Abstract
Several barriers have to be overcome in order to achieve gene expression in target cells, e.g. cellular uptake, endosomal release and translocation to the nucleus. Nuclear localization sequences (NLS) enhance gene delivery by increasing the uptake of plasmid DNA (pDNA) to the nucleus. So far, only monopartite NLS were analysed for non-viral gene delivery. In this study, we examined the characteristics of a novel bipartite NLS like construct, namely NLS Ku70. We synthesized a dimeric structure of a modified NLS from the Ku70 protein (Ku702-NLS), a nuclear transport active mutant of Ku702-NLS (s1Ku702-NLS) and a nuclear transport deficient mutant of Ku702-NLS (s2Ku702). We examined the transfection efficiency of binary Ku702-NLS/DNA and ternary Ku702-NLS/PEI/DNA gene vector complexes in vitro by using standard transfection protocols as well as the magnetofection method. The application of Ku702-NLS and s1Ku702-NLS increased gene transfer efficiency in vitro and in vivo. This study shows for the first time that the use of bipartite NLS compounds alone or in combination with cationic polymers is a promising strategy to enhance the efficiency of non-viral gene transfer.
Introduction
The transfer of nucleic acids into somatic cells offers new perspectives for the treatment of lethal acquired or inherited diseases. To date, effective delivery of the nucleic acids to the target cells is hindered by extracellular and intracellular biological barriers. Regarding efficiency, the most potent carrier systems are based on viral transfection systems like recombinant deficient retrovirus vectors [1], [2].
Non-viral gene delivery is limited by the low endosomal escape after cellular uptake and the low translocation of DNA into the nucleus [3], [4]. It has been shown that the endosomal escape could be improved by compacting DNA with the cationic polymer PEI which compacts and releases DNA efficiently from the endosomes into the cytosol [5]. To further improve non-viral transfection, nuclear localization sequences (NLS) have been synthesized and were used to facilitate nuclear translocation of the DNA. NLS shuttle proteins into the nucleus by binding to nuclear transport proteins such as importin α or importin β through the nuclear pore and are released in the nucleus [6]. The first studies used NLS covalently bound to pDNA. These studies proved that NLS can promote the transport of pDNA into the nucleus [7], [8], but covalently bound NLS interfered with the transgene expression of the pDNA [9]. An easier and less complicated method was developed by Ritter et al. by binding NLS and DNA in an electrostatic way [10].
So far, only monopartite NLS were analysed for non-viral gene delivery. In this study, we examined the characteristics of a novel bipartite NLS like construct, namely NLS Ku70, for the use as a non viral gene carrier.
Materials and Methods
Peptide Synthesis
Three peptides were synthesized by the department of medicine (Institute of Biochemistry, Humboldt-University, Berlin): C-KVTKRKHGAAGAASKRPK-G-KVTKRKHGAAGAASKRPK (Ku702-NLS) as dimeric peptide of the Ku70-NLS, C-ASGSKGARPAKKRKPKRGAAHKHAGAKVRKTVTGAKK (s1Ku702-NLS) as a supposed nuclear transport active mutant of the Ku702-NLS and C-KTAHSKAARGHTPKGKARVVKAKAGKAKGGKAKPRSR (s2Ku702) as transport deficient mutant. As far as the intervening regions of Ku702-NLS are concerned the first and fourth alanine had to be replaced with glycine because 6 alanines cannot be synthesized in series. Synthesis of all peptides started with glycine. The free sulfhydryl groups of the cysteines were modified by dithiopyridin reaction in order to protect them of oxidation [11].
