Abstract
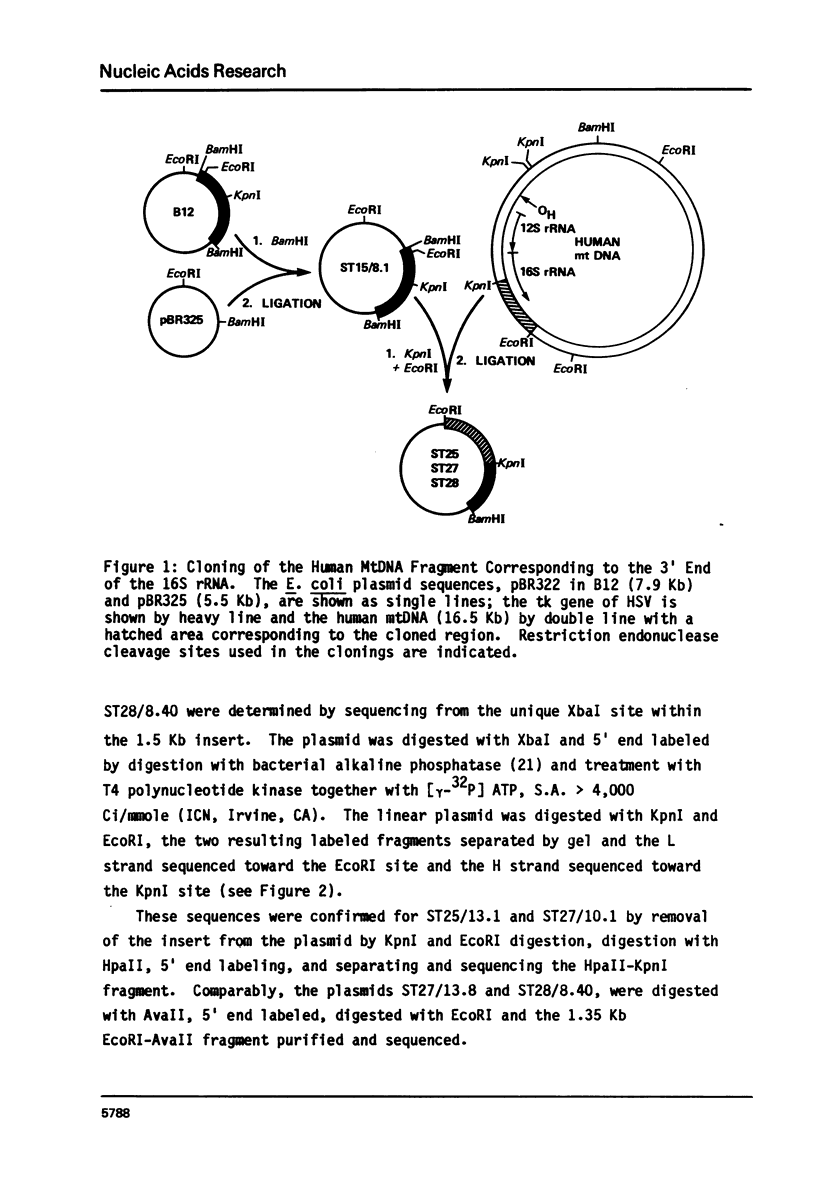
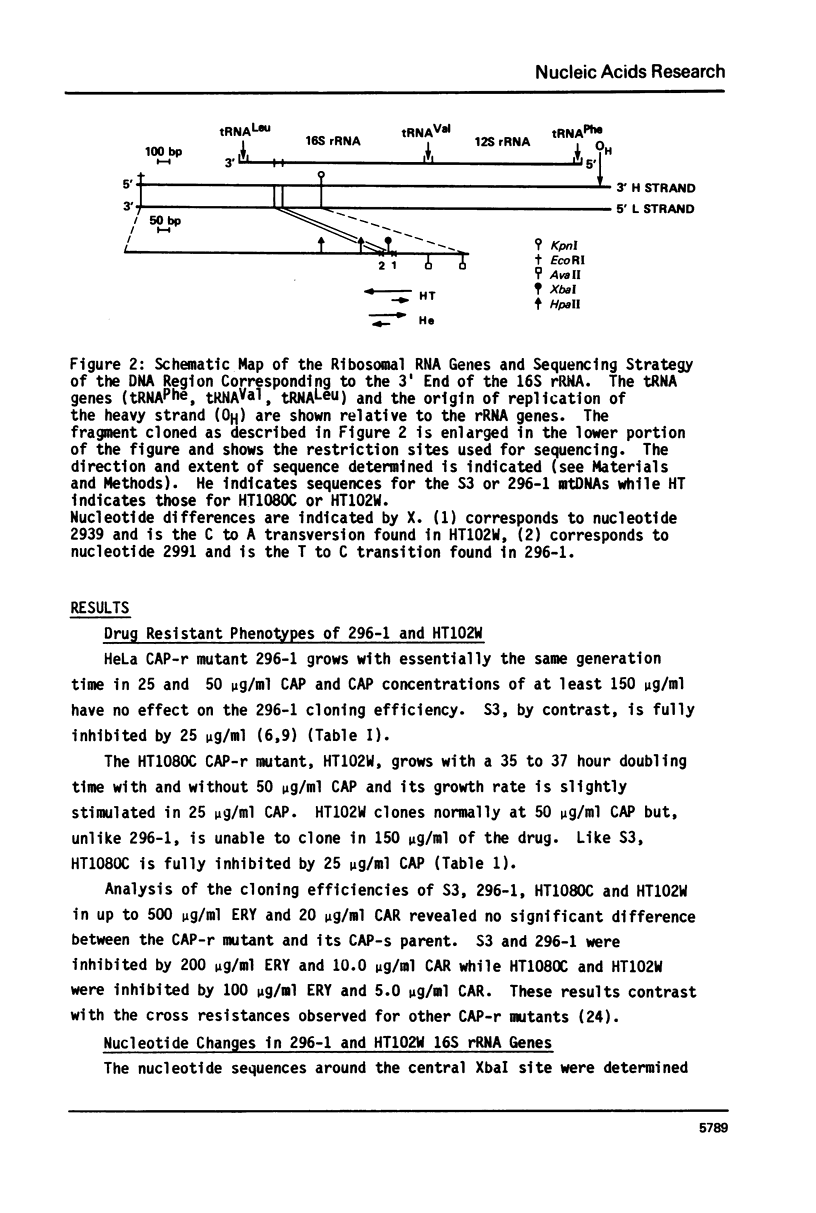
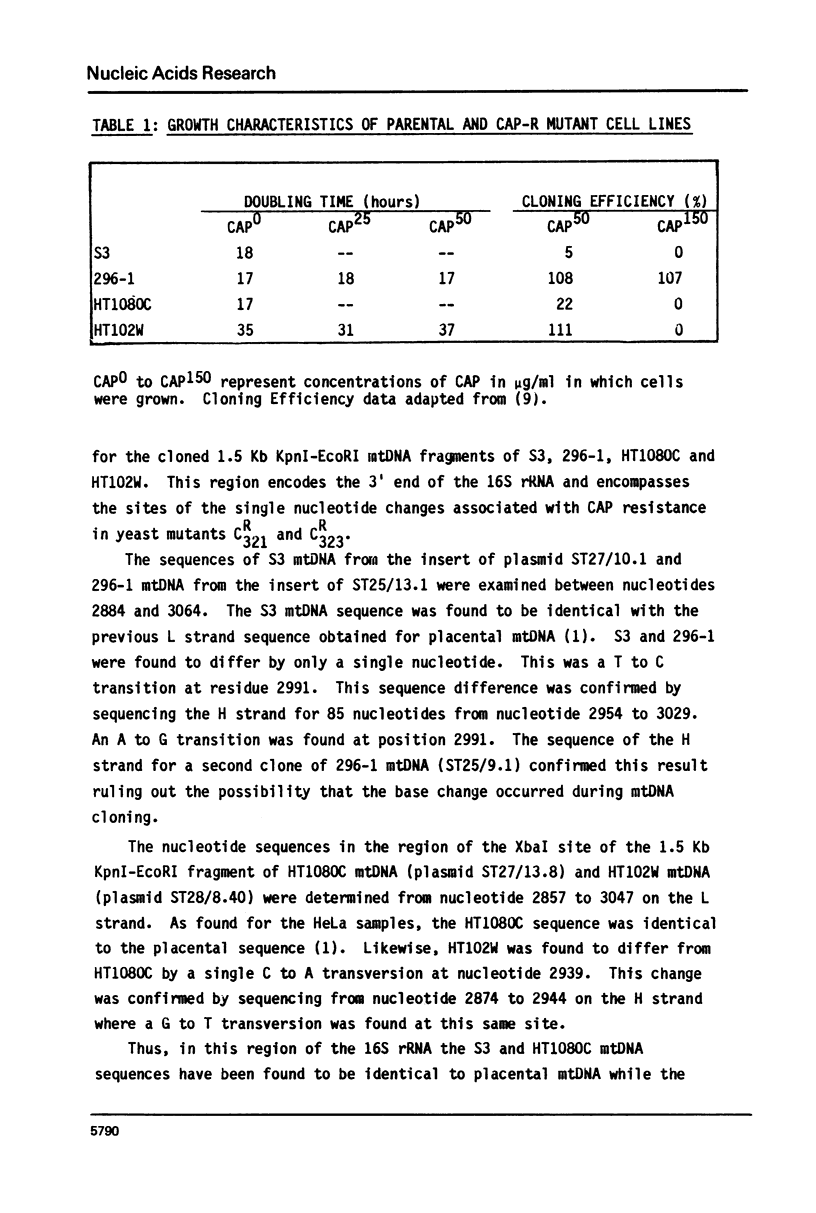
The nucleotide sequence of the mitochondrial DNA (mtDNA) in the region coding for the 3' end of the large rRNA has been determined for two human cell lines bearing independent cytoplasmic chloramphenicol-resistant (CAP-r) mutations. Comparison of the sequences of these two phenotypically different CAP-r mutants with their CAP-sensitive (CAP-s) parental cell lines has revealed a single base change for each in a region which is highly conserved among species. One CAP-r mutation is associated with an A to G transition on the coding strand while the second contains a G to T transversion 52 nucleotides away. Comparable sequence changes in this region had previously been found for mouse and yeast cell mitochondrial CAP-r mutants. Thus, changes in the large rRNA gene eliminate the inhibition of the ribosome by CAP and different nucleotide changes may result in variations in the drug-r phenotype.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baer R. J., Dubin D. T. Methylated regions of hamster mitochondrial ribosomal RNA: structural and functional correlates. Nucleic Acids Res. 1981 Jan 24;9(2):323–337. doi: 10.1093/nar/9.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc H., Wright C. T., Bibb M. J., Wallace D. C., Clayton D. A. Mitochondrial DNA of chloramphenicol-resistant mouse cells contains a single nucleotide change in the region encoding the 3' end of the large ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3789–3793. doi: 10.1073/pnas.78.6.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D. A. