Abstract
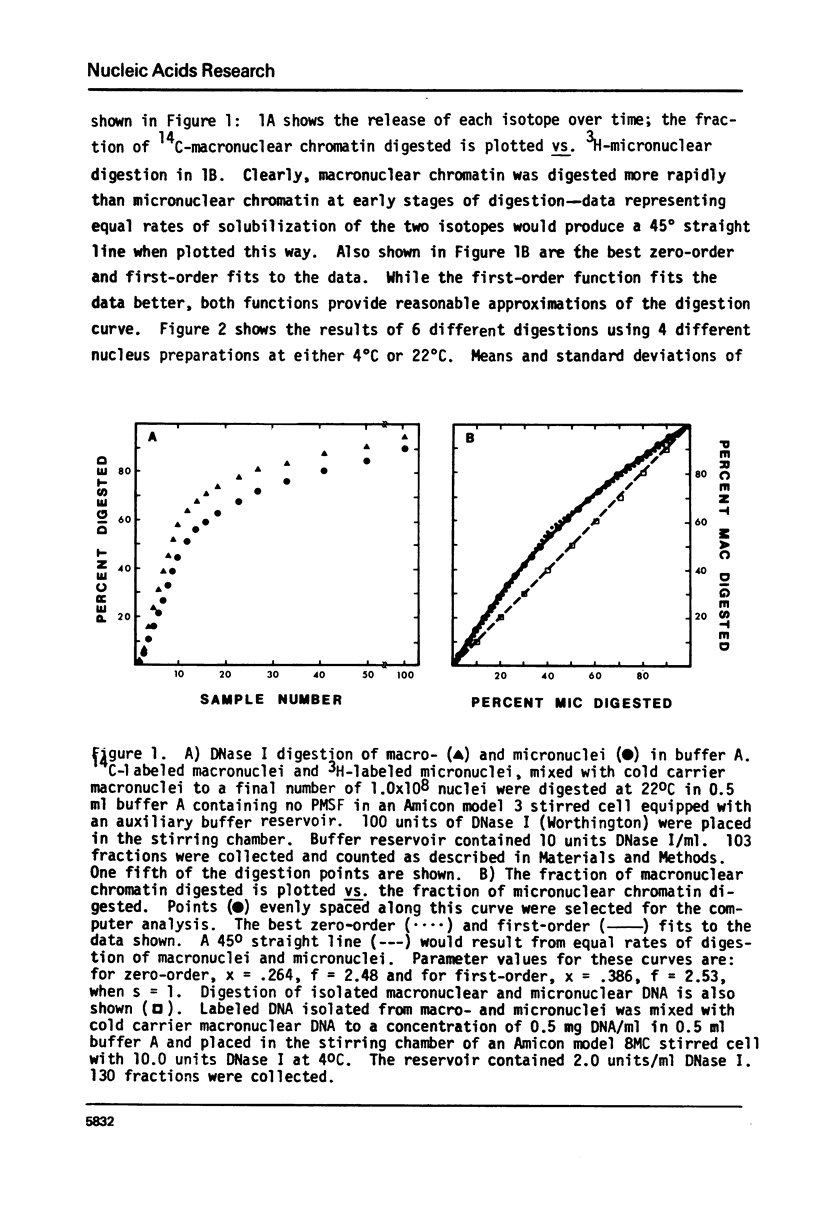
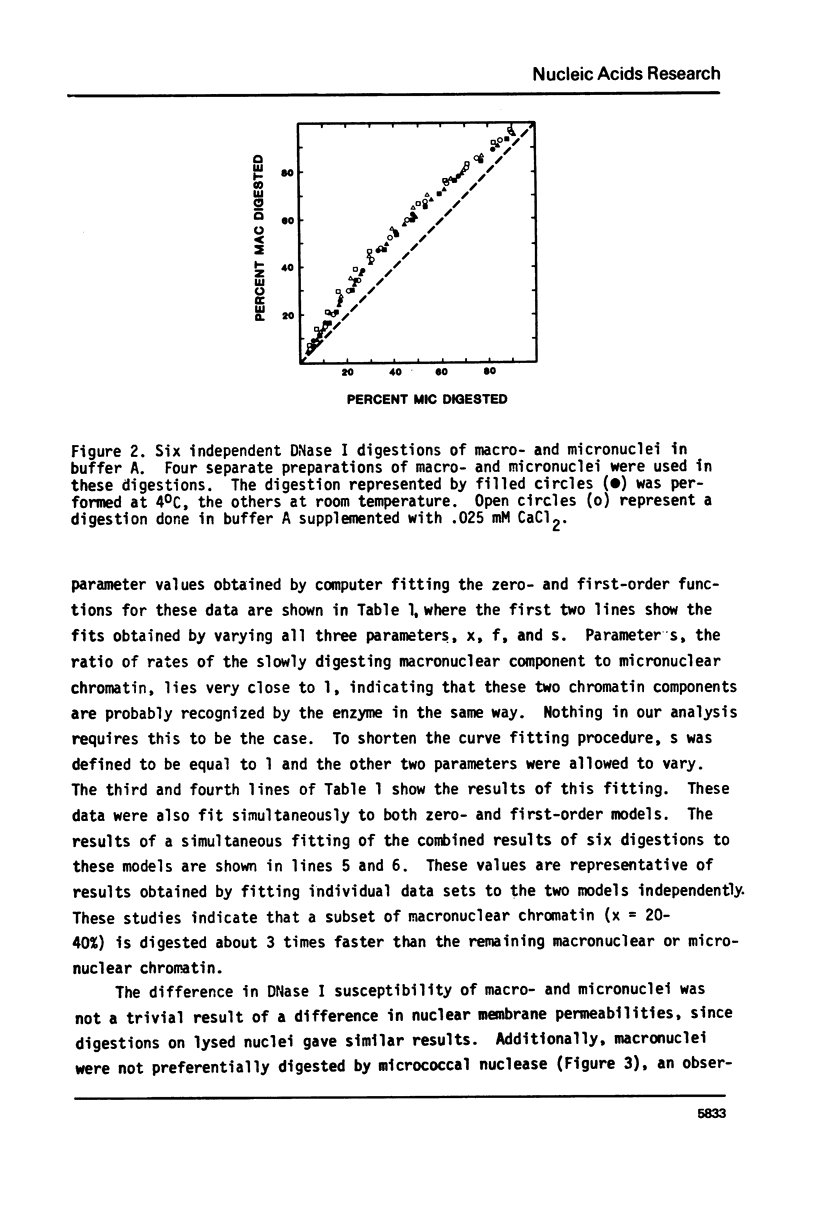
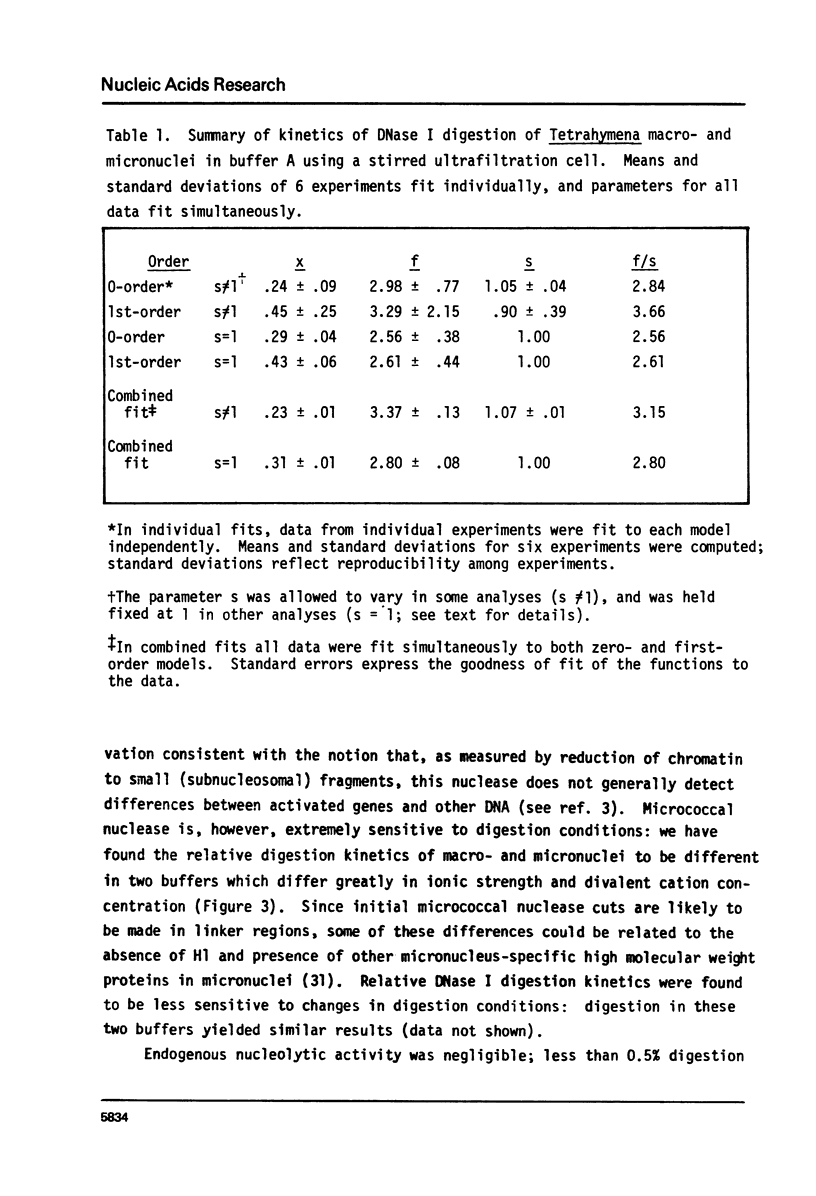
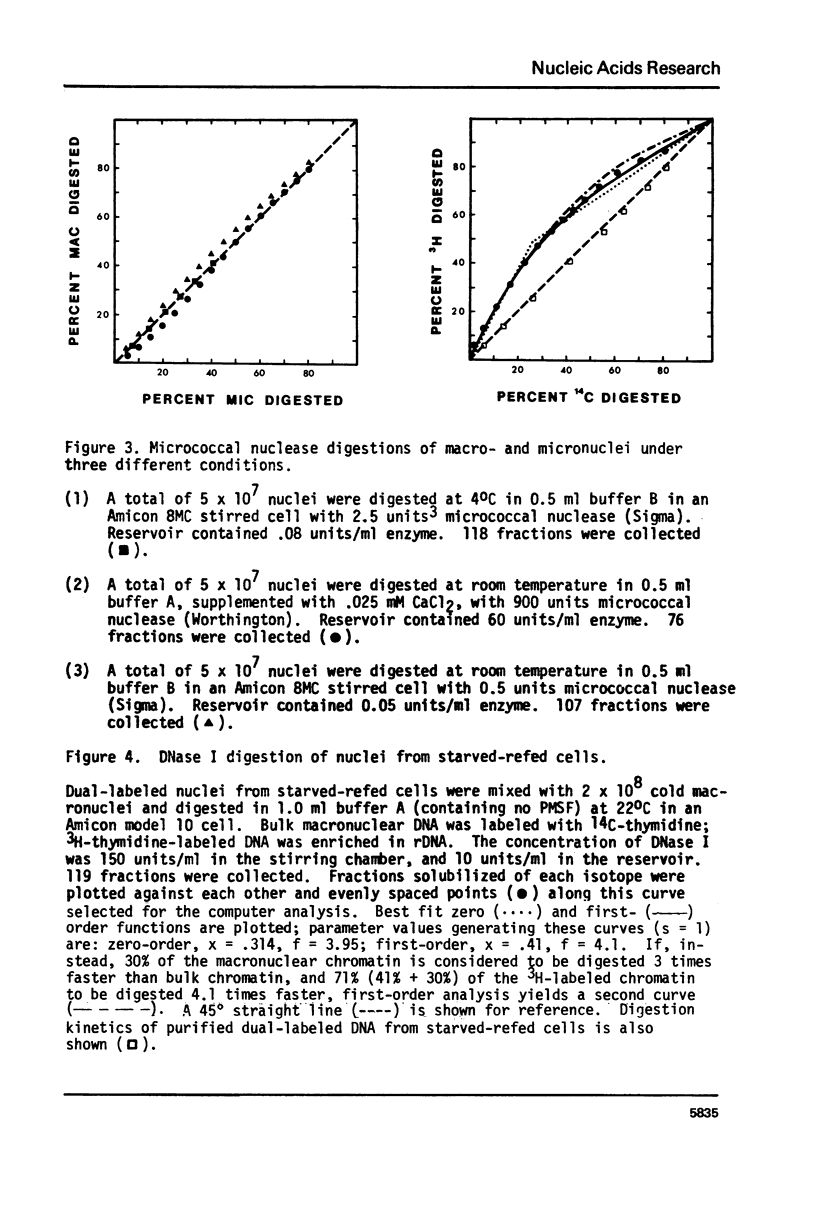
Methods have been developed to analyze the kinetics of digestion of chromatin by nucleases. Radioactively labeled nuclei were incubated with enzyme in an ultrafiltration apparatus and digestion rates of different chromatin samples were computed employing a least-squares curve fitting technique to fit the data to zero-order and/or first-order kinetic models. These methods allow detailed kinetic analyses on small amounts of chromatin. Two biological systems were studied. 1) Tetrahymena thermophila macronuclei and micronuclei were compared; these nuclei differ in their transcriptional activities. 2) Ribosomal DNA (rDNA) of Tetrahymena pyriformis, approximately 60% of which codes for rRNA, can be preferentially labeled during starvation-refeeding; its digestion kinetics relative to bulk chromatin were studied. DNase I digested 20-40% of the macromolecular DNA about 3 times faster than bulk macronuclear or micronuclear DNA, and 60-80% of ribosomal gene-containing chromatin about 5 times faster than bulk chromatin. Filter hybridization studies of the DNAase I sensitivity of tRNA, 5S RNA, and ribosomal genes yielded similar results. These data are consistent with the observation that transcribed genes are especially sensitive to attach by DNase I and suggest that activated chromatin structure as probed by extensive DNase I digestion is the same in higher and lower eucaryotes for genes transcribed by all three RNA polymerases. Digestion