Cloning of β-galactosidase fusion proteins
For subcloning of plasmid DNA coding β-galactosidase fusion proteins, we used pVAX1/lacZ plasmids (Invitrogen. UK). The coding and non-coding strand of Ku702-NLS-, s1Ku702-NLS and s2Ku702 were synthesized by Biomers (Ulm, Germany). All annealed oligonucleotides were cloned into the pVAX1/lacZ plasmid between NheI and BamHI restriction sites. The sequencing of all cloned plasmids showed that between NLS- and β-galactosidase DNA sequence there existed one start codon and one excess nucleotide. Thereby it could not be ensured that the Ku702-NLS-β-Galactosidase fusion protein could be read completely and correctly by DNA polymerase. The excess nucleotide led to a frame shift; therefore the open reading frame of β-galactosidase DNA sequence was disarranged. In order to exclude the nucleotide sequence GATG we conducted a site directed mutagenesis. Hence, we designed a forward primer (5′-TTGAATTCTGCAGATCGAAACATAGATCCCGTCGTTTTACAA-3′) and a reverse primer (5′-TTGTAAAACGACGGGATCTATGTTTCGATCTGCAGAATTCCA-3′) flanking the nucleotide sequence GATG. Using proof reading enzyme PFU II Ultra (Stratagene, CA, USA) we conducted an inverse PCR of the already cloned plasmid DNA by using the following PCR protocol: 2 min at 94°C denaturation, 18 cycles of 20 sec denaturation at 95°C, 20 sec annealing of primer at 45°C, 90 sec elongation at 68°C, and accordingly 3 min proof reading at 68°C. Afterwards, the new plasmids were digested with enzyme DpnI (Fermentas, St. Leon-Rot, Germany). Again, after transformation in E. coli, new plasmids were identified by gel electrophoresis using 1% agarose gel and by sequencing (GATC Biotech AG, Konstanz, Germany). Then, the plasmids pVAX1/lacZ-Ku702-NLS, pVAX1/lacZ-s1Ku702-NLS and pVAX1/lacZ-s2Ku702 were transformated into E. coli strain DH10B (ElectroMAX DH10B Cells, Invitrogen, Karlsruhe, Germany), isolated and purified by using NucleoBond® EF plasmid purification kits (Macherey-Nagel, Düren, Germany).
Plasmid DNA
The pCLuc containing firefly luciferase (a gift by Ernst Wagner, department of pharmacy, University of Munich,) and pEGFP-N1 containing enhanced green fluorescent Protein (Clontech, Palo Alto, CA, USA) were used for in vitro transfections. In vivo experiments were conducted with ccc-pCp-Luc coding for luciferase (Invitrogen, UK). For β-galactosidase experiments we used pVR1411 containing SV40-NLS (Biomers, Ulm, Germany), pVAX1/lacZ (Invitrogen, UK) containing β-galactosidase reporting gene as well as pVAX1/lacZ-Ku702-NLS, pVAX1/lacZ-s1Ku702-NLS and pVAX1/lacZ-s2Ku702.
Size measurement
Particle size was determined by dynamic light scattering (Brookhaven Instruments Corporation, Austria). Gene vector complexes were generated as described above in double-distilled water and PBS. Measurements were performed using the following settings: 10 sub-run measurements per sample; viscosity for water 0.89 cPa; beam mode F(Ka) ¼ 1.50 (Smoluchowsky); and temperature 25°C.
Cell Culture
BEAS-2B cells (ATCC No. CRL-9609) and 16HBE14o− cells (Prof. Dr. Dieter C. Gruenert, University of Vermont, Burlington, VT, USA), a human bronchial epithelial cell line, and HELA (DSMZ No: ACC 57, Germany), a cervical carcinoma cell line, were cultured in minimal essential medium (MEM, Gibco/Invitrogen, Karlsruhe, Germany) containing 10% fetal bovine serum (PAA Laboratories, Austria). All cells were maintained at 37°C in a 5% CO2 humidified air atmosphere.
