Abstract
In mammals, the suprachiasmatic nuclei (SCN) of the hypothalamus control endogenous circadian rhythms and entrainment to the environment. A core SCN region of calbindin (CalB)-containing cells is retinorecipient and the cells therein lack rhythmic expression of clock genes and electrical activity. The core is surrounded by a ‘shell’ of rhythmic oscillator cells. In the present experiments, we studied the spatial arrangement of oscillator cells by examining the spatial and temporal patterns of expression of the canonical clock genes Per1, Per2 and vasopressin mRNA, a clock-controlled gene. Surprisingly, in the SCN shell, the dorsomedial cells were the first to rhythmically express both Per1 and VP mRNA, with gene expression then spreading very slowly through much of the nucleus for the next 12 h then receding to baseline levels. Following a light pulse, Per expression increased after 1 h in the core SCN and after 1.5 h in the shell. Although expression in the shell occurred earlier in light-pulsed animals than in those housed in constant darkness, it still followed the same spatial and temporal expression pattern as was observed in constant darkness. The results suggest that not only is the SCN organized into light-responsive and rhythmic regions but also that the rhythmic region of the SCN itself has an ordered arrangement of SCN oscillator cells.
Keywords: calbindin D-28K, circadian rhythms, in situ hybridization, networks, oscillators, pacemakers, Per1, suprachiasmatic nucleus, Syrian hamster, vasopressin
Introduction
The mammalian circadian clock, located in the suprachiasmatic nucleus (SCN), controls the phase of daily rhythms and synchronizes these rhythmic responses to the local environment. The SCN is made up of bilaterally symmetrical nuclei each containing ≈ 8000–10 000 neurons in rodents (Moore et al., 2002). There is substantial evidence that the SCN can be divided into two broadly distinct regions, namely the ventrolateral or ‘core’ and the dorsomedial or ‘shell’, based on evidence from peptide expression, cytoarchitecture, efferent projections and gene expression (Moore, 1996; Silver et al., 1996; Hamada et al., 2001). Furthermore, in hamsters and mice, some neurons in the core SCN lack circadian rhythmicity in clock gene expression and electrical activity, while the cells in the shell express such rhythms (Hamada et al., 2001; Jobst & Allen, 2002; LeSauter et al., 2003; Karatsoreos et al., 2004). In hamsters, the region of nonrhythmic cells is delineated by a population of calbindin (CalB)-containing neurons that receive direct synaptic retinal input (Bryant et al., 2000). We have recently demonstrated that the expression of CalB protein has a daily rhythm in nuclear localization (Hamada et al., 2003). Importantly, this localization of CalB is related to phase shifting and expression of Per genes in response to a light pulse. During the night, light induction of Per is blocked when expression of CalB is prevented with antisense pretreatment. This evidence is incorporated in a formal model in which the SCN is made of two distinct cell types in which the core gate cells provide a daily organizing signal that maintains phase coherence in the ensemble of shell oscillator cells, while the output of this ensemble regulates the activity of the gate (Antle et al., 2003).
Empirically, the nature of the intra-SCN communication network has been difficult to unravel. It is known that some but not all SCN neurons receive direct retinal input (Moore, 1996; Aioun et al., 1998; Bryant et al., 2000). These core SCN cells communicate with oscillators lying in the shell (Leak et al., 1999), thereby serving to reset oscillator phase. The individual neurons of the SCN are cell-based autonomous oscillators, with differing periods and phases (Welsh et al., 1995; Herzog et al., 1997; Liu et al., 1997; Nakamura et al., 2001; Quintero et al., 2003; Sigworth et al., 2003). Importantly, the tissue as a whole acts in synchrony to generate a coherent rhythm of electrical and metabolic activity (reviewed in Herzog & Schwartz, 2002). Determining the network organization of the individual oscillating units is an important step towards understanding how the tissue as a whole produces this coherent output.
The present study examines the nature of communication within the SCN, focusing on the organization of the SCN shell, by monitoring the spatial and temporal patterns of Per (a clock gene) and VP (a clock-controlled gene) expression under constant conditions, following a light pulse. Individual cells of the SCN rhythmically express Per but out of phase with one another (Quintero et al., 2003). It is not known whether these different populations of oscillating cells within the SCN have a spatially ordered or a random spatial arrangement. The present results indicate highly ordered sequential changes in gene activation within the SCN shell revealing a highly ordered spatial organization of this subregion of the nucleus.
Materials and methods
Animals and housing
Adult male hamsters (Mesocricetus auratus) were given food and water ad libitum. The animal colony room was kept on a 12 : 12-h light : dark cycle (LD), with light intensity of 600 lux. The room was equipped with a white noise generator (91 dB spl) to mask environmental noise. For animals housed in constant darkness, a dim red light (<1 lux; Delta 1, Dallas, TX, USA) allowed for maintenance.
