Abstract
The proepicardium is a transient embryonic structure that is a source of precursors of the epicardium, coronary smooth muscle cells, and may be a source of coronary endothelial cells (EC). To better understand proepicardium development a systematic analysis of EC appearance was performed. Multiple marker analysis showed that EC are present in the mouse proepicardium at E9.0 through E9.75. Distinct populations of EC were found that were associated with the liver bud, and the sinus venosus, as well as a population that do not appear to be associated with either of these structures. There was a temporal increase in the number of EC and temporal changes in the distribution of EC within the different populations during PE development. These findings indicate that EC exist in the proepicardium prior to coronary vasculogenesis, and support a model in which there is a heterogeneous origin for EC in the proepicardium.
Introduction
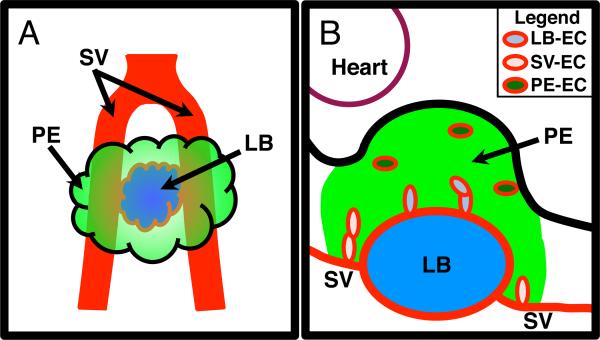
The proepicardium (PE) is a small “grape-like” transient embryonic structure that gives rise to lineage precursors that contribute to the epicardium and the vasculature of the heart. In mouse, the PE is initially present in E9.0 through E9.75 embryos and appears as an outgrowth of mesothelial cells adjacent to the liver bud (LB) and the sinus venosus (SV) all illustrated in Figure 1A. As the embryo develops, the cells of the PE migrate to the surface of the heart where they will begin to form the epicardium. As well as giving rise to the epicardium, lineage-tracing analysis shows that the PE contributes coronary smooth muscle cell progenitors to the developing heart as well as cells that will make up part of the annulus fibrosis (Mikawa and Gourdie, 1996; Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Vrancken Peeters et al., 1999; Guadix et al., 2006; Zhou et al., 2010). It has also been suggested that cells of the PE may contribute cardiomyocytes (Cai et al., 2008; Zhou et al., 2008).
Figure 1.
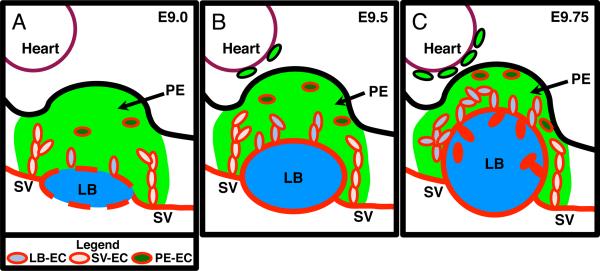
A Illustration displaying the relative location of the PE in comparison to the LB and the SV. Ventral diagram of an E9.5 mouse PE with the PE depicted in green, the SV in red, and the LB in blue. B Illustration displaying the different populations of EC within the mouse E9.5 PE. Depiction of a sagittal section of an E9.5 mouse PE with the green area signifying the PE region, based on WT1 expression. Blue signifies the developing LB. Heart is located in the upper left demarked by a maroon line. Red line demarks EC and vessels. Light blue ovals depict EC associated with the LB (LB-EC), pink ovals depict EC associated with SV (SV-EC) and dark green ovals depict EC not associated with the SV or LB (PE-EC).
The precise origin of coronary endothelial cells (EC) is incompletely understood. Early PE lineage analysis using direct PE labeling and quail/chick chimera suggested that the PE contains cells that contribute to the coronary EC population (Mikawa and Gourdie, 1996; Perez-Pomares et al., 1998b; Perez-Pomares et al., 1998a; Perez-Pomares et al., 2002a; Guadix et al., 2006). Further analysis of chick embryos identified EC in the PE that appear to originate from two highly vascularized adjacent structures: the underlying LB and the adjacent SV (Vrancken Peeters et al., 1997; Lie-Venema et al., 2005). It is unclear whether the EC observed in these studies migrate to the heart along with the rest of the PE cells to contribute to the coronary EC population. Additionally, non-PE origins for coronary EC have also been suggested. A recent study using transgenic lineage analysis in mice identified the endocardium as well as the sinus venosus associated with the atria as major sources for coronary EC and has questioned the role of the PE as a source for coronary EC (Red-Horse et al., 2010).
