Abstract
Stimulation of pituitary gonadotropes by hypothalamic GnRH leads to the rapid expression of several immediate early genes that play key roles in orchestrating the response of the gonadotrope to hypothalamic stimuli. Elucidation of the signaling mechanisms that couple the GnRH receptor to this immediate early gene repertoire is critical for understanding the molecular basis of GnRH action. Here we identify signaling mechanisms that underlie regulation of the orphan nuclear receptor Nur77 as a GnRH-responsive immediate early gene in αT3-1 cells and mouse gonadotropes in culture. Using a variety of approaches, we show that GnRH-induced transcriptional upregulation of Nur77 in αT3-1 cells is dependent on calcium, protein kinase C (PKC), and ERK signaling. Transcriptional activity of Nur77 within the gonadotrope is regulated posttranslationally by GnRH signaling via PKC but not ERK activity. Surprisingly, neither activation of the ERK pathway nor the transcriptional response of Nur77 to GnRH requires the activity of c-Raf kinase. In corroboration of these results, Nur77 responsiveness to GnRH was maintained in gonadotropes from mice with pituitary-targeted ablation of c-Raf kinase. In contrast, gonadotropes from mice with pituitary deficiency of ERK signaling failed to up-regulate Nur77 after GnRH stimulation. These results further clarify the role of ERK and PKC signaling in regulation of the GnRH-induced immediate early gene program as well as GnRH-induced transcription-stimulating activity of Nur77 in the gonadotrope and shed new light on the complex functional organization of this signaling pathway in the pituitary gonadotrope.
In mammals, reproductive function is dependent on the coordinated synthesis and secretion of the gonadotropins LH and FSH by the pituitary gonadotrope. Production of the gonadotropins is largely controlled by the hypothalamic decapeptide GnRH. GnRH is released in pulsatile fashion from the hypothalamus and acts through the GnRH receptor (GnRHR) to stimulate biosynthesis of the gonadotropin subunits as well as the GnRHR itself. The signaling events initiated by the GnRHR coordinate the expression of a diverse set of immediate early response genes, several of which have been shown to regulate gonadotropin biosynthesis (1–5). In the gonadotrope, as in most other cell types, early response genes play a critical role in linking a relatively transitory extracellular stimulus (the pulsatile GnRH signal) with more sustained changes in gene expression that underlie physiologically appropriate cellular responses to that stimulus (such as gonadotropin biosynthesis). Elucidation of the signaling activities that link the GnRH signal with the immediate early gene repertoire is thus important for understanding the molecular basis of gonadotrope function.
The ERK signaling pathway is rapidly activated by GnRH, and ERK activity has been linked to the expression of several genes important for gonadotrope function including the gonadotropin subunit genes as well as the dual specificity MAPK phosphatase MKP2/DUSP4 (1, 6–9). Several ERK-dependent immediate early genes have been shown to play key roles in mediating the effects of GnRH, including early growth response protein 1 (Egr1), c-Fos, and activating transcription factor 3 (ATF3) (1, 4–7). Nur77 (also referred to as NR4A1, NGFIB, NAK1, and TR3) is an immediate early gene belonging to the NR4A family of orphan nuclear receptors. Nur77 is rapidly up-regulated in response to a wide range of extracellular signals and has been shown to play diverse and important roles as a transcriptional regulator in several cell types including pituitary cells (10–18). Microarray analysis showed that Nur77 was strongly up-regulated by GnRH in the murine gonadotrope-derived LβT2 cell line (19); however, the signaling mechanism(s) linked to this regulation by GnRH remain to be fully elucidated. In the LβT2 cell line, GnRH-induced up-regulation of Nur77 has been linked to cAMP/protein kinase A and calcium (20–22). Nur77 was also shown to be expressed in the less differentiated αT3-1 gonadotrope cell line and regulated by cAMP-mediated signaling (23). Interestingly in these studies, Nur77 and steroidogenic factor 1 appear to function antagonistically to modulate GnRH receptor gene regulation.
GnRH-induced Nur77 up-regulation in αT3-1 cells has also been linked to control of the FSHβ subunit gene in this cell line using Nur77 overexpression, chromatin immunoprecipitation studies, and a Nur77 dominant-negative approach (24). These studies are also complicated by the fact that the FSHβ subunit gene is not expressed in αT3-1 cells under normal circumstances; thus, it is difficult to determine the physiological importance of these observations. ERK activity has been shown to be important for agonist-induced up-regulation of Nur77 in several cell types (25–29). Therefore, we set out to examine and more clearly define the role of ERK signaling in GnRH-induced expression of Nur77 in the gonadotrope. Our results establish Nur77 as an ERK-dependent GnRH-responsive immediate early gene and shed unexpected new light on the functional organization of the ERK pathway within the gonadotrope.
Materials and Methods
Cells, reagents, and animals
αT3-1 cells were a generous gift from Dr. Pamela Mellon (University of California at San Diego, San Diego, CA) and were cultured as described previously (30). NIH-3T3 cells were cultured as described previously (31). Primary antibodies against Nur77 (rabbit polyclonal), ERK2, c-Raf, and β-actin along with horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Biotinylated antirabbit secondary antibody for immunocytochemistry was obtained from Vector Laboratories (Burlingame, CA). Antiphospho-ERK was from Sigma Chemical (St. Louis, MO). Mouse monoclonal anti-Nur77 antibody was from PharMingen (San Diego, CA). Rabbit anti-LHβ antibody was from the National Hormone and Peptide Program (National Institute of Diabetes and Digestive and Kidney Diseases, Torrance, CA). Rat monoclonal antihemagglutinin antibody was from Roche (Indianapolis, IN). Buserelin [des-GLY(10d-Ser[t-But]6)-LHRH ethylamide; referred to as a GnRH agonist (GnRHa)], CRH, and the GnRHR antagonist antide, were from Sigma. All other pharmacological inhibitors were from Calbiochem (Gibbstown, NJ).
Plasmid construction and transfection
The pKH3 expression vector, containing three tandem copies of the hemagglutinin epitope tag, was a gift from Dr. Jun-Lin Guan (University of Michigan, Ann Arbor, MI). The Raf-CAAX expression vector was a gift from Dr. Linda Van Aelst (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). To generate the Nur77 luciferase reporter, a 1566-bp fragment of the Nur77 proximal promoter was amplified by PCR from mouse genomic DNA and subcloned into the luciferase expression vector. The c-Fos luciferase reporter has been described previously (32). The fidelity of all constructs was verified by direct nucleotide sequencing. Mouse Nur 77 cDNA was generated by PCR and inserted into a mammalian Gal4 expression vector (kindly provided by Richard Maurer, Oregon Health Sciences University, Portland, OR). The resulting Gal4-Nur77 fusion was confirmed by nucleotide sequence analysis. Transfections were performed using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Carlsbad, CA).