For studies performed under constant conditions, animals were housed in LD for at least 1 week and then placed in constant darkness for at least 1 week in cages equipped with running wheels (diameter, 16 cm) prior to being killed. Locomotor activity was monitored continuously using a computer-based data acquisition system (Data-quest, Data Sciences, St Paul, MN, USA) and this data was used to determine the appropriate circadian time (CT) of treatment and killing.
All handling of animals was done in accordance with the Institutional Animal Care and Use Committee guidelines of Columbia University.
Free-floating digoxigenin (DIG)
Animals were deeply anaesthetized and perfused with 0.1 M phosphate buffer containing 4% paraformaldehyde. Frozen serial sections through the entire extent of the SCN (≈ 15–30 sections; 20–30 μm thick) were collected for in situ hybridization using DIG cRNA probes in free-floating tissue. In situ hybridization was performed as previously described (Hamada et al., 2001).
Quantification of the DIG signal
As previously described, the SCN was divided into four quadrants for analysis (Hamada et al., 2001). The rostral SCN corresponds to the level of this nucleus in fig. 23 of the golden hamster brain atlas (Morin & Wood, 2001), while the central anterior SCN corresponds to fig. 24 and the caudal SCN corresponds to fig. 25. The central-posterior SCN lies between figs 24 and 25 of the atlas, contains the CalB subnucleus, and is characterized by a distinctive ‘X’ shape in VP mRNA staining. For the study of light-induced Per1 and VP mRNA, we focused on the central (anterior and posterior) aspect of the SCN. At this level, both the Per and VP mRNAs are strongly expressed, providing markers for this region, and enabling the detection of both endogenous and phase-shifted expression of these genes. Furthermore, the central–posterior SCN contains the CalB cell population, while the SCN area outside this small population of neurons is devoid of CalB immunoreactivity, providing a highly localized marker within the SCN. Finally, light-induced Per expression is restricted to the CalB region, while rhythmic expression of Per genes is limited to the VP region of the hamster SCN (Hamada et al., 2001), rendering this region suitable for examination of the effect of a light pulse on the Per mRNA and VP mRNA expression.
Sections were photographed on Fuji 35 mm film using a camera (Olympus C-35AD-4) attached to a light microscope (Olympus BH-2) at ≈ 200× and colour prints made. For cell counting, colour photographs were taken of 4 or 5 sections for each animal of the central SCN (anterior and posterior; see Fig. 1), and Per1- or VP-positive cells were counted in each section. Counts were made by an observer blind to the experimental condition of the animal, as follows: first, the mean number of labelled cells was determined for the left and right SCN for each section. Next, the average number of cells for all sections was calculated for each animal. The data is given as mean (± SEM) number of cells per section (30 μm) in the central (anterior and posterior) area of the SCN.
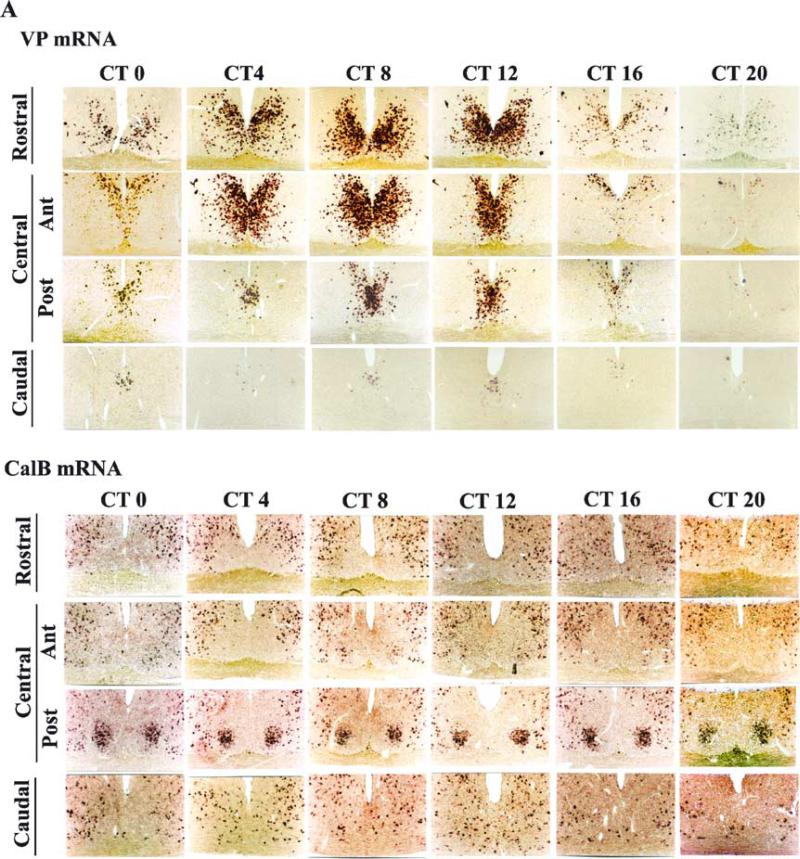
Fig. 1.