Preparation of Gene Vector Complexes
Gene vector complexes were generated in HBS (150 mM NaCl, 10 mM HEPES, pH 7.4) or PBS. For formulating binary gene vector complexes 0.5 µg DNA and a varying amount of GTA depending on the ± ratio were dissolved in 75 µl of solvent. The DNA solution was pipetted to the GTA solution and mixed vigorously by pipetting up and down. The complexes were incubated at room temperature for 20 min before use. Ternary complexes were formulated in the same way, but 0.5 µg of DNA, NLS and PEI (average molecular mass of 25 kDA; Sigma Aldrich, Deisenhofen, Germany; dialyzed against water, 12–14-kDa molecular mass cut-off and adjusted to pH 7) were diluted in 25 µl solvent per GTA. Initially, DNA solution was pipetted to the NLS solution and incubated for 10 min. Accordingly PEI was added, the ternary solution was mixed vigorously and incubated for further 10 min before use.
In vitro electroporation of Ku702-NLS-β-galactosidase fusion proteins
Electroporation was only used for β-galactosidase experiments and performed with BioRad GenPulser II apparatus.
In vitro transfection/magnetofection and luciferase activity measurement
For transfection and magnetofection experiments, cells (10.000/well) were seeded in 96 well plates (Techno Plastic Products AG, Trasadingen, Suisse). For transfection experiments, complexes were pipetted in each well and incubated. 4 h later, the medium was replaced with 200 µl 10% FCS containing MEM supplemented with 0.1% (v/v) penicillin/streptomycin and 0.5% (v/v) gentamycin (Gibco/Invitrogen). 24 h later luciferase activity was measured after cell lysis by addition of 100 µl lysis buffer to each well (250 mM Tris, 0.1% Triton, ph = 7.8) and incubated at room temperature for 15 min. [12]. Luciferase expression was measured with Wallac Victor2/1420 Multilabel Counter (PerkinElmer; Rodgau-Jügesheim, Germany). The protein content was determined by a standard Bio-Rad protein assay (Bradford method). Magnetofection experiments were performed accordingly, but after having pipetted complexes to the wells, 96 well plates were placed on a sintered Nd-Fe-B magnet (NeoDelta; remanence Br, 1080–1150 mT), purchased from IBS Magnet (Berlin, Germany). Dimension of the magnet: cylindrical; d = 6 mm, h = 5 mm, inserted in an acrylic glass template in 96-well microplate format with strictly alternating polarization. Cells were incubated for 20 minutes instead of 4 h for transfection.
FACS analysis
For FACS measurements, 100,000 cells per well were seeded in 24-well plates (TPP, Trasadingen, Switzerland). Magnetofection was performed as described above. For FACS measurements cells were washed with PBS. Then the cells were trypsinized and measured using a Becton Dickinson FACScan (San Jose, USA).
Animal experiments and administration of gene vectors into the lung
Female BALB/c mice were purchased form Janvier (Elevage Janvier, Le Genst St. Isle, France) and maintained under specific pathogen free conditions. All experiments were approved and controlled by local ethic committee and conducted according to the guidelines of the German law of protection of animal life. Animals were anaesthetized by intraperitoneal injection of a mixture of medetomidine, midazolam, and fentanyl. The amount of plasmid DNA administered per mouse was 30 µg of CpG-free pCpGLuc. Gene transfer agents were diluted in double distilled water (Fresenius AG, Bad Homburg, Germany) in a volume of 100 µl per mouse. Firstly, branched PEI (N/P-ratio = 10) and Ku702-NLS-, s1Ku702-NLS and s2Ku702 (+/− ratio = 5) were mixed and incubated for 10min. Afterwards, DNA solution was pipetted directly to PEI/Ku702-NLS solution. Then, 100 µl of gene transfer solution was drop-wise pipetted onto the nose of each mouse. After application, mice were administered an antidote dose which consisted of atipamezol (50 µg/kg), flumazenil (10 µg/kg) and naloxon (24 µg/kg). The mice recovered from anaesthesia within 15 min and no adverse effects of the anaesthesia were observed. 24 h later, the efficiency of the gene vector application was measured by bioluminescence (IVIS 100 imaging system; Xenogen, Almeda, CA). For this purpose, mice were anaesthetized again. Signals were quantified and analyzed using the Living Image Software ver. 2.50. After imaging, anaesthetized mice were killed. Then, the posterior vena cava exit was cut and 1 ml of an isotonic sodium chloride solution was perfused slowly into the right heart in order to wash blood from the lungs and to avoid interference with the subsequent luciferase assay. The lungs were dissected from animals, frozen in liquid nitrogen and stored at −80 C. For the measurement of luciferase activity, minced lungs were each mixed with 400 µl of cell lysis buffer with addition of protease inhibitors (Roche Protease Inhibitor Cocktail Tablets). Samples were vortexed and centrifuged at 10,000 g at 4 C for 10 min. 100 µl of supernatant were measured for luciferase activity in a Lumat LB 9507 instrument (Berthold, Bad Wildbad, Germany) in duplicates by injecting 100 µl luciferin assay buffer. The emitted light was measured over 30 sec. The background was subtracted from the reported values.