To better understand the origin of the different cell populations that make up the PE and to determine whether EC are present within the mouse PE, we have undertaken a careful analysis of PE-associated EC during mouse PE development. Using a cell-type marker analysis approach, our results indicate that three apparently distinct populations of EC exist within the PE: 1) a population associated with the developing LB (LB-EC), 2) a population associated with the SV (SV-EC) and 3) a population of EC that is not associated with either of these two structures which we have termed PE-EC (Figure 1B). The distribution of EC within each of these populations was found to differ from E9.0 through E9.75, suggesting regional differentiation and signaling events occur during PE development. To our knowledge, this is the first analysis in which EC within the developing mouse PE has been described in detail. Our analysis provides important information as to the make up of the cells within the mouse PE.
Results
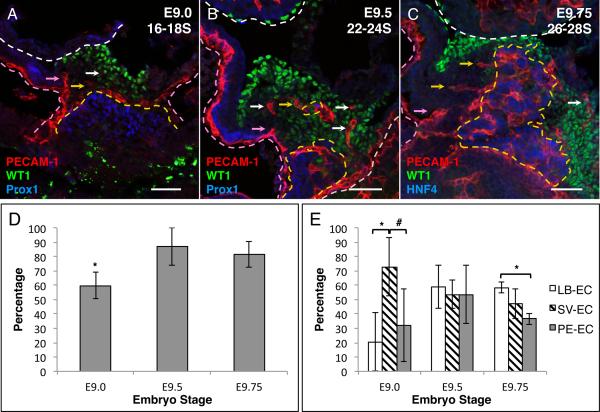
PECAM-1 positive endothelial cells are present in the mouse PE
Platelet endothelial cell adhesion molecule-1 (PECAM-1), a common EC marker expressed on the cell surface, was used to analyze the in vivo pattern of EC within tissue (Baldwin et al., 1994; Drake and Fleming, 2000). To examine the temporal pattern of PECAM-1 expression in the developing PE, we examined embryos at different developmental stages: E9.0 (16–18 somites), E9.5 (22–24 somites) and E9.75 (26–28 somites). The boundaries of the PE are based on the expression of the published PE marker gene WT1 and the LB markers Prox1 for E9.0 and E9.5 and HNF4 for E9.75 (Moore et al., 1999; Li et al., 2000; Burke and Oliver, 2002; Perez-Pomares et al., 2002b; Tremblay and Zaret, 2005). PECAM-1 expression was observed in the PE in E9.0, E9.5 and E9.75 mouse embryos (Figure 2A–C). There were fewer analyzed confocal z-series steps that contained PECAM-1 positive cells in the PE of E9.0 embryos (59.88% ± 9.10) compared to observation in the PE of E9.5 (86.9% ± 13.06; p value = 0.021) or E9.75 (81.56% ± 8.91; p value = 0.034) embryos (Figure 2D). These results established that EC are present in the developing PE. We therefore next wanted to address whether different populations of EC might exist in the PE. Based on the location of PECAM-1 expression, we identified three distinct populations of EC in the PE: 1) EC that are associated with the LB (LB-EC), 2) those associated with the SV (SV-EC), and 3) any EC that are in the PE and not associated with SV or LB (PE-EC) as illustrated in Figure 1B. As seen in Figure 2A, in E9.0 embryos, PECAM-1 positive cells are observed surrounding the developing LB with a small number of cells outspreading from the LB into the PE. EC are also seen extending/spreading from the SV into the PE. Comparison of the total PECAM-1 positive confocal z-series steps between the three populations reveals that at E9.0, PECAM-1 expression is more frequent within the SV-EC population (72.91% ± 20.21) than the LB-EC (20.61% ± 20.29; p value = 0.034) or the PE-EC (32.09% ± 25.25; p value = 0.094) (Figure 2E). In E9.5 embryos, there were EC observed extending from the SV and the LB as well as EC observed in the PE-EC population (Figure 2B). All three populations were roughly equally observed (LB-EC 59.22% ± 15.18, SV-EC 53.75% ± 9.73, and PE-EC 53.50% ± 20.35) (Figure 2E). In E9.75 embryos, PECAM-1 expression is more frequent within the LB-EC population (58.49% ± 3.82) when compared to PE-EC (36.57% ± 4.02; p value = 0.002) (Figure 2C and E). These observations support the idea that three different populations of PECAM-1 positive cells are present during PE formation and migration (E9.0 through E9.75), and that each population appears to have a temporal distribution that varies as a function of developmental stage of the PE. Interestingly, we did not observe any PECAM-1 positive cells on the surface of the heart indicating that at E9.5 or E9.75, these cells either have not yet migrated or do not migrate to the heart with the rest of the cells of the PE.
Figure 2.