The time course and localization of molecular constituents of the SCN, revealing substantial heterogeneity described in the expression of (A) VP, CalB, (B) Per1, and Per2 mRNA in coronal sections of each quadrant of the SCN from its rostral to central (anterior and posterior regions) to caudal aspect. Each column shows serial sections obtained from a single animal.
Experiment 1: temporal and spatial analysis of gene expression in the SCN
To establish the time course of expression of various genes, in situ hybridization for VP, Per1, Per2 and CalB mRNA was performed throughout the SCN at CT 0, 4, 8, 12, 16 and 20. Two series of brains were processed for VP or CalB mRNA, with alternate sections being processed for Per1 mRNA or for Per2 mRNA.
Experiment 2: temporal and spatial analysis of effect of phase resetting cues on induced gene expression in the SCN
To examine gene expression following photic phase shifting, experimental animals were given a light pulse (LP+ 600 lux, 30 min) at CT 19, while control animals were killed at the same time, but not given a light pulse (LP–). For anatomical studies of VP and Per1 mRNA, animals were killed at 1, 1.5, 2, 4, 6 and 9 h after the start of the light pulse. DIG in situ hybridization was performed as described above, with alternate sections being processed for CalB mRNA to allow the CalB region to be delineated in analysed tissue.
Effects of treatments were examined using analysis of variance. When main effects were detected, pair-wise analyses (Fisher's PLSD) were used to determine specific differences.
Results
Experiment 1: temporal and spatial analysis of gene expression in the SCN in DD
The temporal changes in VP mRNA indicated a high amplitude circadian rhythm in the time course of expression (Fig. 1A). The distinct delineation of SCN regions can be seen in the expression of CalB mRNA in the lower panels of this figure, as CalB mRNA expression was high at all circadian times. The initial daily increase in the expression of this clock-controlled gene occurred at ≈ CT 0 and was first seen in the dorsomedial zone in the rostral and central SCN (Fig. 1A rows 1–3, column 1). VP mRNA expression peakrd at ≈ CT 8–12. At this time, it could be detected throughout the rostro-caudal extent of the SCN, with expression occurring more medially at caudal levels. At the time of maximal expression, the distribution of VP mRNA corresponds to the distribution of the VP peptide (Kalsbeek et al., 1993). These results drew our attention to the population of cells lying in the DM area, in the VP region of the rostro-central SCN, as this is the site where the expression of VP was first seen, and the expression of VP mRNA subsequently extended from this point over the next 12 h.
In alternate sections Per1 mRNA expression was first detected at CT 0, peaked at CT 4, was still high at CT 8 and had declined by CT 12. At its peak, some Per1-expressing cells were observed in every section along the rostro-caudal extent of the VP region of the SCN (Fig. 1B). In the rostral half of the nucleus, Per1 was expressed in most of the coronal cross-section, with only the most lateral edges being devoid of endogenous expression. In the caudal half of the SCN, Per1 was expressed primarily in the medial region of the coronal SCN cross-section. During the subjective night, corresponding to the trough of expression, little if any expression was observed in any part of the SCN.
Per2 mRNA was strongly expressed between CT 8 and CT 12 at all rostro-caudal levels of the VP region (Fig. 1B). At peak expression time, Per2 expression was observed in much of the rostral SCN, with only the most lateral and caudal regions being devoid of expression. In the caudal half of the SCN, expression was primarily observed in the dorsomedial region.