Immunocytochemistry and fluorescence microscopy
For immunofluorescent colabeling of ERK2 and LHβ, 5-μm pituitary sections were deparaffinized and then rehydrated through ethanol dilution series to distilled H2O. Antigen retrieval was performed by boiling slides in 0.01 m citrate buffer (pH 6.0). Sections were washed in PBS and blocked with 10% normal rabbit serum/10% nonfat dry milk in PBS for 20 min at room temperature. Sections were then incubated in polyclonal anti-Nur77 primary antibody diluted 1:10 in PBS/1× casein (Vector Laboratories), for 2 h at 37 C. Sections were further washed in PBS and incubated at room temperature for 20 min with biotinylated rabbit-antigoat IgG (Invitrogen). Normal rabbit IgG was used at equivalent concentration as negative control. After incubation with biotinylated donkey-antirabbit IgG and further washing, sections were incubated with Streptavidin Alexa Fluor 488 (Invitrogen) for 20 min at room temperature in the dark. Stained slides were washed further with PBS and stored in distilled water overnight at 4 C. On d 2, sections were blocked with Fab fragment goat antirabbit IgG H&L (Jackson ImmunoResearch, West Grove, PA) diluted 1:50 in PBS for 30 min at 37 C and reblocked for 20 min with 10% goat serum/2× casein in PBS. Rabbit anti-LH primary antibody was reconstituted in PBS at a concentration of 1 μg/μl, and applied at a 1:50 dilution for 2 h at 37 C, substituting normal rabbit IgG at an equivalent concentration (micrograms per milliliter) as a negative control. LHβ was detected with Texas Red goat-antirabbit IgG at a 1:80 dilution in PBS for 20 min at room temperature in the dark. Slides were washed and mounted in Vectashield 4′,6′-diamino-2-phenylindole (Vector Laboratories). Images were obtained on a Nikon E400 epi-fluorescence microscope (Tokyo, Japan) using the appropriate filters.
Luciferase assay
The αT3-1 cells were grown to approximately 60% confluence and were transfected and treated as indicated within 24 h of plating. Media were removed and the cells were collected in 200 μl of 1× passive lysis buffer (Promega, Madison, WI). Cells were lysed by two freeze-thaw cycles and the lysates were clarified by centrifugation. Protein concentration of the lysates was determined by Bradford assay. Aliquots (100 μg) were assayed for luciferase activity in triplicate for each treatment, and all experiments were repeated at least three times.
Immunoprecipitation and immunoblots
For immunoblotting, cells were washed in cold PBS and scraped into PBS containing 5 mm sodium vanadate, 0.2 mm phenylmethylsulfonyl fluoride, and 5 mm benzamidine. Cells were pelleted by centrifugation, and pellets were resuspended in lysis buffer containing 20 mm Tris-HCl (pH 8.0), 140 mm NaCl, 10% glycerol, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), 0.5% deoxycholate, 2 mm EDTA, 5 mm sodium vanadate, 0.2 mm phenylmethylsulfonyl fluoride, and 5 mm benzamidine. Lysates were cleared by centrifugation and protein concentrations of the lysates were determined by Bradford assay. Protein samples were resolved by SDS-PAGE, and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 5% nonfat dry milk in 10 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 0.05% Tween 20 (TBST) and then incubated with primary antibodies overnight in TBST with 5% milk. Membranes were then washed in TBST and incubated in horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. After further washing, proteins were visualized using enhanced chemiluminescence (PerkinElmer, Boston, MA).
For immunoprecipitations, cells were prepared as above, except that the lysis buffer was adjusted to 200 mm NaCl. Aliquots of lysate containing 400 μg protein were adjusted to 5 mm Tris-HCl (pH 8.0), 70 mm NaCl, 5% glycerol, 0.2% Nonidet P-40, 0.01% SDS, 0.1% deoxycholate, 1 mm EDTA, 5 mm sodium vanadate, 0.2 mm phenylmethylsulfonyl fluoride, and 5 mm benzamidine in a total volume of 400 μl. Rabbit polyclonal anti-Nur77 antibody (2 μg per 400 μg cellular protein) was added and the samples were rocked at 4 C overnight. Protein A/G agarose (30 μl of 50% slurry per sample) was added and the samples were rocked at 4 C for an additional hour. Beads were washed three times in wash buffer [5 mm Tris-HCl (pH 8.0), 70 mm NaCl, 1% glycerol, 0.1% Nonidet P-40, 0.01% SDS, 0.1% deoxycholate, 1 mm EDTA] and boiled in SDS load buffer. Immunoprecipitates were blotted as described above. After blocking, membranes were incubated with mouse monoclonal anti-Nur77 antibody at 1:1000 dilution in TBST with 5% milk overnight at 4 C. Membranes were then washed and developed as described above.
Primary pituitary cell culture
Immediately after euthanasia, pituitaries were collected into cold DMEM containing 10% fetal bovine serum (FBS). Pituitaries were digested at 37 C for 10 min in DMEM containing 0.5 mg/ml each of collagenase and hyaluronidase (Sigma). Tissues were dispersed by repeated pipetting and tissue remnants were allowed to settle by gravity. The supernatant was removed and adjusted to 20% FBS. Remaining tissue was resuspended in DMEM containing 0.25 mg/ml each of the same enzymes and digested for an additional 10 min. After additional dispersal, the supernatants were combined. Cells were washed in DMEM containing 10% FBS and plated in the same medium with penicillin and streptomycin on 10-mm-diameter poly-l-lysine-coated dishes at a density of 1 pituitary equivalent per well. Cells were maintained at 37 C in 5% CO2 overnight before treatment.
RNA isolation and quantitative PCR
Total RNA was isolated using the RNeasy kit (QIAGEN, Valencia, CA), according the manufacturer's instructions. Reverse transcription was carried out using the high capacity cDNA archive kit (Applied Biosystems, Carlsbad, CA). Taqman primer-probe sets for mouse Nur77, c-Raf, B-Raf, Egr-1, and β-actin were purchased commercially (Applied Biosystems). Amplification was performed under standard conditions using the ABI Prism 7500 sequence detection system (Applied Biosystems). Transcript levels were normalized to corresponding levels of β-actin and were calibrated to the control group within each experiment.
Data analysis
All studies presented are representative, with individual treatments carried out in triplicate within an experiment. These studies were completed two to three or more times in separate occasions as indicated. Data were analyzed using t test or one-way ANOVA. The overall differences for t test and ANOVA were considered significant at P < 0.05. Post hoc tests involving comparison of multiple treatments against a control were performed using Dunnett's test. All other post hoc tests were performed using Bonferroni's all pairwise comparisons. Differences in post hoc pairwise comparisons were considered significant at P < 0.05.
Results
Nur77 is up-regulated with immediate early gene kinetics in both αT3-1 cells and primary mouse gonadotropes after GnRHa stimulation
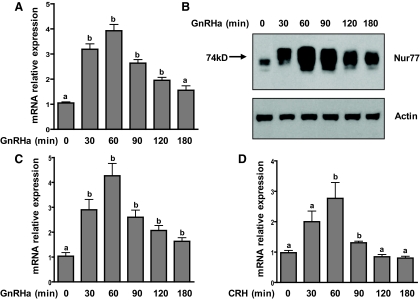
Nur77 has been shown to be expressed in αT3-1 cells; however, its responsiveness to GnRH stimulation has not been reported in this cell line (23). To address this, αT3-1 cells were exposed to the GnRH agonist buserelin (GnRHa) over a 3-h time course, and Nur77 up-regulation was examined at the mRNA level by quantitative PCR (qPCR) and at the protein level by immunoprecipitation followed by immunoblotting. Nur77 transcripts were approximately 4-fold up-regulated after 1 h of GnRHa stimulation and returned to baseline levels within 4 h (Fig. 1A, and data not shown). Nur77 protein levels were also robustly up-regulated by GnRHa, reaching maximal expression at approximately 1 h of GnRHa stimulation (Fig. 1B).
Fig. 1.