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974 Dec 25;249(24):7991–7995. [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunn C. L., Wallace D. C., Eisenstadt J. M. Cytoplasmic inheritance of chloramphenicol resistance in mouse tissue culture cells. Proc Natl Acad Sci U S A. 1974 May;71(5):1681–1685. doi: 10.1073/pnas.71.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen D., Deutsch J., Netter P., Petrochilo E., Slonimski P. P. Mitochondrial genetics. I. Methodology and phenomenology. Symp Soc Exp Biol. 1970;24:449–496. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Edwards K., Kössel H. The rRNA operon from Zea mays chloroplasts: nucleotide sequence of 23S rDNA and its homology with E.coli 23S rDNA. Nucleic Acids Res. 1981 Jun 25;9(12):2853–2869. doi: 10.1093/nar/9.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles R. E., Stroynowski I., Wallace D. C. Characterization of mitochondrial DNA in chloramphenicol-resistant interspecific hybrids and a cybrid. Somatic Cell Genet. 1980 Jul;6(4):543–554. doi: 10.1007/BF01539155. [DOI] [PubMed] [Google Scholar]
- Grivell L. A., Netter P., Borst P., Slonimski P. P. Mitochondrial antibiotic resistance in yeast: ribosomal mutants resistant to chloramphenicol, erythromycin and spiramycin. Biochim Biophys Acta. 1973 Jun 23;312(2):358–367. doi: 10.1016/0005-2787(73)90380-8. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey S. E., Craig I. W. Altered ribosomal RNA genes in mitochondria from mammalian cells with chloramphenicol resistance. Nature. 1981 Apr 16;290(5807):607–608. doi: 10.1038/290607a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meagher R. B., Tait R. C., Betlach M., Boyer H. W. Protein expression in E. coli minicells by recombinant plasmids. Cell. 1977 Mar;10(3):521–536. doi: 10.1016/0092-8674(77)90039-3. [DOI] [PubMed] [Google Scholar]