Experiment 2: temporal and spatial analysis of photically induced gene expression in the SCN
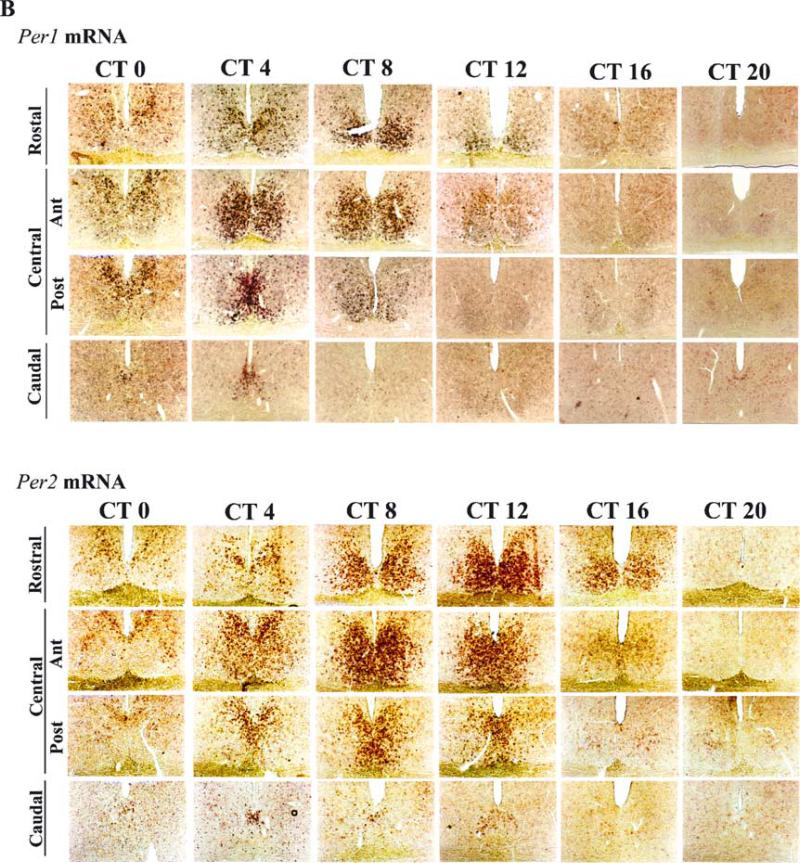
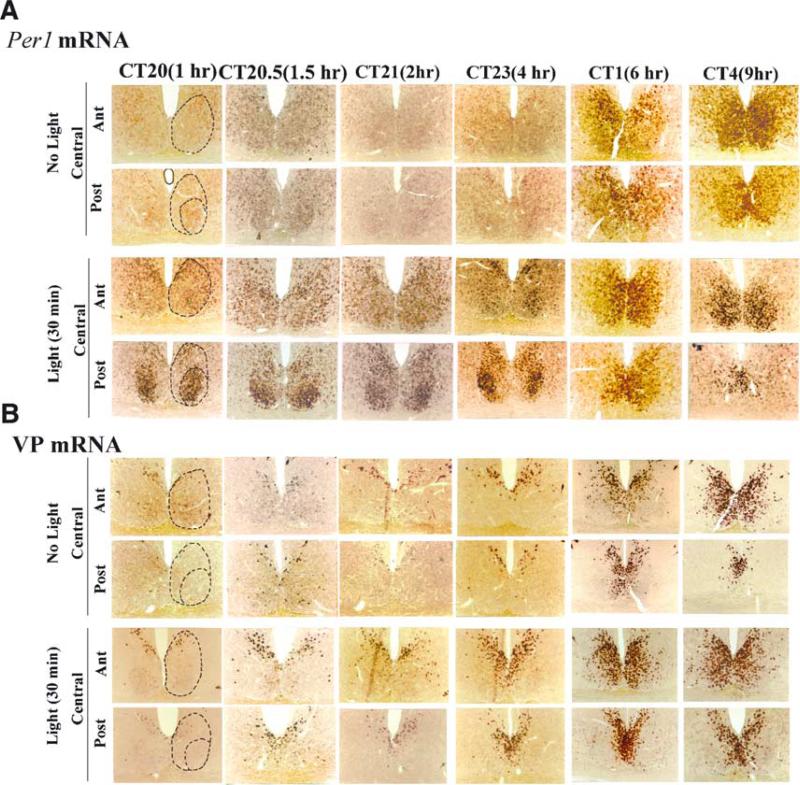
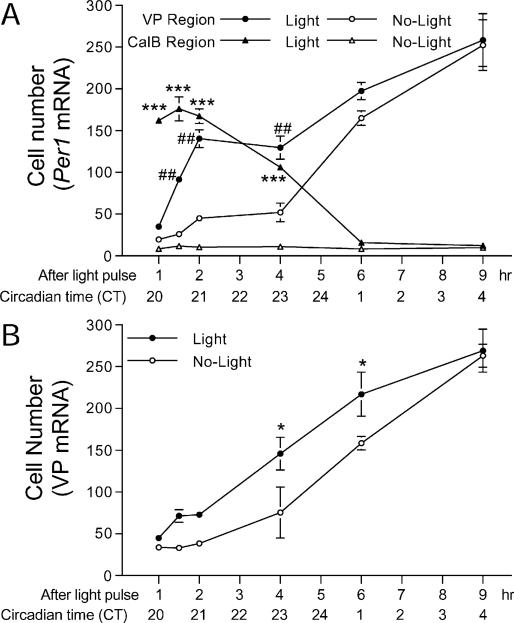
Expression of Per1 mRNA following a light pulse was highest in the CalB region, as previously reported (Hamada et al., 2001; Fig. 2A. Here, the number of Per1-labelled cells increased rapidly and remained high for at least 2 h following the light pulse, fell by 4 h after the light pulse, and reached baseline values by 6 h (F11,15 = 116.37, P < 0.001; Figs 2A and 3A). In contrast, Per1-labelled cells at the central–posterior level of the VP region appeared more slowly, with an increase detectable 1.5 h following the light pulse (F11,15 = 42.26, P < 0.001; Figs 2A and 3A). Although an even greater number were found 6–9 h after the light pulse, the endogenous rhythmic expression of Per1 had started to increase in the control animals by this circadian phase (Figs 2A and 3A).
