Abstract
The dominant family of interspersed repetitive DNA sequences in the human genome has been termed the Alu family. We have found that more than 75% of the lambda phage in a recombinant library representing an African green monkey genome hybridize with a human Alu sequence under stringent conditions. A group of clones selected from the monkey library with probes other than the Alu sequence were analyzed for the presence and distribution of Alu family sequences. The analyses confirm the abundance of Alu sequences and demonstrate that more than one repeat unit is present in some phages. In the clones studied, the Alu units are separated by an average of 8 kilobase pairs of unrelated sequences. The nucleotide sequence of one monkey Alu sequence is reported and shown to resemble the human Alu sequences closely. Hence, the sequence, dispersion pattern, and copy number of the Alu family members are very similar in the African green monkey and human genomes. Among the clones investigated were two that contain segments of the satellite DNA term alpha-component joined to non alpha-component DNA. The experiments indicate that in the monkey genome Alu sequences can occur close to regions of alpha-component DNA.
Full text
PDF



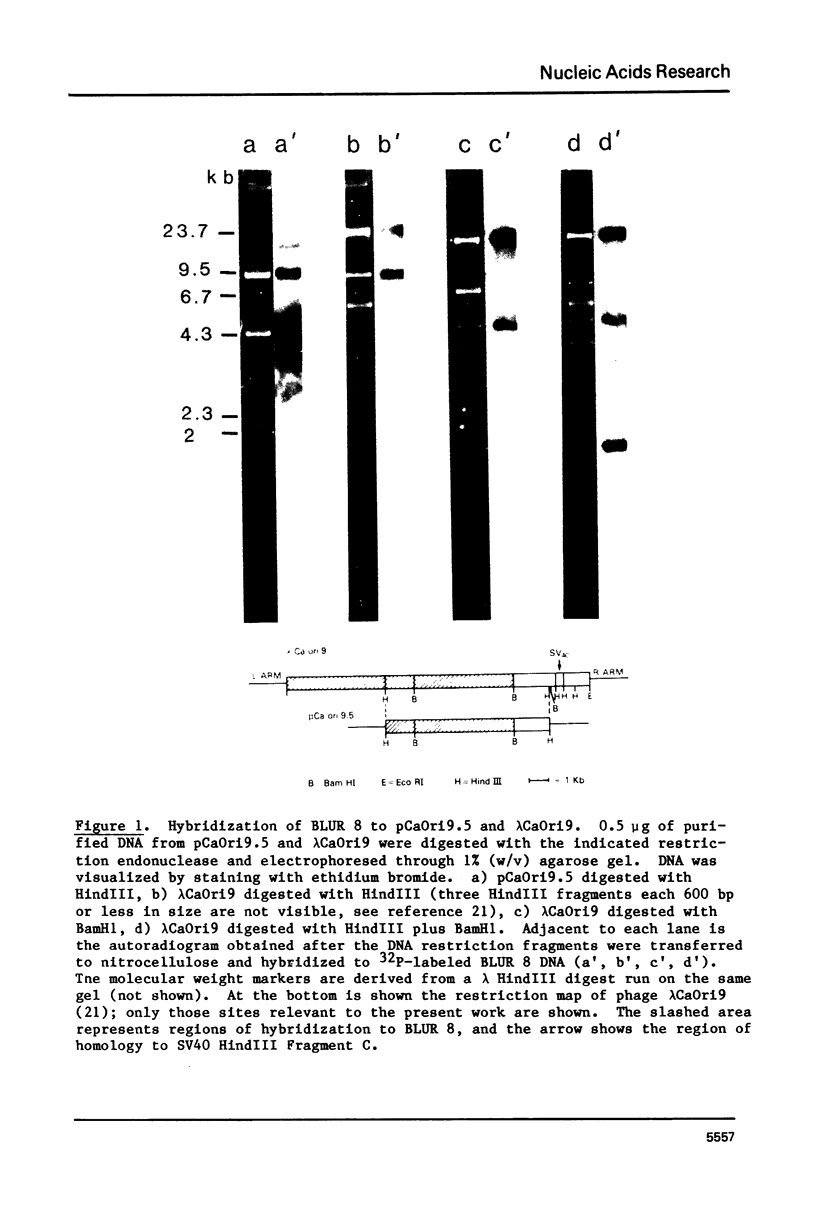
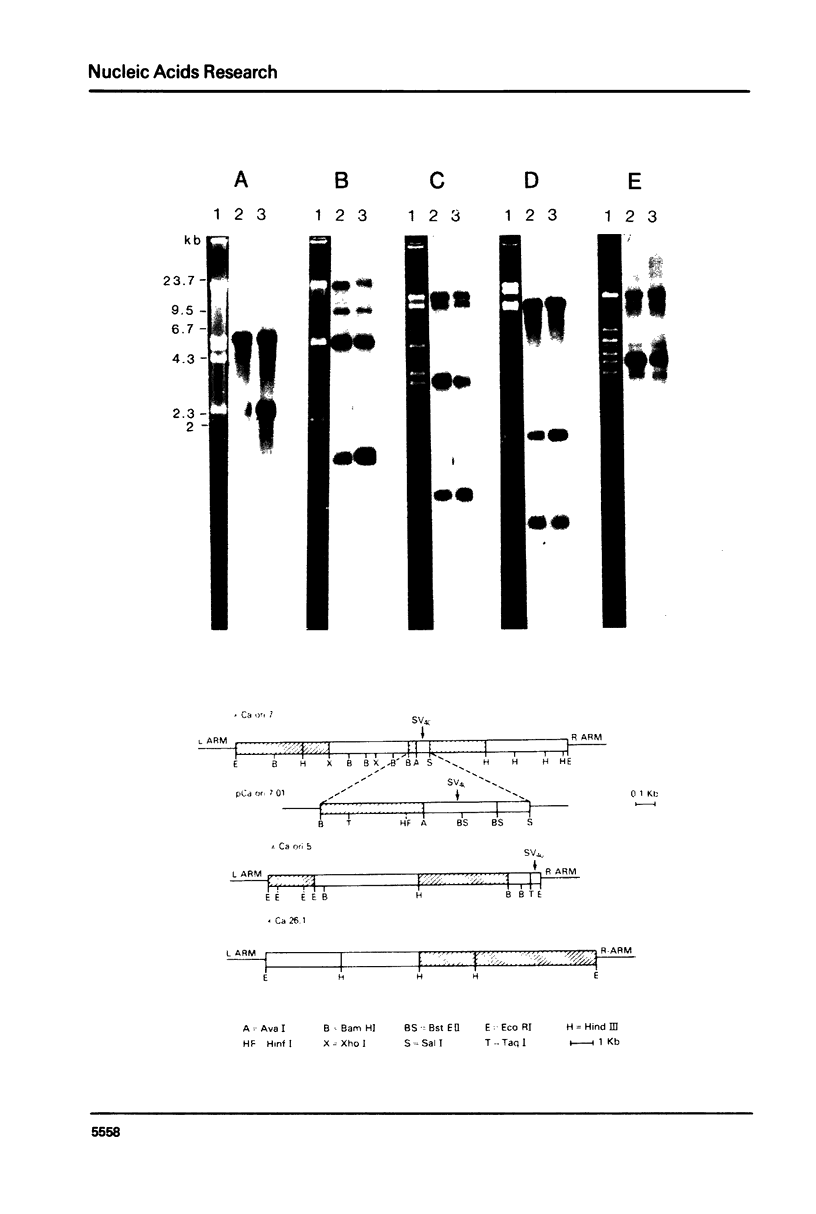

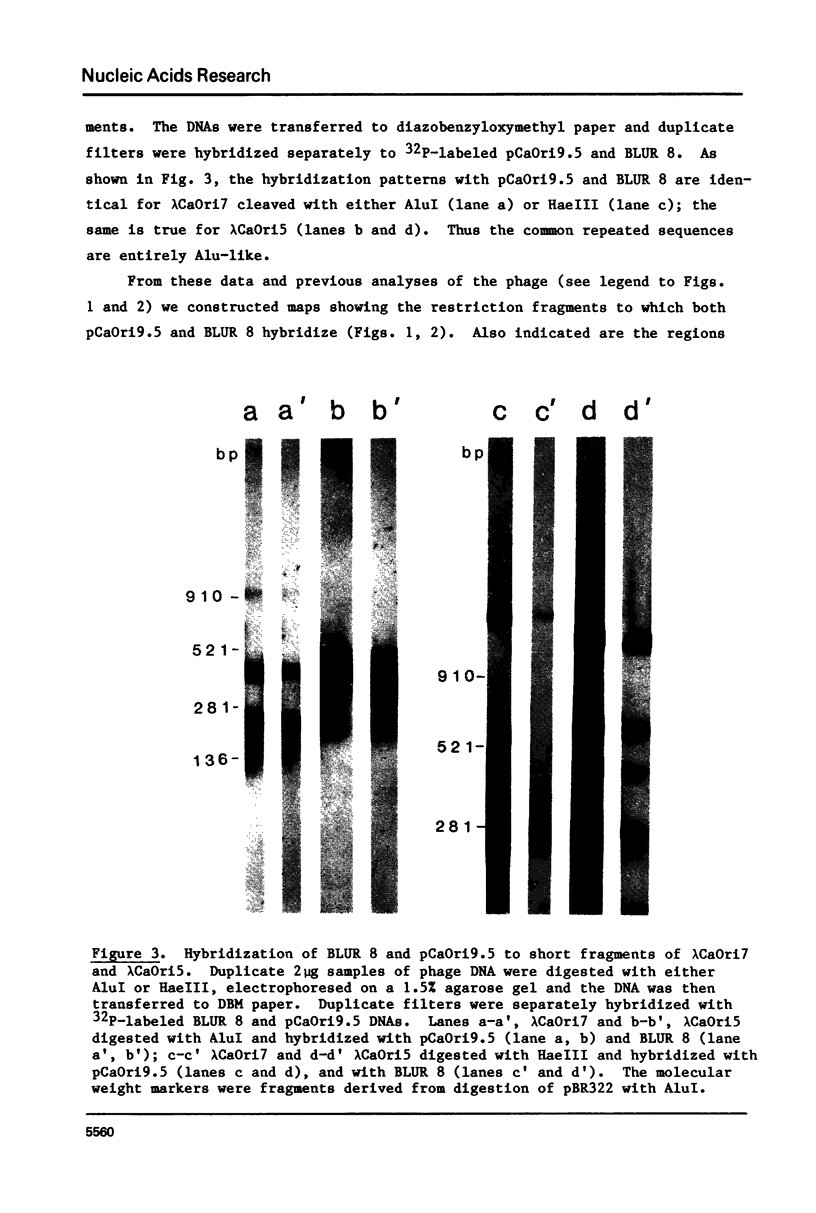








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Baltimore D. Gene conversion: some implications for immunoglobulin genes. Cell. 1981 Jun;24(3):592–594. doi: 10.1016/0092-8674(81)90082-9. [DOI] [PubMed] [Google Scholar]
- Baralle F. E., Shoulders C. C., Goodbourn S., Jeffreys A., Proudfoot N. J. The 5' flanking region of human epsilon-globin gene. Nucleic Acids Res. 1980 Oct 10;8(19):4393–4404. doi: 10.1093/nar/8.19.