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr. Biol. 21:25–33 10.1016/j.cub.2010.12.016 [DOI] [PubMed] [Google Scholar]
- Gershon H., Gershon D. 2000. The budding yeast, Saccharomyces cerevisiae, as a model for aging research: A critical review. Mech. Ageing Dev. 120:1–22 10.1016/S0047-6374(00)00182-2 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 311:1471–1474 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Gillespie C.S., Proctor C.J., Boys R.J., Shanley D.P., Wilkinson D.J., Kirkwood T.B. 2004. A mathematical model of ageing in yeast. J. Theor. Biol. 229:189–196 10.1016/j.jtbi.2004.03.015 [DOI] [PubMed] [Google Scholar]
- Gönczy P. 2008. Mechanisms of asymmetric cell division: Flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 9:355–366 10.1038/nrm2388 [DOI] [PubMed] [Google Scholar]
- Guo S., Kemphues K.J. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 81:611–620 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Haber J.E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561–599 10.1146/annurev.genet.32.1.561 [DOI] [PubMed] [Google Scholar]
- Hahn P.J. 1993. Molecular biology of double-minute chromosomes. Bioessays. 15:477–484 10.1002/bies.950150707 [DOI] [PubMed] [Google Scholar]
- Jansen R.P., Dowzer C., Michaelis C., Galova M., Nasmyth K. 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 84:687–697 10.1016/S0092-8674(00)81047-8 [DOI] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J., Gonzalez C. 2011. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2:243 10.1038/ncomms1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., Kopito R.R. 1998. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Illing M.E., Kopito R.R. 2002. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil. Cytoskeleton. 53:26–38 10.1002/cm.10057 [DOI] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani K.R., Karess R.E., Dostatni N., Doye V. 2008. In vivo dynamics of Drosophila nuclear envelope components. Mol. Biol. Cell. 19:3652–3666 10.1091/mbc.E07-11-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Keller P.J., Lorenz H., Schiebel E., Knop M. 2010. Segregation of yeast nuclear pores. Nature. 466:E1 10.1038/nature09255 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Meurer M., Knop M., Schiebel E. 2011. Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr. Biol. 21:R17–R18 10.1016/j.cub.2010.11.034 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. 2010. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11:849–860 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek K. 2010. Bacterial aging: From mechanistic basis to evolutionary perspective. Cell. Mol. Life Sci. 67:3131–3137 10.1007/s00018-010-0417-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisada T., Yamagishi H., Ogita Z., Kirakawa T., Mitsui Y. 1985. Appearance of extrachromosomal circular DNAs during in vivo and in vitro ageing of mammalian cells. Mech. Ageing Dev. 29:89–99 10.1016/0047-6374(85)90050-8 [DOI] [PubMed] [Google Scholar]
- Kusch J., Liakopoulos D., Barral Y. 2003. Spindle asymmetry: A compass for the cell. Trends Cell Biol. 13:562–569 10.1016/j.tcb.2003.09.008 [DOI] [PubMed] [Google Scholar]
- Leach N.T., Jackson-Cook C. 2004. Micronuclei with multiple copies of the X chromosome: Do chromosomes replicate in micronuclei? Mutat. Res. 554:89–94 10.1016/j.mrfmmm.2004.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom D.L., Leverich C.K., Henderson K.A., Gottschling D.E. 2011. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 7:e1002015 10.1371/journal.pgen.1002015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C.D., Fonte V., Hiester B., Yerg J., Ferguson J., Csontos S., Silverman M.A., Stein G.H. 2006. Conversion of green fluorescent protein into a toxic, aggregation-prone protein by C-terminal addition of a short peptide. J. Biol. Chem. 281:1808–1816 10.1074/jbc.M505581200 [DOI] [PubMed] [Google Scholar]
- Liu B., Larsson L., Caballero A., Hao X., Oling D., Grantham J., Nyström T. 2010. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 140:257–267 10.1016/j.cell.2009.12.031 [DOI] [PubMed] [Google Scholar]
- Luedeke C., Frei S.B., Sbalzarini I., Schwarz H., Spang A., Barral Y. 2005. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169:897–908 10.1083/jcb.200412143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I.G., Mili S. 2008. Polarity and differential inheritance—universal attributes of life? Cell. 135:801–812 10.1016/j.cell.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W., Szostak J.W. 1983. Pedigree analysis of plasmid segregation in yeast. Cell. 34:961–970 10.1016/0092-8674(83)90553-6 [DOI] [PubMed] [Google Scholar]