Fig. 2.
The time course of Per1 and VP mRNA expression after a light pulse.The initial substantial expression of Per1 occurred in the CalB region of the SCN. (A) At ≈ 1–1.5 h after a light pulse, Per1 mRNA subsequently spread to the rest of the VP region. (B) The time course of VP mRNA expression after light pulse.
Fig. 3.
(A) Quantification of the time course of Per1 expression in the central zone of the SCN in animals exposed to a light pulse (LP) and in controls killed at the same circadian times. Comparison of the animals in the LP+ and LP– conditions indicates that significant levels of Per1 mRNA first occurred in the CalB region of the SCN. In this zone, Per1 expression continued to rise for the next hour, then began its gradual decline, reaching baseline values at ≈ 6 h after the LP. In contrast, the light-induced expression of Per1 in the VP region was first detected at 1–1.5 h following the LP and remained at peak values for 2 h following the LP. After this time, the endogenous rhythmic expression of Per1 started to rise (hour 6 or CT 1) and there was no difference between the light-pulsed and control animals. Fisher's test, *P < 0.05, ##P < 0.01, ***P < 0.001. (B) Time-dependent expression of VP mRNA in the central SCN in animals given an LP at CT 19 and in controls killed at the same time but not exposed to an LP. The results indicate that a significant increase in VP mRNA was first detectable at 4 h following the LP, and that this light-induced increase lasted ≈ 2 more hours (to CT 1). After that time (CT 4), endogenous rhythms of VP mRNA were high and no difference was detected between the light-pulsed and control groups.
Following the light pulse, the increase in the number of cells expressing Per1 mRNA was much more rapid than that seen for VP mRNA. While a significant increase in Per1 mRNAwas detected in the VP region at 1.5 h, a significant increase over LP– controls in VP mRNA was found only at 4–6 h following the light pulse (F11,21 = 15.16, P < 0.001; Figs 2B and 3B).
Discussion
This study examines expression of both a canonical clock gene and a clock-controlled gene in the SCN in conditions of constant darkness, and following a light pulse. The main findings are as follows. (i) The daily pattern of changes in Per1 and VP expression within the shell region of the SCN involves a stereotyped sequence of temporal and spatial changes, first seen in the dorsomedial SCN and later in the ventrolateral area. This pattern was particularly striking for VP mRNA expression. It remains to be determined whether this was the product of dorso-ventral differences in individual SCN cells or the result of a gradient of activation of a uniform cell population. (ii) Following a light pulse, the expression of Per1 was first observed in the core SCN region marked by CalB cells. Per1 mRNA was then seen in the shell region marked by VP cells, and was consistent with a phase advance in the expression of this clock gene. (iii) Although gene expression in the SCN shell rose earlier in animals that received a light pulse, the spatial and temporal sequence of changes of this rise in expression were similar to those seen in constant darkness. (iv) The phase of expression of Per1 was advanced with respect to that of VP, consistent with evidence that VP is a clock-controlled gene (Silver et al., 1999; Ripperger et al., 2000; Hamada et al., 2001; Dardente et al., 2002).
