Abstract
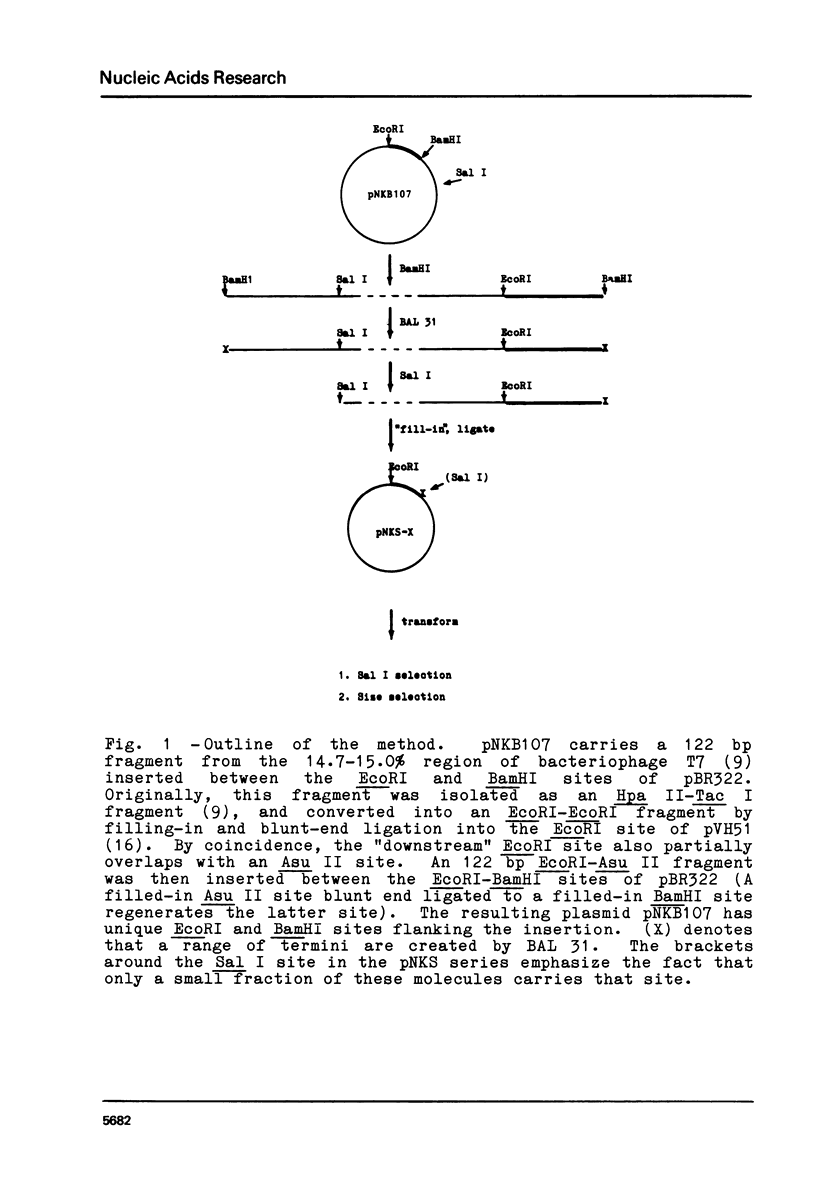
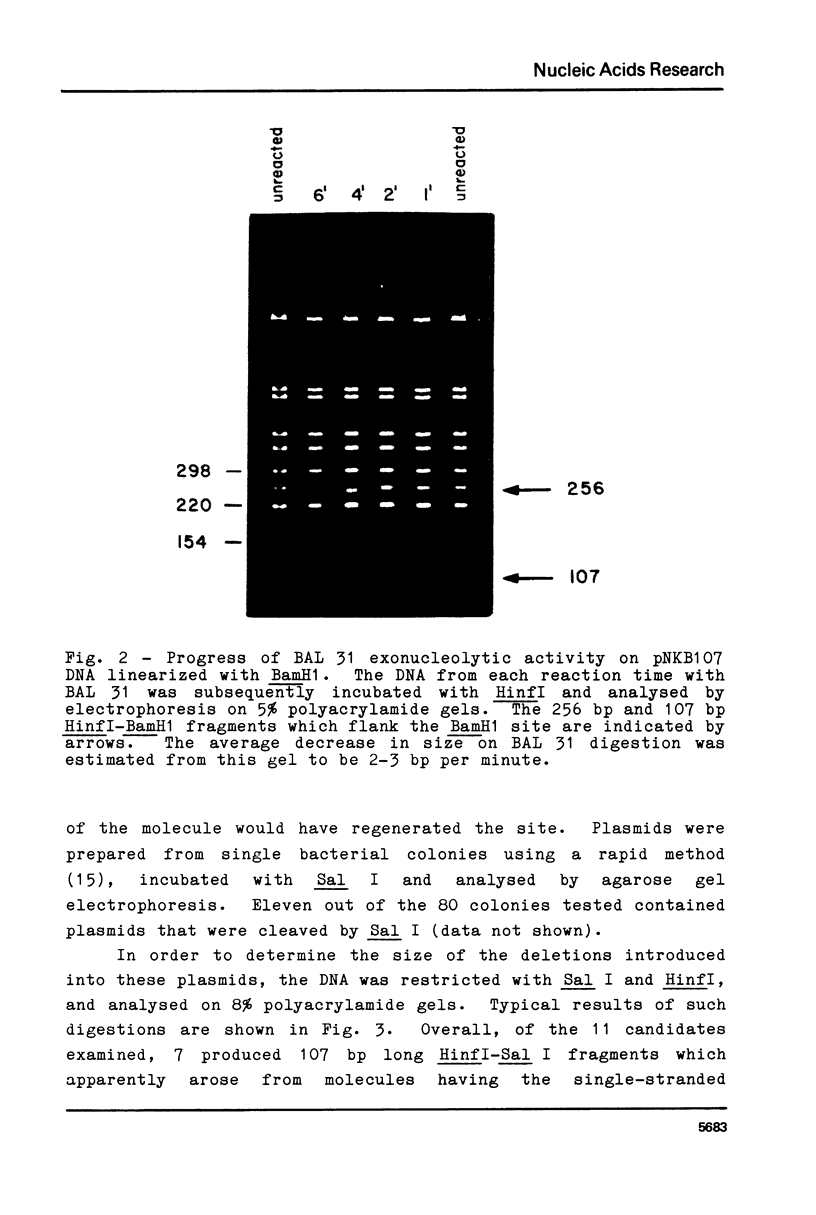
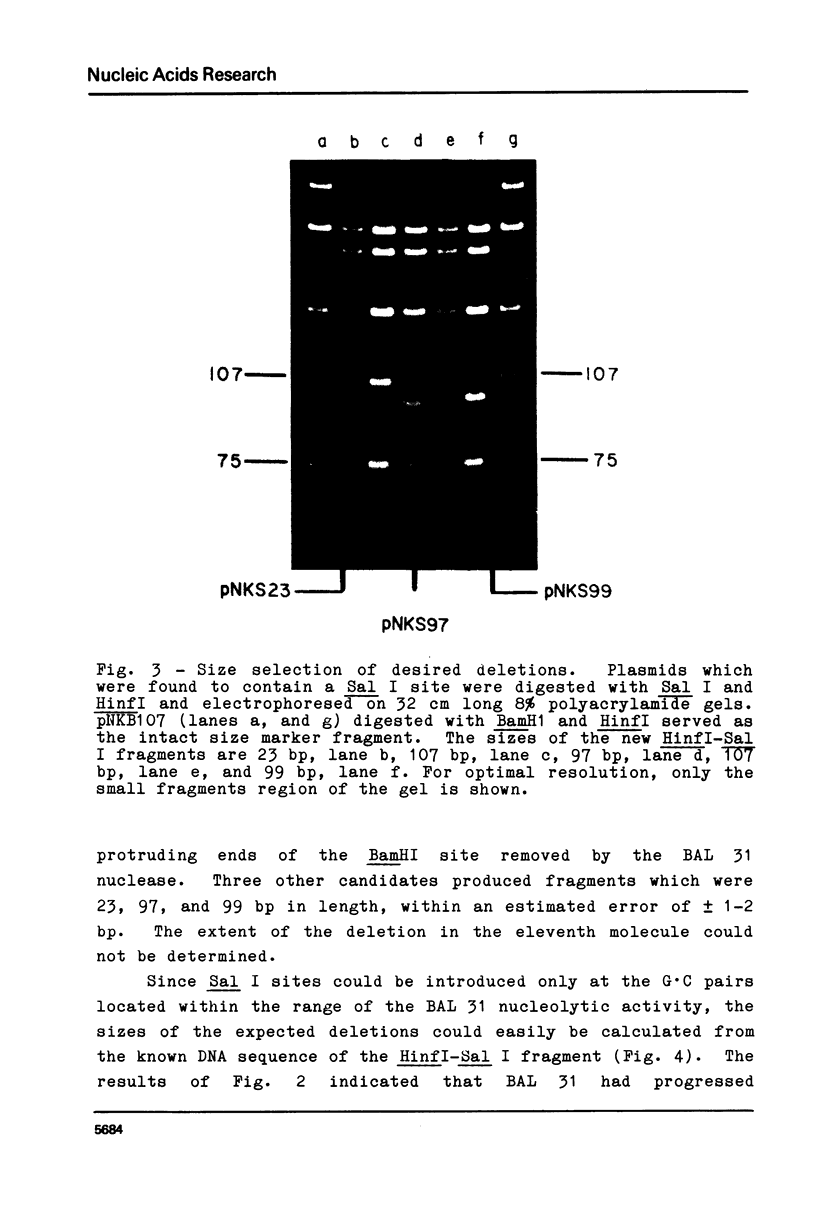
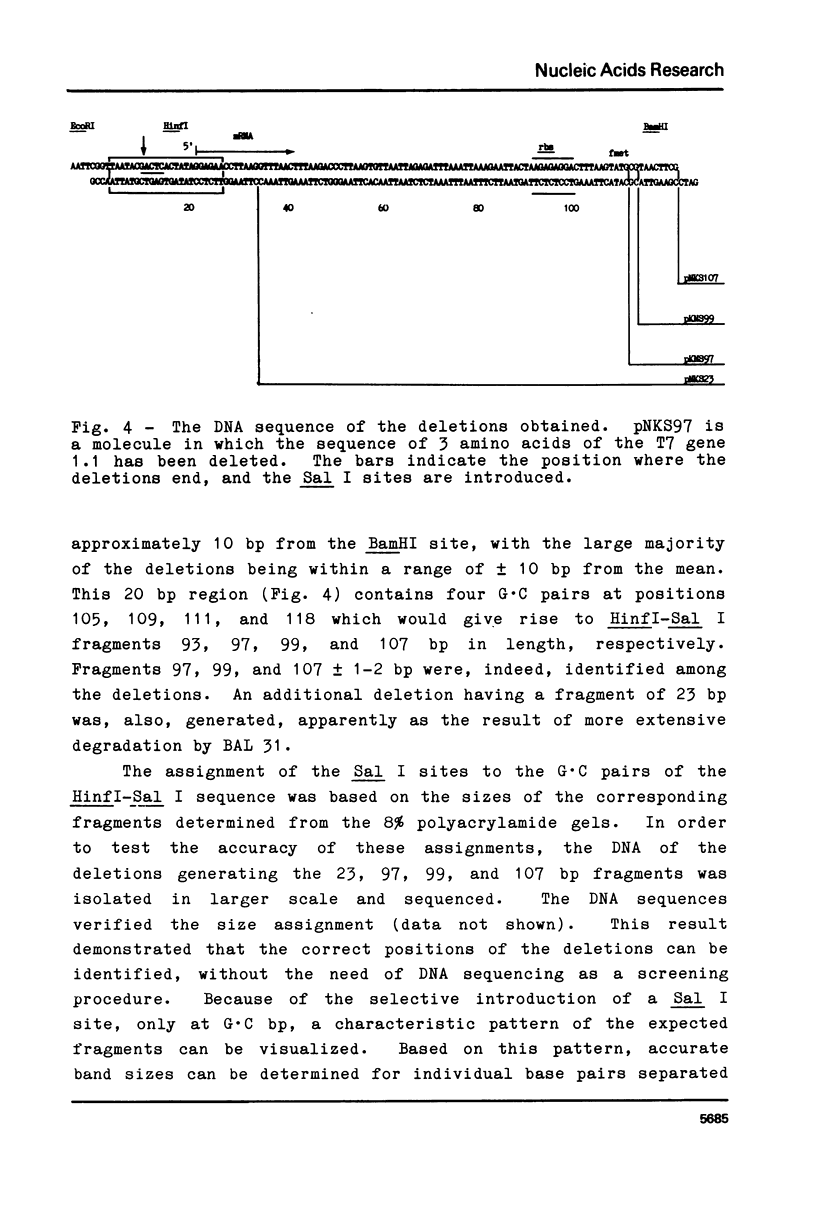
Blunt-end ligation of a "filled-in" HindIII, Sal I, Ava I or Bcl I restriction site with a DNA fragment having A, G, C, or T as the terminal 3' nucleotide regenerates the corresponding restriction site. A combination of this property with the action of BAL 31 nuclease which progressively removes base-pairs from the ends of linear DNA, can generate deletions extending to desired pre-selected nucleotides, and introduces unique restriction sites at those positions. Similarly other restriction sites can be used to select for the deletion of sequences between specific di-, tri-, tetra- and penta-nucleotides. Using this method, 10 base pairs were deleted from the end of a