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., Pictet R., Rutter W. J. Analysis of the regions flanking the human insulin gene and sequence of an Alu family member. Nucleic Acids Res. 1980 Sep 25;8(18):4091–4109. doi: 10.1093/nar/8.18.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bonner J., Garrard W. T., Gottesfeld J., Holmes D. S. Functional organization of the mammalian genome. Cold Spring Harb Symp Quant Biol. 1974;38:303–310. doi: 10.1101/sqb.1974.038.01.034. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Britten R. J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979 Jun 8;204(4397):1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Dhruva B. R., Shenk T., Subramanian K. N. Integration in vivo into simian virus 40 DNA of a sequence that resembles a certain family of genomic interspersed repeated sequences. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4514–4518. doi: 10.1073/pnas.77.8.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Fittler F. Analysis of the alpha-satellite DNA from African green monkey cells by restriction nucleases. Eur J Biochem. 1977 Apr 1;74(2):343–352. doi: 10.1111/j.1432-1033.1977.tb11399.x. [DOI] [PubMed] [Google Scholar]
- Fritsch E. F., Lawn R. M., Maniatis T. Molecular cloning and characterization of the human beta-like globin gene cluster. Cell. 1980 Apr;19(4):959–972. doi: 10.1016/0092-8674(80)90087-2. [DOI] [PubMed] [Google Scholar]
- Haynes S. R., Toomey T. P., Leinwand L., Jelinek W. R. The Chinese hamster Alu-equivalent sequence: a conserved highly repetitious, interspersed deoxyribonucleic acid sequence in mammals has a structure suggestive of a transposable element. Mol Cell Biol. 1981 Jul;1(7):573–583. doi: 10.1128/mcb.1.7.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Schmid C. W. The evolution of a family of short interspersed repeats in primate DNA. J Mol Evol. 1981;17(3):148–155. doi: 10.1007/BF01733908. [DOI] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981 Jul 23;292(5821):306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Maio J. J. DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J Mol Biol. 1971 Mar 28;56(3):579–595. doi: 10.1016/0022-2836(71)90403-7. [DOI] [PubMed] [Google Scholar]