kinetics of micrococcal nuclease were found to depend on the digestion conditions employed. These two biological systems and the methods we have developed should facilitate analyses of the factors responsible for maintaining an active chromatin structure.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Allis C. D., Glover C. V., Gorovsky M. A. Micronuclei of Tetrahymena contain two types of histone H3. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4857–4861. doi: 10.1073/pnas.76.10.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard M., Kuo M. T., Dretzen G., Chambon P. Differential nuclease sensitivity of the ovalbumin and beta-globin chromatin regions in erythrocytes and oviduct cells of laying hen. Nucleic Acids Res. 1980 Jun 25;8(12):2737–2750. doi: 10.1093/nar/8.12.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nilsson J. R., Pearlman R. E., Leick V. Induction of nucleolar and mitochondrial DNA replication in Tetrahymena pyriformis. Proc Natl Acad Sci U S A. 1974 Mar;71(3):894–898. doi: 10.1073/pnas.71.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garel A., Zolan M., Axel R. Genes transcribed at diverse rates have a similar conformation in chromatin. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4867–4871. doi: 10.1073/pnas.74.11.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri C. P., Gorovsky M. A. DNase I sensitivity of ribosomal genes in isolated nucleosome core particles. Nucleic Acids Res. 1980 Jan 11;8(1):197–214. doi: 10.1093/nar/8.1.197-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A., Carlson K., Rosenbaum J. L. Simple method for quantitive densitometry of polyacrylamide gels using fast green. Anal Biochem. 