Statistical Analysis
Statistical analyses were carried out using IBM SPSS 19.0 (Chicago, IL, U.S.A.). Means and one standard deviation of a representative experiment performed in triplicates are reported. A p value<0.05 was considered to be significant. Intergroup analyses were carried out using the Mann-Whitney U test as the data were not normally distributed in all groups.
Results
Proof of nuclear localization activity
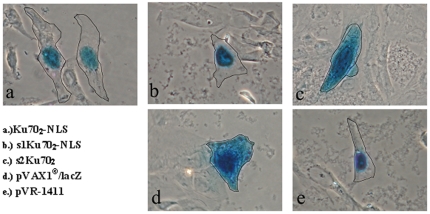
First, we analyzed the nuclear localization activity of the newly synthesized bipartite peptides Ku702-NLS, s1Ku702-NLS, and s2Ku702. These examinations had to be conducted because Ku702-NLS was synthesized as a dimer and the intervening region of Ku702-NLS had been changed which could have influenced the nuclear localization activity. The analysed cells showed the typical blue β-galactosidase coloration after staining with β-galactosidase staining solution [13]. If the tested peptides had characteristics of a nuclear localization sequence only the nucleus should stain blue. Otherwise the complete cytosol would show the blue staining. Using this method, we could show that Ku702-NLS and s1Ku702-NLS had nuclear localization activity, whereas s2Ku702 did not show any nuclear localization activity (Figure 1).
Figure 1. Intracellular localization of β-galactosidase fusion proteins.
BEAS-2B cells were electroporated with 10 µg β-galactosidase coding plasmid DNA. All images show the typical blue β-galactosidase staining. Images (a) and (b) point that Ku702-NLS and s1Ku702-NLS show nuclear localization signal activity. The exclusive blue staining of nuclei is clearly visible. The s2Ku702 shows staining of the cytosol. The NLS large antigen (e) was used as positive control for nuclear localisation and β-galactosidase without NLS (d) as negative control.
Biophysical properties of binary and ternary Ku702–NLS gene vector complexes
The size of particles is influenced by the solvent in such a way that ionic solvents enlarge particle size, and non-ionic solvents like distilled water result in a smaller size of the complexes [11], [14], [15], [16], [17]. Analysing the synthesized gene vector complexes for a period of 10 minutes, we could show that Ku702-NLS/DNA, s1Ku702-NLS/DNA and s2Ku702/DNA complexes (±5) that were generated in distilled water had a constant size of about 80 nm and transMagPEI/DNA complexes of about 190 nm. Ternary Ku702-NLS/transMagPEI/DNA, s1Ku702-NLS/transMagPEI/DNA and s2Ku702/transMagPEI/DNA are just as large as transMagPEI/DNA complexes and constant in size for about 10 minutes. Ternary gene vector complexes which were generated in PBS increased continuously during 20 minutes of measurement. All complexes showed a comparable size of about 300 to 400 nm.