restriction fragment carrying the late promoter for bacteriophage T7 gene 1.1, to create a molecule with a unique restriction site at the initiation codon for translation.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dunn J. J., Studier F. W. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J Mol Biol. 1981 Jun 5;148(4):303–330. doi: 10.1016/0022-2836(81)90178-9. [DOI] [PubMed] [Google Scholar]
- Green C., Tibbetts C. Targeted deletions of sequences from closed circular DNA. Proc Natl Acad Sci U S A. 1980 May;77(5):2455–2459. doi: 10.1073/pnas.77.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., So M., McCarthy B. J. In vitro mutagenesis of a circular DNA molecule by using synthetic restriction sites. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6012–6016. doi: 10.1073/pnas.75.12.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cloning and localization of the in vitro functional origin of replication of bacteriophage T7 DNA. J Biol Chem. 1979 Jun 25;254(12):5555–5561. [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Recognition and initiation site for four late promoters of phage T7 is a 22-base pair DNA sequence. Nature. 1979 Jul 5;280(5717):35–39. doi: 10.1038/280035a0. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Lauer G. D. Maximizing gene expression on a plasmid using recombination in vitro. Methods Enzymol. 1979;68:473–482. doi: 10.1016/0076-6879(79)68036-9. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Stahl S., Gilbert W. Eukaryotic signal sequence transports insulin antigen in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3369–3373. doi: 10.1073/pnas.77.6.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]