1970 Jun;35(2):359–370. doi: 10.1016/0003-2697(70)90196-x. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Gorovsky M. A. Numbers of 5S and tRNA genes in macro- and micronuclei of Tetrahymena pyriformis. Chromosoma. 1976 Mar 10;54(4):327–337. doi: 10.1007/BF00292813. [DOI] [PubMed] [Google Scholar]
- Kuo M. T., Mandel J. L., Chambon P. DNA methylation: correlation with DNase I sensitivity of chicken ovalbumin and conalbumin chromatin. Nucleic Acids Res. 1979 Dec 20;7(8):2105–2113. doi: 10.1093/nar/7.8.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D. J., Gorovsky M. A. Structure of rDNA-containing chromatin of Tetrahymena pyriformis analyzed by nuclease digestion. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):773–778. doi: 10.1101/sqb.1978.042.01.077. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Gorovsky M. A. Subunit structure of rDNA-containing chromatin. Biochemistry. 1976 Feb 24;15(4):750–755. doi: 10.1021/bi00649a005. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Oudet P., Wasylyk B., Chambon P. Effect of histone acetylation on structure and in vitro transcription of chromatin. Nucleic Acids Res. 1978 Oct;5(10):3523–3547. doi: 10.1093/nar/5.10.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. A., Perry M., Sealy L., Chalkley R. DNAse I preferentially digests chromatin containing hyperacetylated histones. Biochem Biophys Res Commun. 1978 Jun 29;82(4):1346–1353. doi: 10.1016/0006-291x(78)90337-6. [DOI] [PubMed] [Google Scholar]