Comparison of the bipartite Ku702–NLS with monomeric nuclear localization sequences
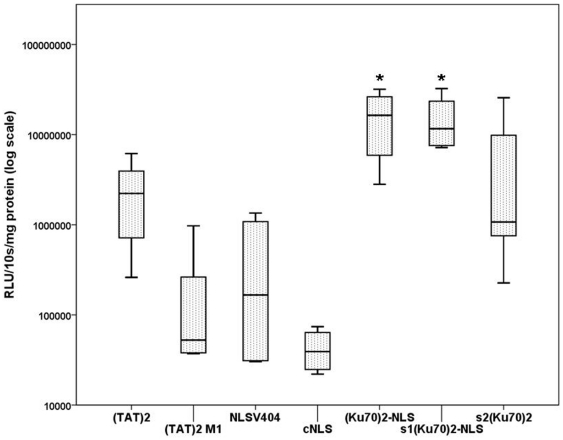
Ku702-NLS, s1Ku702-NLS were analyzed in comparison to monopartite nuclear localization sequences. We used NLSV404 (origin of SV40 virus) and TAT2, and their analogous nuclear transport deficient mutants cNLS and TAT2M1, as well as s2Ku702. Gene vector complexes were generated with charge ratio of ± = 5 and transfected to BEAS-2B cells. Gene transfer efficiency mediated by Ku702-NLS was significantly higher as compared to TAT2 or to NLS404. The same was the case for s1Ku702-NLS (Figure 2). This study clearly shows an advantage of the bipartite NLS over monopartite NLS.
Figure 2. Ku702-NLS in comparison to monopartite nuclear localization sequences.
BEAS-2B cells were transfected with Ku702-NLS/DNA complexes or the monopartite active NLS (TAT)2 or NLSV404, and their nuclear transport deficient sequences (control) NLS (TAT2M1 or cNLS). All the complexes were generated in HBS (charge ± = 5). Transgene expression of Ku702-NLS and s1Ku702-NLS was significantly higher (p<0.05, Mann-Whitney-U-Test) compared to the respective monopartite NLS (TAT)2 or NLSV404. Transgene expression of Ku702-NLS and s1Ku702-NLS compared to the control peptide s2Ku702 was close to significance. This study clearly suggests an advantage of the bipartite NLS over monopartite NLS.
Transfection efficiency depending on the +/−ratio
Plasmid DNA was complexed with Ku702-NLS or with s2Ku702. Transfections were performed with different charge ratios at a DNA dose of 0.5 µg. Using different +/−ratios in BEAS-2B cells and 16-HBE cells, we found that gene transfer efficiency at a ratio of ± = 5 was highest and future used.
Ternary gene vector complexes: combination of Ku702–NLS with polyethylenimine (PEI)
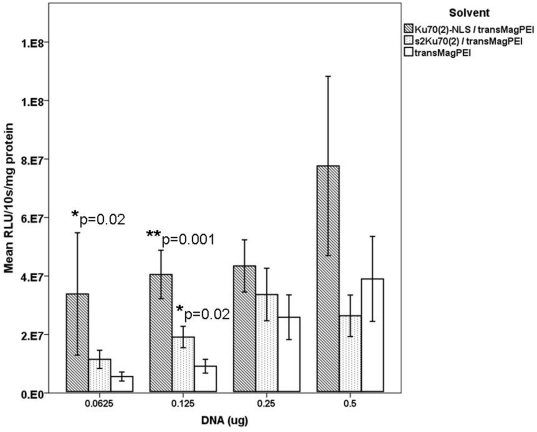
In a first step, we formulated ternary gene vector complexes with a DNA dose of 0.25 µg at a ± ratios of 5, 2.5, 1.25, 0.625 and 0.313. As a control, binary PEI/DNA (± = 10) complexes were used. Ku702-NLS/PEI/DNA and s2Ku702/PEI/DNA mediated higher gene transfer efficiency than binary PEI/DNA complexes. Transgene expression mediated by Ku702-NLS/PEI/DNA is about 1.7-fold to 8-fold higher as compared to transgene expression mediated by PEI/DNA. Using ± ratio of 5 Ku702-NLS/PEI/DNA complexes showed the most efficient gene delivery compared to PEI/DNA. ± ratios of 2,5 and 5 tested for Ku702-NLS/PEI are about 1.7-fold to 3.4-fold higher compared to s2Ku702/PEI (data not shown). Furthermore, we analyzed the transfection efficiency using the magnetofection method [18]. [19]. Gene vector complexes were formulated in PBS. DNA doses 0.03125, 0.0625, 0.125, 0.25 and 0.5 µg were analyzed. Ku702-NLS/transMagPei/DNA complexes solvented in PBS mediated 1.7-fold to 6-fold higher transgene expression compared to transMagPei/DNA. Transfection efficiency of Ku702-NLS/transMagPei/DNA compared to s2Ku702/PEI/DNA is about 1.1-fold to 3-fold higher (Figure 3).