Up-regulation of Nur77 in αT3-1 and primary mouse pituitary cells after stimulation with GnRHa. A, αT3-1 cells were exposed to the GnRH agonist buserelin (GnRHa; 10 nm) for the indicated times and Nur77 mRNA levels were measured by qPCR. Bars represent mean ± sem of three experimental replicates. Bars with different letter designations (a and b) represent mean values that are statistically different from the zero time point (P < 0.05). The experiment was repeated three times with similar results. B, Up-regulation of Nur77 protein was examined by immunoprecipitation (IP) followed by immunoblotting. Actin is shown as a protein loading control for the IP, reflecting similar amounts of protein at the start of each IP. This study was carried out on three separate occasions with similar results. C and D, Whole mouse pituitaries were dispersed into primary culture. After a 2-h serum starvation, cultures were exposed to either GnRHa (C) or CRH (10 nm; D) for the indicated time courses. Nur77 mRNA levels were measured within each experimental replicate by qPCR. For each graph, bars represent mean ± sem for three replicates representing a single experiment. These studies were conducted twice (each in triplicate) with similar results. Bars with different letter designations (a and b) represent mean values that are statistically different from the zero time point (P < 0.05).
Nur77 has been shown to be induced by CRH in pituitary corticotropes in which it plays a role in the transcriptional up-regulation of the proopiomelanocortin gene (27). To determine whether Nur77 is induced by GnRHa in primary mouse gonadotropes and to compare its induction kinetics between gonadotropes and corticotropes, whole mouse pituitaries were dispersed into primary culture and then stimulated with vehicle, GnRHa, or CRH. GnRHa stimulation led to a rapid 4-fold up-regulation of Nur77 transcript levels within mixed primary pituitary cell cultures (Fig. 1C), the kinetic profile of which was similar to that observed in the αT3-1 cell line. CRH stimulation led to a more modest (∼3-fold) increase in Nur77 transcript levels, with maximal expression observed after 60 min of agonist exposure (Fig. 1D). CRH-induced Nur77 transcript levels returned to baseline within 120 min of agonist exposure, whereas the transcript up-regulation induced by GnRHa was more sustained, remaining significantly elevated above baseline levels for at least 3 h after agonist exposure (Fig. 1C).
GnRHa-induced up-regulation of Nur77 in αT3-1 cells is dependent on the classical calcium-protein kinase C (PKC)-ERK signaling pathway.
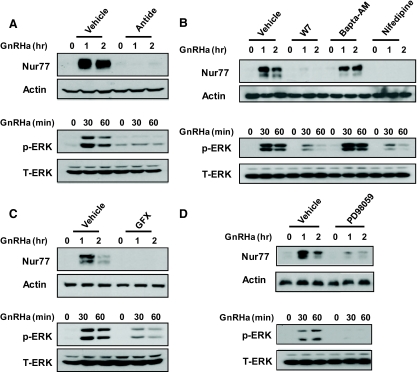
We next used a cadre of pharmacological inhibitors of specific signaling events known to be induced by GnRH and analyzed their effects on both GnRHa-induced Nur77 expression and ERK activation. Pretreatment of cells with the GnRHR-specific antagonist antide abrogated both ERK activation and Nur77 up-regulation after GnRHa treatment (Fig. 2A). We have previously demonstrated that calcium influx through voltage-dependent calcium channels is required for ERK activation by GnRH (21, 22). Inhibition of both calmodulin and calcium flux through L-type voltage gated calcium channels with the compounds W7 and nifedipine, respectively, blocked GnRH-induced Nur77 expression coincident with a blunted ERK response (Fig. 2B). In contrast, inhibition of intracellular calcium fluxes with the intracellular calcium chelator bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid, tetra(acetoxymethyl)-ester (Bapta-AM) did not appear to affect GnRHa-induced Nur77 levels and was without effect on ERK activation (Fig. 2B). As a positive control, this dose of Bapta-AM was sufficient to reduce GnRHa-induced c-Jun N-terminal kinase activity in these same studies (data not shown) as previously shown (22). Inhibition of PKC isozymes with GF109203X (GFX) similarly attenuated both Nur77 induction and ERK activation (Fig. 2C). Finally, inhibition of MAPK kinase (MEK)-1/2 with the specific inhibitor PD98059 decreased GnRHa-induced Nur77 up-regulation and predictably abolished ERK activation (Fig. 2D). Together these results reinforce the requirement for ERK signaling for GnRH-induced Nur77 up-regulation in αT3-1 cells and provide further demonstration of the roles of PKC, extracellular calcium influx, and calmodulin in mediating the effects of GnRH action on this immediate early gene.
Fig. 2.
Analysis of signaling requirements for Nur77 up-regulation in αT3-1 cells by GnRHa. Serum-starved (2 h) αT3-1 cells were pretreated for 30 min with either vehicle or the indicated pharmacological agents. Cells were then exposed to a time course (either 1 or 2 h, as indicated) of GnRHa (10 nm) treatment. Lysates from cells exposed to a 2-h time course of GnRHa were analyzed by immunoprecipitation (IP) followed by immunoblotting for Nur77 up-regulation (upper portion of each panel). Actin levels within the lysates are shown as a control for the input of the IP. Lysates from cells treated in parallel over a 1-h time course were analyzed by immunoblot for levels of activated ERK (p-ERK; lower portion of panels). Total ERK (T-ERK) immunoreactivity is shown as a lane loading control for these samples. Each experiment was repeated at least three times with similar results. A, Effects of the GnRH receptor antagonist antide (100 nm). B, Effects of calmodulin inhibition (W7; 15 μm), chelation of intracellular calcium (Bapta-AM; 50 μm), or blockade of voltage-dependent calcium channels (nifedipine; 1 μm). C, Effects of PKC blockade by GFX109203X (GFX; 1 μm). D, Effects of the MEK inhibitor, PD98059 (50 μm).
Our pharmacological data indicate that PKC activation, calcium, calmodulin, and ERK signaling are required for GnRHa-induced up-regulation of Nur77 in αT3-1 cells. However, in this cell line, as in other systems, ERK activation itself has been shown to be dependent on PKC, calcium, and calmodulin (1, 21, 22, 30, 33). Therefore, to determine whether ERK signaling alone is sufficient for GnRHa-induced transcriptional up-regulation of Nur77 in αT3-1 cells, we used an expression vector encoding the c-Raf kinase fused to the CAAX membrane-targeting sequence from the small GTP-binding protein ras (Raf-CAAX). Raf-CAAX associates constitutively with the plasma membrane in which it induces sustained and high level activation of the ERK pathway independent of extracellular stimuli.
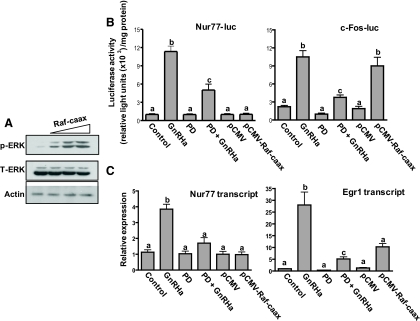
Transient overexpression of Raf-CAAX in αT3-1 cells led to phosphorylation of ERK1/2 in a dose-dependent manner (Fig. 3A). Cells were then transfected with a luciferase reporter containing 1566 bp of the mouse Nur77 proximal promoter (Nur77-luc). In addition, some cells were transfected with either pCMV-Raf-CAAX or the empty control vector (pCMV). After transfection, cells were exposed to either vehicle or the MEK inhibitor PD98059, followed by treatment with either vehicle or GnRHa. GnRHa stimulation led to activation of the Nur77 luciferase reporter; however, this effect was significantly reduced by pharmacological inhibition of ERK signaling (Fig. 3B). Significant increases in Nur77 luciferase reporter activity were not observed after transfection of Raf-CAAX alone (Fig. 3B). To verify the ability of Raf-CAAX to regulate a transcriptional response in this experimental paradigm, we repeated the study using a c-Fos luciferase reporter (c-Fos-luc; Fig. 3B). GnRHa induced an increase in the activity of the c-Fos-luc reporter, which was largely abrogated by inhibition of MEK1/2 with PD98059. Raf-CAAX also induced an increase in c-Fos-luc activity demonstrating the ability of Raf-CAAX to activate a MEK-ERK-responsive promoter (Fig. 3B); importantly, this effect was blunted by pretreatment with PD98059 (not shown).