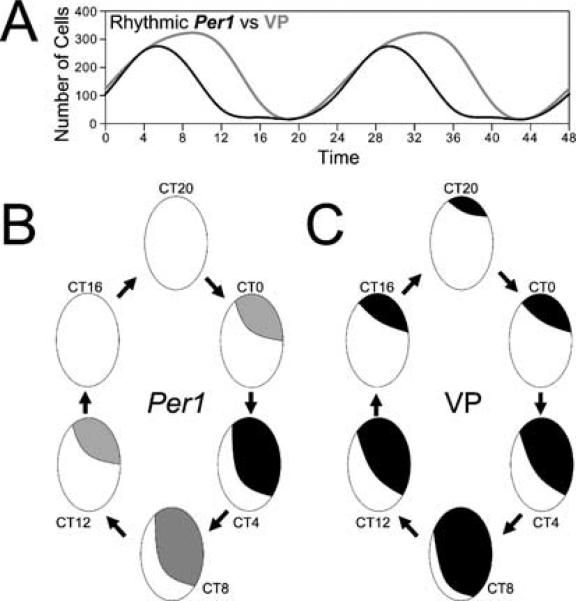
The coordinated temporal and spatial sequences of changes are summarized schematically (Fig. 4). In constant darkness, Per1 and VP expression first rose at CT 0 in the dorsomedial SCN. For both genes, the expression spread ventrally and laterally (Fig. 4D and E). The number of Per1-expressing cells peaked at CT 4, while the VP cells peaked at CT 8. After their respective peaks, expression of both genes declined rapidly, first ventrally and then dorsally. The trough of expression occurred from CT 12–20 for Per1 and from CT 16–20 for VP mRNA. A temporally and spatially organized sequence of changes in Per1 and Per2 mRNA has been described in the rat (Yan & Okamura, 2002; Nagano et al., 2003; Yamaguchi et al., 2003). In these studies, expression was initially observed in the dorsomedial part of the SCN, and subsequently in its ventral and lateral extent. The present study extends the ®ndings of Yan & Okamura (2002) in showing the absence of Per1 expression in the CalB region in DD at all times. Furthermore, this work also documents a robust rhythm of VP mRNA that parallels that of Per1. The similar expression pattern in the hamster and rat of rhythmic Per1 and VP mRNA suggests that this organization is a common property of mammalian SCN.
Fig. 4.
Summary of experimental findings depicting both temporal and spatial expression patterns. (A) Temporal pattern of rhythmic Per1 and VP mRNA levels over two circadian cycles, starting at CT 0. The number of cells expressing each gene initially increased together, although VP expression continued to increase longer than did Per1. (B) Spatial expression of Per1 over the course of a circadian day. One SCN is depicted for each phase (ventricle to the right of each SCN). Low, moderate and high signal density are represented by light grey, medium grey and black shading, respectively. (C) Spatial expression of VP mRNA over the course a circadian day. One SCN is depicted for each phase (ventricle to the right of each SCN).
Following a light pulse (Fig. 3A), Per1 was induced within 60 min in the core SCN region of CalB-expressing cells (Hamada et al., 2001), and was elevated for ≈ 4 h. Per1 expression in the SCN shell occurs later than in the core; it occurred first in the dorsomedial shell at ≈ 90 min after the LP with expression increasing and then spread ventrally for 6 h following the light pulse. The rise in Per1 and VP expression in the shell followed the same temporal and spatial pattern with and without a LP, though the phase was advanced in the former case (Fig. 3A and B), indicating that in the shell region the same sequence of events unfolds in free-running and phase-shifted conditions.
The lack of a difference at CT 4 in the levels of both VP and Per1 mRNA between the LP+ and LP– groups could be due to a broad (≈ 2-h) peak in the expression profile, an explanation we favour given the results observed in Fig. 1A and B. Another contributing factor may be the sampling frequency used, with samples taken from LP– and LP+ animals just before and just after their expression peaks, respectively. Presumably, the decline in expression is also advanced following a light pulse, though this was not tested directly.
Mechanisms of slow signal spread within the shell
We have previously demonstrated that the SCN can be subdivided into a rhythmic and a nonrhythmic region (Hamada et al., 2001). Within the rhythmic region of the SCN, delineated by cells containing VP, it is widely assumed that individual cells are independent weakly coupled oscillators, and that properties of SCN cells are not related to their position in the tissue (Welsh et al., 1995; Herzog et al., 1997; Liu et al., 1997; Nakamura et al., 2001; Herzog & Schwartz, 2002). The present data, particularly that examining VP mRNA expression, is not consistent with this assumption and suggests instead that there is an important structural organization within the SCN shell. In both constant conditions (reflecting endogenous rhythmicity) and following a light pulse (reflecting entrainment), the initial site of increased Per1 and VP mRNA is in a small population of dorsomedial cells. If the population of SCN cells were entirely comprised of uniform, weakly coupled oscillators, one might anticipate random start sites for the daily initial gene expression, and that such expression would spread from this random initiation site (Strogatz & Stewart, 1993). We show instead that cells in the dorsal region continue to express VP and Per1 mRNA for many more hours than do cells in the ventral SCN, as a slowly moving ‘front’ of gene expression first spreads and then retracts through the nucleus.