Figure 3. Transfection efficiency of ternary gene vector complexes using transMagPEI and different solvents.
BEAS-2B cells were transfected with gene vector complexes. Transgene expression of Ku702-NLS/transMagPei/DNA is more efficient than s2Ku702/transMagPei and transMagPei/DNA. *p = 0.02 and **p = 0.001.
Quantification of the number of transfected cells
The number of transfected cells was investigated with FACS analysis. 16HBE14o− and BEAS-2B cells were transfected using magnetofection. The highest number of EGFP positive 16HBE14o− cells was found in the following order: Ku702-NLS/transMagPEI>s2Ku702/transMagPEI>transMagPEI (10.4%>8.2%>2.2%). The incremental factor of luciferase expression (Ku702-NLS/transMagPEI: 8-fold, s2Ku702/transMagPEI: 2.4-fold) compared to incremental factor of a number of transfected cells (Ku702-NLS/transMagPEI: 1.5-fold, s2Ku702/transMagPEI: 1.5-fold) clarifies that the Ku702-NLS/transMagPEI mediated luciferase expression is 5.3-fold (s2Ku702/transMagPEI: 1.6-fold) compared to the number of transfected cells. In this respect Ku702-NLS/transMagPEI enhances gene transfer by enhancing transgene expression of each cell, but not by the number of transfected cells.
In vivo application of ternary Ku702-NLS/PEI/DNA and s1Ku702-NLS/PEI/DNA complexes using nasal instillation
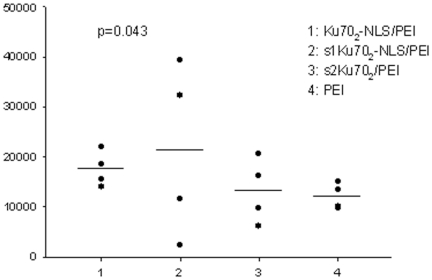
Using the IVIS in vivo imaging method it could be established that every type of gene vector complex was able to mediate gene transfer. Luciferin luminescence was measurable over all segments of the lungs of the tested groups of mice. In some contrast to the in vitro data, the Ku702-NLS/PEI/DNA mediated gene transfer was about 20% lower than PEI/DNA, but this effect was not statistically significant. s1Ku702-NLS/PEI/DNA mediated gene transfer was about 12% and s2Ku702/PEI/DNA mediated gene transfer about 28% higher compared to PEI/DNA (n.s.). In addition to the in vivo measurements with IVIS, lung homogenisates were analyzed for the presence of luciferase activity. In this analysis, the Ku702-NLS/PEI mediated gene transfer was about 46% higher, s1Ku702-NLS/PEI/DNA about 77% and s2Ku702/DNA about 9% higher compared to PEI/DNA. The values are significantly different from control (p≤0.043; n = 4) (Figure 4). Although the IVIS measurements may not have reflected the in vitro results, the analyses in lung homogenisates partly confirmed the in vitro data.