Fig. 3.
ERK signaling is required, but not sufficient, for GnRH-induced transcriptional up-regulation of Nur77 in αT3-1 cells. A, αT3-1 cells were transfected for 4 h with increasing quantities (0.25, 1.0, or 2.5 μg) of Raf-CAAX expression vector. After an additional 4 h incubation in complete media, cell lysates were analyzed by immunoblot for levels of activated ERK1/2 (p-ERK). Total ERK (T-ERK) and actin immunoreactivity are shown as lane loading controls. B, αT3-1 cells were transfected with a Nur77-luciferase reporter (Nur77-luc, left panel) or a c-Fos-luciferase reporter (c-Fos-luc, right panel). In addition, some cells were transfected with empty control vector or a Raf-CAAX expression vector to stimulate hormone-independent activation of the ERK pathway. Cells were pretreated or not as indicated with the MEK inhibitor PD90859 (PD; 50 μm), and/or the GnRH agonist buserelin (GnRHa; 10 nm). Bars represent mean ± sem normalized luciferase activity of three replicates within a single experiment. Each experiment was repeated at least three times with similar results. Bars with different letter designations represent mean values that are statistically significantly different at P < 0.05. C, αT3-1 cells were pretreated or not with PD98059 or were transfected with empty vector or Raf-CAAX as indicated and were then treated with either vehicle or GnRHa as in A. Bars represent mean ± sem (n = 3) endogenous Nur77 (left panel) or Egr1 (right panel) transcript levels as measured by qPCR. Bars with different letter designations represent mean values that are statistically significantly different at P < 0.05. These experiments were repeated at least three times with similar results.
To assess the effect of ERK signaling on the endogenous Nur77 gene in αT3-1 cells, we treated cells as described above (Fig. 3B) and then measured Nur77 transcript levels by quantitative RT-PCR (Fig. 3C). The inductive effect of GnRHa on Nur77 transcript levels was blocked by pharmacological inhibition of the ERK pathway (Fig. 3C). Consistent with our observations on the Nur77 luciferase reporter, overexpression of Raf-CAAX had no effect on endogenous Nur77 transcript levels (Fig. 3C). As a positive control, we measured endogenous transcript levels of the ERK-responsive immediate early gene Egr1 in these samples. Treatment with GnRHa led to an approximately 27-fold increase in Egr1 transcript levels within 1 h; however, this induction was blocked by inhibition of MEK1/2 (Fig. 3C). Raf-CAAX led to a more modest (∼10-fold) elevation in Egr1 transcript levels; however, this change in expression level did not reach statistical significance. Collectively, these results provide evidence that ERK signaling is required, but alone does not appear to be sufficient, for GnRHa-induced transcriptional up-regulation of Nur77 in αT3-1 cells.
Nur77 is expressed in mouse pituitary gonadotropes
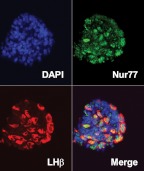
To assess the lineage-specific expression and subcellular localization of Nur77 in primary mouse pituitary cells, we examined pituitary sections from randomly cycling adult female mice using fluorescence immunolabeling of Nur77 and LHβ. Nur77 was expressed in many cells throughout the anterior pituitary parenchyma and colocalized extensively with LHβ immunoreactivity. Within LHβ-positive gonadotropes, Nur77 was localized predominantly to the nucleus (Fig. 4).
Fig. 4.
Nur77 is expressed in mouse pituitary gonadotropes. Expression of Nur77 in gonadotropes was examined by immunofluorescent colabeling of sections on mouse pituitary using fluorescein isothiocyanate (FITC)-conjugated antibodies against Nur77 and Texas Red-conjugated antibodies against LHβ. Individual labeling of Nur77 (green) and LHβ (red) as well as simultaneous visualization of both wavelengths (merge) and 4′,6′-diamino-2-phenylindole (DAPI) is shown.
Our initial studies provided important evidence that Nur77 protein was induced by GnRHa administration. Induced Nur77 immunoreactivity appeared as multiple closely spaced bands on the immunoblot, suggesting posttranslational modification (Fig. 1B). Calf intestine-derived phosphatase treatment of the immunoprecipitates resolved the Nur77 immunoreactivity into a more uniform size of approximately 74 kDa (data not shown), supporting the hypothesis that posttranslational modification by phosphorylation may be central to modulating Nur77 activity. In an effort to examine this possibility, we used a Gal4 DNA binding domain-Nur77 fusion protein. For these studies, an expression vector for Gal4-Nur77 was cotransfected in αT3-1 cells along with a luciferase reporter gene carrying five Gal4 DNA binding sites upstream of a minimal promoter (34). GnRHa administration induced a robust up-regulation of Gal4-Nur77 transcriptional activity in αT3-1 cells (Fig. 5). This response was due to the presence of Nur77 in this fusion protein as similar control studies revealed that the Gal4 DNA binding domain alone was unresponsive to GnRHa administration (data not shown). The effects of GnRHa on Gal4-Nur77 were blocked by pretreatment with GFX but not by PD98059. Thus, Gal4-Nur77 transcription stimulating capabilities induced by GnRH are dependent on GnRH-induced PKC activity and not signaling through the ERK pathway, clearly separating mechanisms involved with GnRH-induced Nur77 biosynthesis with posttranslational modifications necessary for Nur77 activity.
Fig. 5.
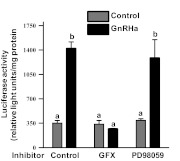
Nur77 transcription stimulating activity induced by GnRH requires PKC but not ERK signaling. An expression vector encoding a Gal4 DNA binding domain fused in frame with full length Nur77 was cotransfected into αT3-1 cells along with a luciferase reporter containing five Gal4 DNA binding sites upstream of luciferase. Transfected cells were pretreated with dimethylsulfoxide (vehicle), GFX109203X (GFX; 1 μm) or PD98059 (50 μm) for 30 min followed by application of either saline or GnRHa for 6 h. Luciferase activity was then determined in cell lysates. Bars represent mean ± sem normalized luciferase activity carried out in triplicate within a single experiment. Each experiment was repeated three times with similar results. Bars with different letter designations represent mean values that are statistically significantly different at P < 0.05.