- Manning J. E., Schmid C. W., Davidson N. Interspersion of repetitive and nonrepetitive DNA sequences in the Drosophila melanogaster genome. Cell. 1975 Feb;4(2):141–155. doi: 10.1016/0092-8674(75)90121-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T. F., Singer M. F. DNA sequences similar to those around the simian virus 40 origin of replication are present in the monkey genome. Proc Natl Acad Sci U S A. 1981 Jan;78(1):95–99. doi: 10.1073/pnas.78.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. R., Beauchamp R. S., Bostock C. J. A study of sequence homologies in four satellite DNAs of man. J Mol Biol. 1979 Nov 25;135(1):127–149. doi: 10.1016/0022-2836(79)90344-9. [DOI] [PubMed] [Google Scholar]
- Musich P. R., Brown F. L., Maio J. J. Highly repetitive component alpha and related alphoid DNAs in man and monkeys. Chromosoma. 1980;80(3):331–348. doi: 10.1007/BF00292688. [DOI] [PubMed] [Google Scholar]
- Pan J., Elder J. T., Duncan C. H., Weissman S. M. Structural analysis of interspersed repetitive polymerase III transcription units in human DNA. Nucleic Acids Res. 1981 Mar 11;9(5):1151–1170. [PMC free article] [PubMed] [Google Scholar]
- Papamatheakis J., Lee T. N., Thayer R. E., Singer M. F. Recurring defective variants of simian virus 40 containing monkey DNA segments. J Virol. 1981 Jan;37(1):295–306. doi: 10.1128/jvi.37.1.295-306.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosenberg H., Singer M., Rosenberg M. Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978 Apr 28;200(4340):394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Houck C. M., Deininger P. L., Friedmann T., Schmid C. W. Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature. 