Figure 4. Analysing luciferase expression in lung homogenisates after nasal instillation of gene transfer agents.
1, Ku702-NLS/PEI/DNA; 2, s1Ku702-NLS/PEI/DNA; 3, s2Ku702/PEI/DNA; 4, PEI/DNA. Gene vector complexes were formulated as follows: 30 µg DNA; NLS: ± ratio = 5; PEI: N/P ratio = 10. Luciferase activity was measured in lung homogenisates 24 h after application of gene vectors. Values between Ku702-NLS/PEI and PEI significantly different (p≤0.043; n = 4). Statistical analysis was performed using the Mann-Whithney-U test.
Discussion
It is known that the nuclear membrane in eukaryotic cells is a major barrier for efficient gene transfer using non-viral vectors. Based on this information we pursued a new strategy of using a bipartite nuclear localization sequence of the Ku70 protein in order to develop a more efficient non-viral gene transfer system compared to classical non viral gene transfer agents. This Ku70 protein is a subunit of the Ku protein which was found in patients with systemic lupus erythematosus and scleroderma-polymyositis overlap syndrome. This protein is involved in DNA double-strand break repair and transcription. The Ku70 subunit consists of two basic subregions and a nonbasic intervening region [20], [21]. Insofar, this NLS was interesting for our examination because the intervening region consisting of the aminoacids DNEGSG can be substituted by six alanines without any loss of NLS-functionality [21]. Substituting the negatively charged aminoacids could improve the binding intensity between NLS and pDNA. Rudolph et al. observed that the application of dimeric NLS reached the most efficient gene delivery in their study [11]. In analogy to this study, we characterized this dimeric structure of the Ku70 NLS for the use as a non viral gene carrier.
Firstly, NLS activity of the newly synthesized Ku702-NLS and s1Ku702-NLS was confirmed by using β-galactosidase fusionproteins. Comparing the transfection efficacy of the newly synthesized bipartite NLS with standard polyfection (PEI/DNA) we found better transfection results of Ku702-NLS when using lower DNA doses (0.125 µg–0.25 µg). In contrast, PEI/DNA mediated 2.5-fold to 7-fold higher transfection efficiency in the higher dose range of DNA (0.125 µg–0.5 µg). As a result of better transfection efficiencies using lower DNA doses, the amount of DNA, peptides and PEI could be reduced considerably. Lower DNA doses lead to a reduction of toxic effects by gene transfer complexes. Therefore we used a DNA dose of 0.25 µg in the following transfections.
Using FACS analysis it was clearly visible that by transfecting Ku702-NLS/transMagPei/DNA complexes the number of transfected cells was higher compared to binary transMagPei/DNA complexes. This result could be confirmed on both cell lines. Ku702-NLS enhanced gene expression stronger by enhancing transgene expression per cell than enhancing number of transfected cells. This result was not observed after transfections with s2Ku702. The better transfection results of Ku702-NLS compared to s2Ku702 rely on the fact that Ku702-NLS is a NLS.
The results of in vitro transfections revealed that Ku702-NLS/PEI/DNA particles are able to transfect effectively human bronchoepithelial cells. For this reason an in vivo application of Ku702-NLS/PEI/DNA and s1Ku702-NLS/PEI/DNA complexes was conducted. In these experiments an enhancement of gene transfer mediated from Ku702-NLS (46%) or s1Ku702-NLS (77%) compared to PEI/DNA complexes was observed.