Generation and validation of a pituitary specific c-Raf knockout mouse
Although the αT3-1 cell studies outlined above are compelling, we sought to examine the link between ERK signaling and Nur77 expression in differentiated gonadotropes. We developed a model by which to compare the roles of c-Raf and ERK activity in mediating the Nur77 transcriptional response to GnRH. Previously we described a mouse model with pituitary targeted ablation of ERK1 and ERK2 that resulted in female infertility due to LH deficiency (5). Using a similar genetic approach, we generated mice with pituitary-targeted conditional ablation of c-Raf kinase (c-Raf CKO; see Supplemental Methods, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org). We had previously demonstrated an involvement of c-Raf kinase within the GnRH signaling network leading to ERK activation in αT3-1 cells (21, 22) as well as participation of c-Raf kinase within membrane rafts necessary for productive GnRHR signaling to the ERK pathway (35, 36). However, in none of these studies did we investigate a required role for c-Raf kinase leading to activation of ERK signaling via a canonical Raf-MEK-ERK pathway. Mice homozygous for a floxed mutation at the c-Raf locus (c-Raf fl/fl), with αGSU:Cre mice, in which Cre recombinase is expressed under the regulatory control of a 4.6-kb fragment of the murine glycoprotein hormone α-subunit (αGSU) promoter. Cre-dependent recombination at the c-Raf locus was assessed by PCR in c-Raf fl/fl,CRE+, and c-Raf fl/fl,CRE− animals using primers flanking the floxed region of the c-Raf locus (Supplemental Fig. 1A). Products indicating the excision of the floxed region of the c-Raf gene were produced only from pituitary genomic DNA of Cre+ individuals, indicating tissue-specific susceptibility of the c-Raf locus to Cre-mediated recombination (Supplemental Fig. 1B). Similar PCR studies using genomic DNA from ovaries and testis in Cre+ animals did not demonstrate evidence for gene recombination (data not shown). To further verify this transgenic model, we compared pituitary transcript levels of c-Raf in c-Raf fl/fl, CRE+, and c-Raf fl/fl,CRE− animals using qPCR. Transcript levels of c-Raf were significantly lower in the pituitaries of Cre-expressing animals as compared with controls (Supplemental Fig. 1C). In contrast, B-Raf transcript levels were not significantly different between c-Raf CKO and control mice (Supplemental Fig. 1C). The degree of reduction in c-Raf transcript levels is consistent with the fact that only a minority of cells within the pituitary express the αGSU and would be expected to be rendered c-Raf deficient in this model.
Pituitary-specific c-Raf fl/fl,CRE+ CKO males and females were viable and fertile. They were born at expected Mendelian frequencies, grew at the same rate as control littermates (Supplemental Fig. 2A), and were grossly and histologically indistinguishable from either control littermates or wild-type mice at adulthood (data not shown). Litter sizes from matings between control animals were not significantly different from litter sizes from matings between c-Raf fl/fl,CRE+ CKO animals (Supplemental Fig. 3). Interestingly, these observations support the conclusion that c-Raf is not required for the function of differentiated gonadotropes.
Pharmacological inhibition of c-Raf kinase does not inhibit GnRHa-induced activation of ERK1/2 in αT3-1 cells
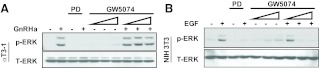
The genetic studies outlined above provide important evidence that c-Raf kinase conditional deletion within gonadotropes does not affect fertility. This is in stark contrast to ERK1 and ERK2 CKO using the identical Cre genetic background (5). Surprisingly, the differences in fertility comparing these two systems suggest that c-Raf kinase is not required for GnRH-induced ERK activity in gonadotropes. In an effort to examine this hypothesis, studies were carried out using the c-Raf kinase inhibitor GW5074 (Fig. 6). Pretreatment of αT3-1 cells with increasing doses of GW5074 (1–10 μm) had no appreciable effect on GnRHa-induced ERK activity, whereas pretreatment with PD98059 abolished ERK activation by GnRH. To confirm the efficacy of the GW5074 compound, NIH-3T3 cells were pretreated with either PD98059 or GW5074 followed by administration of epidermal growth factor (EGF). EGF receptor activation in NIH-3T3 cells resulted in increased ERK phosphorylation that was blocked by both PD98059 and GW5074, suggesting that, indeed, the c-Raf kinase inhibitor functioned as expected with growth factor stimulation (Fig. 6). These pharmacological experiments yielded the unexpected finding that phosphorylation of ERK1/2 is unaffected by pretreatment of αT3-1 cells with the c-Raf inhibitor GW5074. This supported the intriguing hypothesis that GnRH-induced activation of the ERK pathway was independent of c-Raf catalytic activity.
Fig. 6.
Comparative effects of c-Raf inhibition on GnRH- and EGF-induced ERK activation in αT3-1 and NIH-3T3 cells. Serum-starved (2 h) αT3-1 (A) or NIH-3T3 (B) cells were pretreated for 30 min as indicated with vehicle, PD98059 (PD), or increasing concentrations (1, 5, or 10 μm) of the c-Raf inhibitor GW5074. Cells were then exposed to vehicle, GnRHa, or EGF for 10 min. Cell lysates were analyzed by immunoblot for levels of activated ERK (p-ERK). Total ERK immunoreactivity (T-ERK) is shown as a lane loading control. Experiments were performed at least three times with similar results.
Basal and GnRHa-induced transcriptional up-regulation of Nur77 in primary mouse gonadotropes requires ERK signaling but is independent of c-Raf
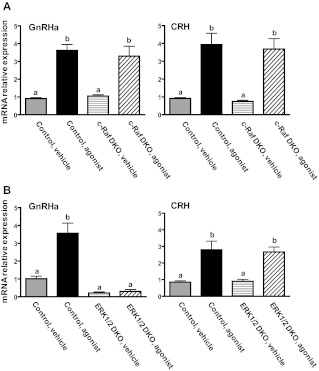
To address the requirements for c-Raf and ERK activity for GnRH-induced expression of Nur77 transcript levels in differentiated gonadotropes, we dispersed whole pituitaries from mice with conditional pituitary deficiency of c-Raf, ERK1/2 or their respective control littermates into primary culture and stimulated the cells with either GnRHa or CRH. Nur77 transcript levels were then measured by qPCR. In pituitary cell cultures from mice with pituitary deficiency of c-Raf, both GnRHa and CRH led to significant increases in Nur77 transcript levels, which were indistinguishable from the response of the controls (Fig. 7A). CRH stimulation led to significant increases in Nur77 transcript abundance in both control mice and mice with pituitary deficiency of ERK1/2 (Fig. 7B). In contrast, although GnRHa stimulation induced significant increases in Nur77 transcript levels in control mice, the effect of GnRHa was completely blocked in ERK-deficient pituitary cells (Fig. 7B). Together these results establish Nur77 as a GnRH-responsive, ERK-dependent immediate early gene in the gonadotrope and further corroborate that c-Raf kinase is not an obligate intermediary in the GnRH-ERK signaling pathway.
Fig. 7.
GnRHa- or CRH-induced transcriptional up-regulation of Nur77 in primary mixed pituitary cell cultures from mice with pituitary-targeted ablation of either c-Raf or ERK1/2. A, Whole pituitaries from mice with αGSU-CRE-mediated pituitary-specific disruption of c-Raf (c-Raf conditional knockout, c-Raf CKO) were dispersed into primary culture. After serum starvation (2 h), cultures were exposed to either vehicle and GnRHa or CRH for 60 min. Nur77 mRNA levels were measured by qPCR. Bars represent mean ± sem for three replicates within an individual experiment. This experiment was conducted on three separate occasions. Bars with different letter designations represent mean values that are statistically significantly different at P < 0.05. B, Whole pituitaries from mice with pituitary-specific compound ablation of ERK1 and ERK2 (ERK1/2 double knockout, ERK1/2 DKO) were dispersed into primary culture and treated as in A. Bars represent mean ± sem for three replicates within an individual experiment. This experiment was conducted on two separate occasions. Bars with different letter designations represent mean values that are statistically significantly different at P < 0.05.