The mechanism mediating the slow spread of gene expression in the SCN may be the product of differences in individual SCN oscillator cell characteristics, or a property of SCN tissue organization. A cell-based mechanism could rest on a dorsal–ventral gradient of individual cell characteristics. This hypothesis currently has no empirical support (Welsh et al., 1995; Herzog et al., 1997; Nakamura et al., 2001). At the level of SCN tissue organization, there is no evidence of either a gradient in intra-SCN connections (Guldner, 1976; Castel et al., 1990) or diffusible gas signalling or paracrine transmission (Miche & Colwell, 2001; Shirakawa et al., 2001). An alternative hypothesis is that there is a gradient of some molecular property resulting in regional differences in drive on gene promoters. While no data is available to address any of these alternatives, each is empirically testable.
Communication from core to shell
The sequential changes in gene expression following a light pulse can be understood in the context of the network organization of the SCN. The earliest assumption was that the SCN had a linear input–output organization with photic input acting directly on oscillators, which in turn sent a signal to targets (Eskin, 1979). It is known, however, from electron-microscopic studies, that some but not all hamster SCN neurons receive direct retinal input (Aioun et al., 1998; Bryant et al., 2000). How the consequence of photic input to directly retinorecipient SCN cells is communicated to those cells that are not directly retinorecipient is less well documented. Tracing studies indicate that core neurons project to shell, but the reverse does not occur (Leak et al., 1999). Data from expression of gene induction in the SCN following a brief light pulse (fos, Per1, Per2) indicates that initial activation occurs in a core retinorecipient population of neurons within the ventral SCN and later in the rest of the nucleus (Silver et al., 1996; Hamada et al., 2001; Yan & Silver, 2002).
While early views of the SCN as comprised of identical resettable oscillators was convenient in its simplicity, evidence is rapidly accumulating that rhythmicity in the nucleus is the product of a highly organized network of heterogeneous cells. The present results indicate that specialized network organization is found not only in the core–shell relationship, but also within the shell itself. Analysis of the network of SCN oscillators presents a unique opportunity to understand the relationship between cellular and molecular events, intercellular connections, and function at the tissue and behavioural level. The availability of molecular tools to understand network properties of a nucleus whose function is so well characterized presents a unique opportunity for analysis of nervous system organization.
Acknowledgements
We thank Dr Joseph LeSauter for helpful comments on an earlier draft of this manuscript, and Carrie Wright for technical assistance. This research was supported by an NIH grant NS37919 (R.S.), in part by the Japanese Society for the Promotion of Science (T.H.) and by a Natural Sciences and Engineering Research Council of Canada fellowship (M.C.A.).
Abbreviations
- CalB
calbindin
- CT
circadian time
- DIG
digoxigenin
- LP
light pulse
- SCN
suprachiasmatic nucleus
References
- Aioun J, Chambille I, Peytevin J, Martinet L. Neurons containing gastrin-releasing peptide and vasoactive intestinal polypeptide are involved in the reception of the photic signal in the suprachiasmatic nucleus of the Syrian hamster: an immunocytochemical ultrastructural study. Cell Tissue Res. 1998;291:239–253. doi: 10.1007/s004410050994. [DOI] [PubMed] [Google Scholar]
- Antle M, Foley D, Foley N, Silver R. Gates and oscillators: a network model of the brain clock. J. Biol. Rhythms. 2003;18:339–350. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, LeSauter J, Silver R, Romero M. Retinal innervation of calbindin-D28K cells in the hamster suprachiasmatic nucleus: ultrastructural characterization. J. Biol. Rhythms. 2000;15:103–111. doi: 10.1177/074873040001500204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel M, Feinstein N, Cohen S, Harari N. Vasopressinergic innervation of the mouse suprachiasmatic nucleus: an immuno-electron microscopic analysis. J. Comp. Neurol. 1990;298:172–187. doi: 10.1002/cne.902980204. [DOI] [PubMed] [Google Scholar]
- Dardente H, Poirel V, Klosen P, Pevet P, Masson-Pevet M. Per and neuropeptide expression in the rat suprachiasmatic nuclei: compartmentalization and differential cellular induction by light. Brain Res. 2002;958:261–271. doi: 10.1016/s0006-8993(02)03563-1. [DOI] [PubMed] [Google Scholar]
- Eskin A. Identification and physiology of circadian pacemakers. Introduction. Fed. Proc. 1979;38:2570–2572. [PubMed] [Google Scholar]