- Montoya J., Ojala D., Attardi G. Distinctive features of the 5'-terminal sequences of the human mitochondrial mRNAs. Nature. 1981 Apr 9;290(5806):465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- Nierhaus D., Nierhaus K. H. Identification of the chloramphenicol-binding protein in Escherichia coli ribosomes by partial reconstitution. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2224–2228. doi: 10.1073/pnas.70.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiyama H., Sakaki Y., Takagi Y. Nucleotide sequence of a ribosomal RNA gene intron from slime mold Physarum polycephalum. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1376–1380. doi: 10.1073/pnas.78.3.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981 Apr 9;290(5806):470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer. 1974 Apr;33(4):1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Jeffreys A. J., Sly W., Craig I. W. Isolation and detailed characterization of human cell lines resistant to D-threo-chloramphenicol. Exp Cell Res. 1976 Oct 15;102(2):298–310. doi: 10.1016/0014-4827(76)90045-8. [DOI] [PubMed] [Google Scholar]
- Spolsky C. M., Eisenstadt J. M. Chloramphenicol-resistant mutants of human HeLa cells. FEBS Lett. 1972 Sep 15;25(2):319–324. doi: 10.1016/0014-5793(72)80514-3. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Assignment of the chloramphenicol resistance gene to mitochondrial deoxyribonucleic acid and analysis of its expression in cultured human cells. Mol Cell Biol. 1981 Aug;1(8):697–710. doi: 10.1128/mcb.1.8.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Bunn C. L., Eisenstadt J. M. Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. J Cell Biol. 1975 Oct;67(1):174–188. doi: 10.1083/jcb.67.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]