Discussion
Using both cell lines and gonadotropes isolated from mouse models, we show here that the orphan nuclear receptor Nur77 is rapidly and robustly up-regulated in gonadotropes after GnRH stimulation. Activation of the ERK signaling pathway is required but alone is not sufficient for Nur77 transcript induction. These conclusions are based on studies using PD98059 (Fig. 2D), overexpression of the activated form of c-Raf kinase (Raf-CAAX; Fig. 3), and studies carried out in our mouse model of conditional deletion of ERK isoforms in the gonadotrope (Fig. 7B). In these studies, the kinetics and magnitude of Nur77 induction were highly similar between the αT3-1 cell line and primary pituitary gonadotropes (Fig. 1, A and C). This contrasts with a recent report demonstrating sustained expression of Nur77 in the LβT2 cell line after GnRH stimulation (20). The overall correspondence between αT3-1 cells and primary gonadotropes supports the validity of this cell line as a model for study of GnRH-induced Nur77 expression in the gonadotrope.
Interestingly, c-Raf kinase does not appear to be required for GnRH-induced transcriptional up-regulation of Nur77; furthermore, our data reveal that c-Raf does not appear to be the primary MAPK-activating kinase kinase of the ERK pathway within these cells. Our original view of the architecture of the GnRH signaling pathway leading to ERK activation was based on the conical Raf-MEK-ERK cascade (recently reviewed in Refs. 37 and 38). Our previous studies implicating c-Raf kinase signaling within the GnRH pathway was largely based on the effects of Raf-CAAX overexpression and GnRH-induced electrophoretic mobility shifts of c-Raf kinase in immunoblot analyses of αT3-1 cell lysates suggestive of phosphorylation and putative activation (21). The present studies are the first to directly examine the role of c-Raf kinase in mediating GnRH-induced ERK activation. Our observations that GnRH-induced ERK activation is independent of c-Raf in the gonadotrope are consistent with many contemporary reports on the cellular functions of c-Raf kinase (39–41). A growing body of evidence indicates that MEK kinase activation is not a primary function of c-Raf. Gene ablation studies showed that c-Raf-deficient mice die at approximately embryonic d 10.5 of gestation due to widespread apoptosis; however, growth factor-induced ERK activation remained unimpaired in c-Raf-deficient embryonic stem cells, suggesting that the developmental defect in these mice was unrelated to the loss of a critical MEK kinase (42). This interesting finding is in contrast to the present data (Fig. 6B), indicating c-Raf kinase is indeed required for EGF-induced ERK activation in NIH-3T3 cells. This difference may be due putatively to a different intracellular context comparing embryonic stem cells to fibroblasts in culture.
The lethal phenotype in the c-Raf-deficient mouse was rescued by reintroduction of a catalytically inactive form of c-Raf showing that the requirement for c-Raf during development is independent of its MEK kinase catalytic activity (43). Recent reports using conditional targeted gene ablation approaches indicate instead that c-Raf may serve a primary role as a negative regulator of apoptosis in vivo (41, 44–46). The mechanism underlying down-regulation of sensitivity to apoptotic stimuli has been linked to direct c-Raf mediated inhibition of the proapoptotic proteins apoptosis signal-regulating kinase 1 or mammalian STE20-like protein kinase (41, 47). In our examination of mice with pituitary targeted ablation of c-Raf, we found no differences in gonadotrope cell numbers between c-Raf deficient animals and littermate controls (data not shown). Thus, targeted ablation of c-Raf in pituitary gonadotropes appears to have no effect on apparent cellular phenotype. However, gonadotropes are highly differentiated postmitotic cells and, given the critical importance of these cells for reproductive function, it may be reasonable to speculate that they have evolved mechanisms for multifaceted resistance to apoptotic stimuli. We cannot rule out the possibility of incomplete Cre-mediated recombination and that residual expression of c-Raf kinase is sufficient to avert a more severe phenotype. This seems unlikely because the use of pharmacological inhibition of c-Raf kinase did not interfere with GnRH-induced ERK activation in vitro. Moreover, our studies clearly cannot rule out a novel role for other Raf family members playing a redundant role in GnRH signaling to ERK. Little is known regarding the expression pattern and signaling role of B-Raf and A-Raf in gonadotropes. Based on the present studies, B-Raf is expressed within the mouse pituitary (see Supplemental Fig. 1), at least at the transcript level. In AtT20 cells (a corticotrope cell line), B-Raf is expressed and appears to play an important role in the regulation of ERK activity via CRH in this cell line (27). Future studies will be necessary to clarify a potential role of other Raf family members serving as an MAPK-activating kinase kinase in the gonadotrope.
Nur77 has been shown to serve a variety of physiological roles in different cells types but functions most commonly as a transcriptional regulator. Monomeric Nur77 binds to a consensus octanucleotide response element (nerve growth factor inhibitor-B response element) consisting of sequence AAAGGTCA and may function as a transcriptional activator or repressor (12, 48–50). Nur77 may also dimerize with other members of the NR4A family (Nor-1 or Nurr1) and bind to a bipartite target DNA motif, the Nur response element (51). Alternatively, Nur77 may dimerize with retinoid X receptor and bind to a DR5 element in a retinoic acid-dependent manner (51, 52). A recent genomic analysis revealed 483 genes in the human genome that possess a nerve growth factor inhibitor-B response element within their proximal promoter, highlighting the possibility that Nur77 may exert broad transcriptional regulatory influence in various contexts (53). Clarification of the physiological role(s) of Nur77 has been confounded by the observation that Nur77-deficient mice are viable and fertile and display only relatively subtle phenotypic abnormalities associated with dysregulation of central dopaminergic neuronal pathways (10). However, a high degree of functional redundancy between Nur77 and the closely related NR4A family member Nor-1 may underlie the disparities between reported gene regulatory roles of Nur77 and the lack of overt phenotypic abnormalities in the Nur77-deficient mouse (54, 55). Elucidation of physiologically relevant targets of Nur77 in the gonadotrope after a GnRH stimulus will likely require a genetic approach involving tissue-specific compound deletion of both Nur77 and Nor-1.
Supplementary Material
Acknowledgments
We thank Dr. J. Wakshlag (Cornell University) for the NIH 3T3 cells and Dr. P. Mellon (University of California at San Diego) for the αT3-1 cells. Appreciation is also expressed for the generosity of Drs. J. L. Guan (University of Michigan, Ann Arbor, MI), L. Van Aelst (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY), G. Romero (University of Pittsburgh, Pittsburgh, PA), and R. Maurer (Oregon Health Sciences University, Portland, OR) for providing plasmid reagents for these studies.
This work was supported by National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development Grants F32 HD044379 (to S.P.B.) and R01HD34722 (to M.S.R.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- Bapta-AM
- Bis-(o-aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid, tetra(acetoxymethyl)-ester
- EGF
- epidermal growth factor
- FBS
- fetal bovine serum
- GFX
- GF109203X
- GnRHa
- GnRH agonist
- GnRHR
- GnRH receptor
- αGSU
- glycoprotein hormone α-subunit
- MEK
- MAPK kinase
- PKC
- protein kinase C
- qPCR
- quantitative PCR
- Raf-CAAX
- c-Raf kinase fused to the CAAX membrane-targeting sequence from the small GTP-binding protein ras
- SDS
- sodium dodecyl sulfate
- TBST
- Tris-HCl, NaCl, and Tween 20.