- Guldner F. Synaptology of the rat suprachiasmatic nucleus. Cell Tissue Res. 1976;165:509–544. doi: 10.1007/BF00224478. [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Lokshin MM-TR, Yan L, Venuti J, S. Calbindin influences response to photic input in suprachiasmatic nucleus. J. Neurosci. 2003;23:8820–8826. doi: 10.1523/JNEUROSCI.23-26-08820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti J, Silver R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J. Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog E, Geusz M, Khalsa S, Straume M, Block G. Circadian rhythms in mouse suprachiasmatic nucleus explants on multimicroelectrode plates. Brain Res. 1997;757:285–290. doi: 10.1016/s0006-8993(97)00337-5. [DOI] [PubMed] [Google Scholar]
- Herzog E, Schwartz W. A neural clockwork for encoding circadian time. J. Appl. Physiol. 2002;92:401–408. doi: 10.1152/japplphysiol.00836.2001. [DOI] [PubMed] [Google Scholar]
- Jobst EE, Allen CN. Calbindin neurons in the hamster suprachiasmatic nucleus do not exhibit a circadian variation in spontaneous firing rate. Eur. J. Neurosci. 2002;16:2469–2474. doi: 10.1046/j.1460-9568.2002.02309.x. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Teclemariam-Mesbah R, Pevet P. Efferent projections of the suprachiasmatic nucleus in the golden hamster (Mesocricetus auratus). J. Comp. Neurol. 1993;332:293–314. doi: 10.1002/cne.903320304. [DOI] [PubMed] [Google Scholar]
- Karatsoreos I, Yan L, LeSauter J, Silver R. Phenotype matters: identification of light-responsive cells in mouse SCN. J. Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leak R, Card J, Moore R. Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res. 1999;819:23–32. doi: 10.1016/s0006-8993(98)01317-1. [DOI] [PubMed] [Google Scholar]
- LeSauter J, Yan L, Vishnubhotla B, Quintero J, Kuhlman S, McMahon D, Silver R. A short half-life GFP mouse model for analysis of suprachiasmatic nucleus organization. Brain Res. 2003;964:279–287. doi: 10.1016/s0006-8993(02)04084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- Miche S, Colwell C. Cellular communication and coupling within the suprachiasmatic nucleus. Chronobiol. Int. 2001;18:579–600. doi: 10.1081/cbi-100106074. [DOI] [PubMed] [Google Scholar]
- Moore R. Entrainment pathways and the functional organization of the circadian system. Prog. Brain Res. 1996;111:103–119. doi: 10.1016/s0079-6123(08)60403-3. [DOI] [PubMed] [Google Scholar]
- Moore R, Speh J, Leak R. Suprachiasmatic nucleus organization. Cell Tissue Res. 2002;309:89–98. doi: 10.1007/s00441-002-0575-2. [DOI] [PubMed] [Google Scholar]
- Morin L, Wood R. A Stereotaxic Atlas of the Golden Hamster Brain. Academic Press; New York: 2001. [Google Scholar]
- Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, Sehgal A, Shigeyoshi Y. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J. Neurosci. 2003;23:6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T, Honma K. Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 2001;14:666–674. doi: 10.1046/j.0953-816x.2001.01684.x. [DOI] [PubMed] [Google Scholar]
- Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J. Neurosci. 2003;23:8070–8076. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger J, Shearman L, Reppert S, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Shirakawa T, Honma S, Honma K. Multiple oscillators in the suprachiasmatic nucleus. Chronobiol. Int. 2001;18:371–387. doi: 10.1081/cbi-100103962. [DOI] [PubMed] [Google Scholar]
- Sigworth L, Liao L, Chandler T, Geusz M. Luciferase expression controlled by a viral gene promoter in a mammalian circadian pacemaker. Neuroreport. 2003;14:443–447. doi: 10.1097/00001756-200303030-00029. [DOI] [PubMed] [Google Scholar]
- Silver R, Romero M, Besmer H, Leak R, Nunez J, LeSauter J. Calbindin-D28K cells in the hamster SCN express light-induced Fos. Neuroreport. 1996;7:1224–1228. doi: 10.1097/00001756-199604260-00026. [DOI] [PubMed] [Google Scholar]
- Silver R, Sookhoo A, LeSauter J, Stevens P, Jansen H, Lehman M. Multiple regulatory elements result in regional specificity in circadian rhythms of neuropeptide expression in mouse SCN. Neuroreport. 1999;10:3165–3174. doi: 10.1097/00001756-199910190-00008. [DOI] [PubMed] [Google Scholar]
- Strogatz SH, Stewart I. Coupled oscillators and biological synchronization. Sci. Am. 1993;269:102–109. doi: 10.1038/scientificamerican1293-102. [DOI] [PubMed] [Google Scholar]
- Welsh D, Logothetis D, Meister M, Reppert S. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- Yan L, Silver R. Differential induction and localization of mPer1 and mPer2 during advancing and delaying phase shifts. Eur. J. Neurosci. 2002;16:1531–1540. doi: 10.1046/j.1460-9568.2002.02224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]