- Nilsson J. R., Leick V. Nucleolar organization and ribosome formation in Tetrahymena pyriformis GL. Exp Cell Res. 1970 Jun;60(3):361–372. doi: 10.1016/0014-4827(70)90529-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Stalder J., Seebeck T., Braun R. Degradation of the ribosomal genes by DNAse I in Physarum polycephalum. Eur J Biochem. 1978 Oct;90(2):391–395. doi: 10.1111/j.1432-1033.1978.tb12616.x. [DOI] [PubMed] [Google Scholar]
- VANECKO S., LASKOWSKI M., Sr Studies of the specificity of deoxyribonuclease I. II. Hydrolysis of oligonucleotides carrying a monoesterified phosphate on carbon 3'. J Biol Chem. 1961 Apr;236:1135–1140. [PubMed] [Google Scholar]
- Vidali G., Boffa L. C., Bradbury E. M., Allfrey V. G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci U S A. 1978 May;75(5):2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Recognition of specific DNA sequences in eukaryotic chromosomes. Nucleic Acids Res. 1980 Oct 24;8(20):4745–4753. doi: 10.1093/nar/8.20.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S., Groudine M., Weintraub H. Interaction of HMG 14 and 17 with actively transcribed genes. Cell. 1980 Jan;19(1):289–301. doi: 10.1016/0092-8674(80)90410-9. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):630–634. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Gilbert W. Tissue-specific exposure of chromatin structure at the 5' terminus of the rat preproinsulin II gene. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1577–1580. doi: 10.1073/pnas.78.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Gorovsky M. A. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48(1):1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]