References
- 1. Liu F, Austin DA, Mellon PL, Olefsky JM, Webster NJ. 2002. GnRH activates ERK1/2 leading to the induction of c-fos and LHβ protein expression in LβT2 cells. Mol Endocrinol 16:419–434 [DOI] [PubMed] [Google Scholar]
- 2. Salisbury TB, Binder AK, Nilson JH. 2008. Welcoming β-catenin to the gonadotropin-releasing hormone transcriptional network in gonadotropes. Mol Endocrinol 22:1295–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Call GB, Wolfe MW. 1999. Gonadotropin-releasing hormone activates the equine luteinizing hormone β promoter through a protein kinase C/mitogen-activated protein kinase pathway. Biol Reprod 61:715–723 [DOI] [PubMed] [Google Scholar]
- 4. Xie J, Bliss SP, Nett TM, Ebersole BJ, Sealfon SC, Roberson MS. 2005. Transcript profiling of immediate early genes reveals a unique role for activating transcription factor 3 in mediating activation of the glycoprotein hormone α-subunit promoter by gonadotropin-releasing hormone. Mol Endocrinol 19:2624–2638 [DOI] [PubMed] [Google Scholar]
- 5. Bliss SP, Miller A, Navratil AM, Xie J, McDonough SP, Fisher PJ, Landreth GE, Roberson MS. 2009. ERK signaling in the pituitary is required for female but not male fertility. Mol Endocrinol 23:1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanasaki H, Bedecarrats GY, Kam KY, Xu S, Kaiser UB. 2005. Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LβT2 cells. Endocrinology 146:5503–5513 [DOI] [PubMed] [Google Scholar]
- 7. Roberson MS, Misra-Press A, Laurance ME, Stork PJ, Maurer RA. 1995. A role for mitogen-activated protein kinase in mediating activation of the glycoprotein hormone α-subunit promoter by gonadotropin-releasing hormone. Mol Cell Biol 15:3531–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang T, Mulvaney JM, Roberson MS. 2001. Activation of mitogen-activated protein kinase phosphatase 2 by gonadotropin-releasing hormone. Mol Cell Endocrinol 172:79–89 [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Fortin J, Lamba P, Bonomi M, Persani L, Roberson MS, Bernard DJ. 2008. Activator protein-1 and smad proteins synergistically regulate human follicle-stimulating hormone β-promoter activity. Endocrinology 149:5577–5591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. St. Hilaire M, Bourhis E, Lévesque D, Rouillard C. 2006. Impaired behavioural and molecular adaptations to dopamine denervation and repeated L-DOPA treatment in Nur77-knockout mice. Eur J Neurosci 24:795–805 [DOI] [PubMed] [Google Scholar]
- 11. Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. 2006. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med 12:1048–1055 [DOI] [PubMed] [Google Scholar]
- 12. Pols TW, Ottenhoff R, Vos M, Levels JH, Quax PH, Meijers JC, Pannekoek H, Groen AK, de Vries CJ. 2008. Nur77 modulates hepatic lipid metabolism through suppression of SREBP1c activity. Biochem Biophys Res Commun 366:910–916 [DOI] [PubMed] [Google Scholar]
- 13. Zeng H, Qin L, Zhao D, Tan X, Manseau EJ, Van Hoang M, Senger DR, Brown LF, Nagy JA, Dvorak HF. 2006. Orphan nuclear receptor TR3/Nur77 regulates VEGF-A-induced angiogenesis through its transcriptional activity. J Exp Med 203:719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonta PI, van Tiel CM, Vos M, Pols TW, van Thienen JV, Ferreira V, Arkenbout EK, Seppen J, Spek CA, van der Poll T, Pannekoek H, de Vries CJ. 2006. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol 26:2288–2294 [DOI] [PubMed] [Google Scholar]
- 15. Fernandez PM, Brunel F, Jimenez MA, Saez JM, Cereghini S, Zakin MM. 2000. Nuclear receptors Nor1 and NGFI-B/Nur77 play similar, albeit distinct, roles in the hypothalamo-pituitary-adrenal axis. Endocrinology 141:2392–2400 [DOI] [PubMed] [Google Scholar]
- 16. Kovalovsky D, Paez Pereda M, Labeur M, Renner U, Holsboer F, Stalla GK, Arzt E. 2004. Nur77 induction and activation are necessary for interleukin-1 stimulation of proopiomelanocortin in AtT-20 corticotrophs. FEBS Lett 563:229–233 [DOI] [PubMed] [Google Scholar]
- 17. Martin LJ, Tremblay JJ. 2005. The human 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase type 2 promoter is a novel target for the immediate early orphan nuclear receptor Nur77 in steroidogenic cells. Endocrinology 146:861–869 [DOI] [PubMed] [Google Scholar]
- 18. Song KH, Park JI, Lee MO, Soh J, Lee K, Choi HS. 2001. LH induces orphan nuclear receptor Nur77 gene expression in testicular Leydig cells. Endocrinology 142:5116–5123 [DOI] [PubMed] [Google Scholar]
- 19. Wurmbach E, Yuen T, Ebersole BJ, Sealfon SC. 2001. Gonadotropin-releasing hormone receptor-coupled gene network organization. J Biol Chem 276:47195–47201 [DOI] [PubMed] [Google Scholar]
- 20. Hamid T, Malik MT, Millar RP, Kakar SS. 2008. Protein kinase A serves as a primary pathway in activation of Nur77 expression by gonadotropin-releasing hormone in the LβT2 mouse pituitary gonadotroph tumor cell line. Int J Oncol 33:1055–1064 [PubMed] [Google Scholar]
- 21. Mulvaney JM, Zhang T, Fewtrell C, Roberson MS. 1999. Calcium influx through L-type channels is required for selective activation of extracellular signal-regulated kinase by gonadotropin-releasing hormone. J Biol Chem 274:29796–29804 [DOI] [PubMed] [Google Scholar]
- 22. Mulvaney JM, Roberson MS. 2000. Divergent signaling pathways requiring discrete calcium signals mediate concurrent activation of two mitogen-activated protein kinases by gonadotropin-releasing hormone. J Biol Chem 275:14182–14189 [DOI] [PubMed] [Google Scholar]
- 23. Sadie H, Styger G, Hapgood J. 2003. Expression of the mouse gonadotropin-releasing hormone receptor gene in αT3-1 gonadotrope cells is stimulated by cyclic 3′,5′-adenosine monophosphate and protein kinase A, and is modulated by steroidogenic factor-1 and Nur77. Endocrinology 144:1958–1971 [DOI] [PubMed] [Google Scholar]
- 24. Lim S, Luo M, Koh M, Yang M, bin Abdul Kadir MN, Tan JH, Ye Z, Wang W, Melamed P. 2007. Distinct mechanisms involving diverse histone deacetylases repress expression of the two gonadotropin β-subunit genes in immature gonadotropes, and their actions are overcome by gonadotropin-releasing hormone. Mol Cell Biol 27:4105–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Darragh J, Soloaga A, Beardmore VA, Wingate AD, Wiggin GR, Peggie M, Arthur JS. 2005. MSKs are required for the transcription of the nuclear orphan receptors Nur77, Nurr1 and Nor1 downstream of MAPK signalling. Biochem J 390:749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bourhis E, Maheux J, Rouillard C, Lévesque D. 2008. Extracellular signal-regulated kinases (ERK) and protein kinase C (PKC) activities are involved in the modulation of Nur77 and Nor-1 expression by dopaminergic drugs. J Neurochem 106:875–888 [DOI] [PubMed] [Google Scholar]
- 27. Kovalovsky D, Refojo D, Liberman AC, Hochbaum D, Pereda MP, Coso OA, Stalla GK, Holsboer F, Arzt E. 2002. Activation and induction of NUR77/NURR1 in corticotrophs by CRH/cAMP: involvement of calcium, protein kinase A, and MAPK pathways. Mol Endocrinol 16:1638–1651 [DOI] [PubMed] [Google Scholar]