In conclusion, ternary gene vector complexes consisting of Ku702-NLS, PEI and DNA represent an effective gene delivery system. A clear enhancement of transgene expression was observed compared to PEI/DNA. Differences between Ku702-NLS and s1Ku702-NLS are marginal, compared to the nuclear transport-deficient s2Ku702. Gene transfer efficiency using the bipartite NLS Ku702-NLS improves transgene expression compared to monopartite NLS. For in vivo applications Ku702-NLS and s1Ku702-NLS have to be further optimized. Ku702-NLS and s1Ku702-NLS promise gene transfer agents in the field of non-viral gene delivery.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was supported by the the German Research Foundation (DFG) (Deutsche Forschungsgemeinschaft) RO 994/2-4. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 2.Honigman A, Zeira E, Ohana P, Abramovitz R, Tavor E, et al. Imaging transgene expression in live animals. Mol Ther. 2001;4:239–249. doi: 10.1006/mthe.2001.0437. [DOI] [PubMed] [Google Scholar]
- 3.Plank C, Oberhauser B, Mechtler K, Koch C, Wagner E. The influence of endosome-disruptive peptides on gene transfer using synthetic virus-like gene transfer systems. J Biol Chem. 1994;269:12918–12924. [PubMed] [Google Scholar]
- 4.Pollard H, Remy JS, Loussouarn G, Demolombe S, Behr JP, et al. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J Biol Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 5.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 7.Ciolina C, Byk G, Blanche F, Thuillier V, Scherman D, et al. Coupling of nuclear localization signals to plasmid DNA and specific interaction of the conjugates with importin alpha. Bioconjug Chem. 1999;10:49–55. doi: 10.1021/bc980061a. [DOI] [PubMed] [Google Scholar]
- 8.Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci U S A. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebestyen MG, Ludtke JJ, Bassik MC, Zhang G, Budker V, et al. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nat Biotechnol. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 10.Ritter W, Plank C, Lausier J, Rudolph C, Zink D, et al. A novel transfecting peptide comprising a tetrameric nuclear localization sequence. J Mol Med. 2003;81:708–717. doi: 10.1007/s00109-003-0483-2. [DOI] [PubMed] [Google Scholar]
- 11.Rudolph C, Plank C, Lausier J, Schillinger U, Muller RH, et al. Oligomers of the arginine-rich motif of the HIV-1 TAT protein are capable of transferring plasmid DNA into cells. J Biol Chem. 2003;278:11411–11418. doi: 10.1074/jbc.M211891200. [DOI] [PubMed] [Google Scholar]
- 12.Surovoy A, Flechsler I, Gaunitz F, Papke M, Jung G. Simple and fast microscale procedure for transfection and quantification of reporter gene expression in eukaryotic cells. Adv Exp Med Biol. 1998;451:457–460. doi: 10.1007/978-1-4615-5357-1_70. [DOI] [PubMed] [Google Scholar]
- 13.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, et al. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci U S A. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erbacher P, Bettinger T, Belguise-Valladier P, Zou S, Coll JL, et al. Transfection and physical properties of various saccharide, poly(ethylene glycol), and antibody-derivatized polyethylenimines (PEI). J Gene Med. 1999;1:210–222. doi: 10.1002/(SICI)1521-2254(199905/06)1:3<210::AID-JGM30>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 15.Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS Pharm Sci. 2001;3:E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, et al. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 17.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J Gene Med. 2001;3:362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 18.Plank C, Schillinger U, Scherer F, Bergemann C, Remy JS, et al. The magnetofection method: using magnetic force to enhance gene delivery. Biol Chem. 2003;384:737–747. doi: 10.1515/BC.2003.082. [DOI] [PubMed] [Google Scholar]
- 19.Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, et al. Magnetofection: enhancing and targeting gene delivery by magnetic force in vitro and in vivo. Gene Ther. 2002;9:102–109. doi: 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- 20.Koike M. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J Radiat Res (Tokyo) 2002;43:223–236. doi: 10.1269/jrr.43.223. [DOI] [PubMed] [Google Scholar]
- 21.Koike M, Ikuta T, Miyasaka T, Shiomi T. The nuclear localization signal of the human Ku70 is a variant bipartite type recognized by the two components of nuclear pore-targeting complex. Exp Cell Res. 1999;250:401–413. doi: 10.1006/excr.1999.4507. [DOI] [PubMed] [Google Scholar]