1980 Mar 27;284(5754):372–374. doi: 10.1038/284372a0. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Deininger P. L. Sequence organization of the human genome. Cell. 1975 Nov;6(3):345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Singer D. S. Arrangement of a highly repeated DNA sequence in the genome and chromatin of the African green monkey. J Biol Chem. 1979 Jun 25;254(12):5506–5514. [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tashima M., Calabretta B., Torelli G., Scofield M., Maizel A., Saunders G. F. Presence of a highly repetitive and widely dispersed DNA sequence in the human genome. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1508–1512. doi: 10.1073/pnas.78.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer R. E., Singer M. F., McCutchan T. F. Sequence relationships between single repeat units of highly reiterated African Green monkey DNA. Nucleic Acids Res. 1981 Jan 10;9(1):169–181. doi: 10.1093/nar/9.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamiya T., McCutchan T., Rosenberg M., Singer M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. I. The structure of variant CVPS/1/P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3584–3591. [PubMed] [Google Scholar]
- Wensink P. C., Tabata S., Pachl C. The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell. 1979 Dec;18(4):1231–1246. doi: 10.1016/0092-8674(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Winocour E., Singer M., Kuff E. Rapid detection, isolation, and amplification of host-substituted SV40 variants. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):621–628. doi: 10.1101/sqb.1980.044.01.065. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Manuelidis L. Sequence definition and organization of a human repeated DNA. J Mol Biol. 1980 Sep 25;142(3):363–386. doi: 10.1016/0022-2836(80)90277-6. [DOI] [PubMed] [Google Scholar]