- 28. Sakaue M, Adachi H, Dawson M, Jetten AM. 2001. Induction of Egr-1 expression by the retinoid AHPN in human lung carcinoma cells is dependent on activated ERK1/2. Cell Death Differ 8:411–424 [DOI] [PubMed] [Google Scholar]
- 29. van den Brink MR, Kapeller R, Pratt JC, Chang JH, Burakoff SJ. 1999. The extracellular signal-regulated kinase pathway is required for activation-induced cell death of T cells. J Biol Chem 274:11178–11185 [DOI] [PubMed] [Google Scholar]
- 30. Roberson MS, Bliss SP, Xie J, Navratil AM, Farmerie TA, Wolfe MW, Clay CM. 2005. Gonadotropin-releasing hormone induction of extracellular-signal regulated kinase is blocked by inhibition of calmodulin. Mol Endocrinol 19:2412–2423 [DOI] [PubMed] [Google Scholar]
- 31. Antonyak MA, McNeill CJ, Wakshlag JJ, Boehm JE, Cerione RA. 2003. Activation of the Ras-ERK pathway inhibits retinoic acid-induced stimulation of tissue transglutaminase expression in NIH3T3 cells. J Biol Chem 278:15859–15866 [DOI] [PubMed] [Google Scholar]
- 32. Xie J, Roberson MS. 2008. 3′, 5′-Cyclic adenosine 5′-monophosphate response element-dependent transcriptional regulation of the secretogranin II gene promoter depends on gonadotropin-releasing hormone-induced mitogen-activated protein kinase activation and the transactivator activating transcription factor 3. Endocrinology 149:783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stocco CO, Lau LF, Gibori G. 2002. A calcium/calmodulin-dependent activation of ERK1/2 mediates JunD phosphorylation and induction of nur77 and 20α-HSD genes by prostaglandin F2α in ovarian cells. J Biol Chem 277:3293–3302 [DOI] [PubMed] [Google Scholar]
- 34. Sun P, Schoderbek WE, Maurer RA. 1992. Phosphorylation of cyclic adenosine 3′,5′-monophosphate (cAMP) response element-binding protein isoforms by the cAMP-dependent protein kinase. Mol Endocrinol 6:1858–1866 [DOI] [PubMed] [Google Scholar]
- 35. Navratil AM, Bliss SP, Berghorn KA, Haughian JM, Farmerie TA, Graham JK, Clay CM, Roberson MS. 2003. Constitutive localization of the gonadotropin-releasing hormone (GnRH) receptor to low density membrane microdomains is necessary for GnRH signaling to ERK. J Biol Chem 278:31593–31602 [DOI] [PubMed] [Google Scholar]
- 36. Bliss SP, Navratil AM, Breed M, Skinner DC, Clay CM, Roberson MS. 2007. Signaling complexes associated with the type I gonadotropin-releasing hormone (GnRH) receptor: colocalization of extracellularly regulated kinase 2 and GnRH receptor within membrane rafts. Mol Endocrinol 21:538–549 [DOI] [PubMed] [Google Scholar]
- 37. Navratil AM, Bliss SP, Roberson MS. 2010. Membrane rafts and GnRH receptor signaling. Brain Res 1364:53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bliss SP, Navratil AM, Xie J, Roberson MS. 2010. GnRH signaling, the gonadotrope and endocrine control of fertility. Front Neuroendocrinol 31:322–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baccarini M. 2005. Second nature: biological functions of the Raf-1 “kinase.” FEBS Lett 579:3271–3277 [DOI] [PubMed] [Google Scholar]
- 40. Hindley A, Kolch W. 2002. Extracellular signal regulated kinase (ERK)/mitogen activated protein kinase (MAPK)-independent functions of Raf kinases. J Cell Sci 115:1575–1581 [DOI] [PubMed] [Google Scholar]
- 41. O'Neill E, Rushworth L, Baccarini M, Kolch W. 2004. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 306:2267–2270 [DOI] [PubMed] [Google Scholar]
- 42. Mikula M, Schreiber M, Husak Z, Kucerova L, Rüth J, Wieser R, Zatloukal K, Beug H, Wagner EF, Baccarini M. 2001. Embryonic lethality and fetal liver apoptosis in mice lacking the c-raf-1 gene. EMBO J 20:1952–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hüser M, Luckett J, Chiloeches A, Mercer K, Iwobi M, Giblett S, Sun XM, Brown J, Marais R, Pritchard C. 2001. MEK kinase activity is not necessary for Raf-1 function. EMBO J 20:1940–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jesenberger V, Procyk KJ, Rüth J, Schreiber M, Theussl HC, Wagner EF, Baccarini M. 2001. Protective role of Raf-1 in Salmonella-induced macrophage apoptosis. J Exp Med 193:353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lamberti A, Longo O, Marra M, Tagliaferri P, Bismuto E, Fiengo A, Viscomi C, Budillon A, Rapp UR, Wang E, Venuta S, Abbruzzese A, Arcari P, Caraglia M. 2007. C-Raf antagonizes apoptosis induced by IFN-α in human lung cancer cells by phosphorylation and increase of the intracellular content of elongation factor 1A. Cell Death Differ 14:952–962 [DOI] [PubMed] [Google Scholar]
- 46. Yamaguchi O, Watanabe T, Nishida K, Kashiwase K, Higuchi Y, Takeda T, Hikoso S, Hirotani S, Asahi M, Taniike M, Nakai A, Tsujimoto I, Matsumura Y, Miyazaki J, Chien KR, Matsuzawa A, Sadamitsu C, Ichijo H, Baccarini M, Hori M, Otsu K. 2004. Cardiac-specific disruption of the c-raf-1 gene induces cardiac dysfunction and apoptosis. J Clin Invest 114:937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen J, Fujii K, Zhang L, Roberts T, Fu H. 2001. Raf-1 promotes cell survival by antagonizing apoptosis signal-regulating kinase 1 through a MEK-ERK independent mechanism. Proc Natl Acad Sci USA 98:7783–7788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rajpal A, Cho YA, Yelent B, Koza-Taylor PH, Li D, Chen E, Whang M, Kang C, Turi TG, Winoto A. 2003. Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor. EMBO J 22:6526–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gruber F, Hufnagl P, Hofer-Warbinek R, Schmid JA, Breuss JM, Huber-Beckmann R, Lucerna M, Papac N, Harant H, Lindley I, de Martin R, Binder BR. 2003. Direct binding of Nur77/NAK-1 to the plasminogen activator inhibitor 1 (PAI-1) promoter regulates TNFα-induced PAI-1 expression. Blood 101:3042–3048 [DOI] [PubMed] [Google Scholar]
- 50. Harant H, Lindley IJ. 2004. Negative cross-talk between the human orphan nuclear receptor Nur77/NAK-1/TR3 and nuclear factor-κB. Nucleic Acids Res 32:5280–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maira M, Martens C, Philips A, Drouin J. 1999. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol 19:7549–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Morita K, Kawana K, Sodeyama M, Shimomura I, Kagechika H, Makishima M. 2005. Selective allosteric ligand activation of the retinoid X receptor heterodimers of NGFI-B and Nurr1. Biochem Pharmacol 71:98–107 [DOI] [PubMed] [Google Scholar]
- 53. Zhao Y, Liu Y, Zheng D. 2008. α1-Antichymotrypsin/serpinA3 is a novel target of orphan nuclear receptor Nur77. FEBS J 275:1025–1038 [DOI] [PubMed] [Google Scholar]
- 54. Kanzleiter T, Schneider T, Walter I, Bolze F, Eickhorst C, Heldmaier G, Klaus S, Klingenspor M. 2005. Evidence for Nr4a1 as a cold-induced effector of brown fat thermogenesis. Physiol Genomics 24:37–44 [DOI] [PubMed] [Google Scholar]
- 55. Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, Conneely OM. 2007. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med 13:730–735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.