- Neumüller R.A., Knoblich J.A. 2009. Dividing cellular asymmetry: Asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 23:2675–2699 10.1101/gad.1850809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes M., Aris J.P., Brockenbrough J.S., Wai H., Vu L., Nomura M. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 143:23–34 10.1083/jcb.143.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowicz C.W., Meiringer C.T., Ungermann C. 2008. Yeast vacuole fusion: A model system for eukaryotic endomembrane dynamics. Autophagy. 4:5–19 [DOI] [PubMed] [Google Scholar]
- Schmid M., Jaedicke A., Du T.G., Jansen R.P. 2006. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 16:1538–1543 10.1016/j.cub.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Schubert C.M., Lin R., de Vries C.J., Plasterk R.H., Priess J.R. 2000. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol. Cell. 5:671–682 10.1016/S1097-2765(00)80246-4 [DOI] [PubMed] [Google Scholar]
- Scott-Drew S., Wong C.M., Murray J.A. 2002. DNA plasmid transmission in yeast is associated with specific sub-nuclear localisation during cell division. Cell Biol. Int. 26:393–405 10.1006/cbir.2002.0867 [DOI] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S.B., Gonnet G., Barral Y. 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature. 454:728–734 [DOI] [PubMed] [Google Scholar]
- Shepard K.A., Gerber A.P., Jambhekar A., Takizawa P.A., Brown P.O., Herschlag D., DeRisi J.L., Vale R.D. 2003. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA. 100:11429–11434 10.1073/pnas.2033246100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N. 2011. Molecular mechanisms of the origin of micronuclei from extrachromosomal elements. Mutagenesis. 26:119–123 10.1093/mutage/geq053 [DOI] [PubMed] [Google Scholar]
- Siller K.H., Cabernard C., Doe C.Q. 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8:594–600 10.1038/ncb1412 [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Guarente L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 91:1033–1042 10.1016/S0092-8674(00)80493-6 [DOI] [PubMed] [Google Scholar]
- Sinclair D.A., Mills K., Guarente L. 1998. Molecular mechanisms of yeast aging. Trends Biochem. Sci. 23:131–134 10.1016/S0968-0004(98)01188-8 [DOI] [PubMed] [Google Scholar]
- Stevens N.R., Raposo A.A., Basto R., St Johnston D., Raff J.W. 2007. From stem cell to embryo without centrioles. Curr. Biol. 17:1498–1503 10.1016/j.cub.2007.07.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Gönczy P. 2008. Mechanisms of procentriole formation. Trends Cell Biol. 18:389–396 10.1016/j.tcb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Strome S. 2005. Specification of the germ line. WormBook. 1–10 10.1895/wormbook.1.9.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., DeRisi J.L., Wilhelm J.E., Vale R.D. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 290:341–344 10.1126/science.290.5490.341 [DOI] [PubMed] [Google Scholar]
- Tenlen J.R., Molk J.N., London N., Page B.D., Priess J.R. 2008. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 135:3665–3675 10.1242/dev.027060 [DOI] [PubMed] [Google Scholar]
- Unal E., Kinde B., Amon A. 2011. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science. 332:1554–1557 10.1126/science.1204349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike D.L., Hachey S.J., Kreher J., Strome S. 2011. P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192:939–948 10.1083/jcb.201010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tsai J.-W., Imai J.H., Lian W.-N., Vallee R.B., Shi S.-H. 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 461:947–955 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman L.S. 2003. Yeast vacuole inheritance and dynamics. Annu. Rev. Genet. 37:435–460 10.1146/annurev.genet.37.050203.103207 [DOI] [PubMed] [Google Scholar]
- Weisman L.S. 2006. Organelles on the move: Insights from yeast vacuole inheritance. Nat. Rev. Mol. Cell Biol. 7:243–252 10.1038/nrm1892 [DOI] [PubMed] [Google Scholar]
- Wigley W.C., Fabunmi R.P., Lee M.G., Marino C.R., Muallem S., DeMartino G.N., Thomas P.J. 1999. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145:481–490 10.1083/jcb.145.3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Fuller M.T. 2008. Asymmetric centrosome behavior and the mechanisms of stem cell division. J. Cell Biol. 180:261–266 10.1083/jcb.200707083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Mahowald A.P., Perlin J.R., Fuller M.T. 2007. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 315:518–521 10.1126/science.1134910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 136:308–321 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tian E., Zhang H. 2009. Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis. Autophagy. 5:717–719 10.4161/auto.5.5.8552 [DOI] [PubMed] [Google Scholar]
- Zhou C., Slaughter B.D., Unruh J.R., Eldakak A., Rubinstein B., Li R. 2011. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell. 147:1186–1196 10.1016/j.cell.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]