PECAM-1 positive cells are present in the developing mouse PE. A–C Immunofluorescence analysis of sagittal sections. A E9.0 16–18 somite (16–18S), B E9.5 22–24 somite (22–24S) and C E9.75 26–28 somite (26–28S) mouse embryos. Sections were stained with the PE marker WT1 (green), EC marker PECAM-1 (red) and LB markers Prox1 or HNF4 (blue). D Percentage of PE confocal step that contain PECAM-1 positive cells in developing mouse PE. E Percentage of PECAM-1 positive confocal steps that contain one or more PECAM-1 positive cells in the three different EC populations (LB-EC, SV-EC and PE-EC). White dashed line demarks the border of the heart. Yellow dashed line demarks the border of the LB. Pink dashed line demarks the SV. Yellow arrows indicate examples of LB-EC, pink arrow indicate examples of SV-EC and white arrows indicate examples of PE-EC. Scale bars = 50μm * p value ≤ 0.05 # p value ≤ 0.1
To address the possibility that an increase in EC proliferation could account for the increase in observed EC in the PE, we examined the proliferation of PECAM-1 positive cells using phosphohistone H3 (PH3) staining in E9.5 mouse embryos. The proliferation rate of EC was found to be very low, with an average of 1–3 PECAM-1/PH3 double positive cells per 20μm section (data not shown). These data indicate that the increase in PECAM-1 positive cells most likely arises either from de novo differentiation of EC progenitor (angioblasts) or from migration into the PE from the LB or the SV.
ERG positive endothelial cells are present in the mouse PE
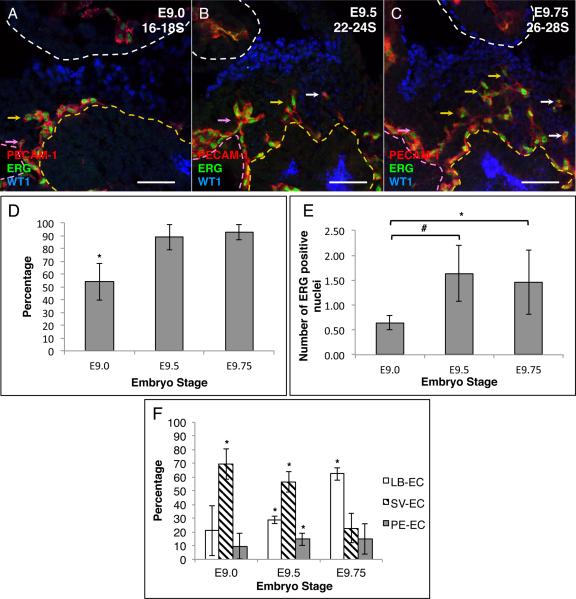
While PECAM-1 or any other EC surface marker are ideal for describing the pattern of EC streaming/migration and differentiation, they are problematic for accurate cell number quantification since EC morphology can vary. To accurately quantitate the number of EC within each population, we used a nuclear endothelial cell marker ETS-related gene (ERG) (Vlaeminck-Guillem et al., 2000; McLaughlin et al., 2001; Mohamed et al., 2010). The boundaries of the PE were also based on the expression of the published PE marker gene WT1 and the LB markers Prox1 for E9.0 and E9.5 and HNF4 for E9.75 (Moore et al., 1999; Li et al., 2000; Burke and Oliver, 2002; Perez-Pomares et al., 2002b; Tremblay and Zaret, 2005). ERG was observed to extensively co-express with PECAM-1 within the developing PE. Therefore we used ERG in conjunction with PECAM-1 to accurately place individual EC into the same three different populations. ERG expression was observed in the PE and in all three populations in E9.0, E9.5 and E9.75 mouse embryos (Figure 3A–C and F). Fewer ERG positive cells were observed in the PE in E9.0 embryos (53.90% ± 14.21) compared to E9.5 (88.76% ± 9.74; p value = 0.025) or E9.75 (92.92% ± 6.09; p value = 0.012) embryos (Figure 3D). The average number of ERG positive cells per μm of PE showed a similar increase over time, with fewer average ERG positive cells observed in E9.0 embryos (0.65 ± 0.15) compared to E9.5 (1.63 ± 0.56; p value = 0.042) or E9.75 (1.46 ± 0.64; p value = 0.100) embryos (Figure 3E).
Figure 3.
ERG positive cells are found in the developing mouse PE. A–C Immunofluorescence analysis of sagittal sections. A E9.0 16–18 somite (16–18S), B E9.5 22–24 somite (22–24S) and C E9.75 26–28 somite (26–28S) mouse embryos. Sections were stained with the PE marker WT1 (blue), EC markers ERG (green) and PECAM-1 (red). The expression of Prox1 and HNF4 was omitted in these figures to more clearly visualize the co-expression of PECAM-1 and ERG along with the PE marker WT1. D Percentage of PE confocal steps that contain ERG positive cells in developing mouse PE. E Average number of ERG positive cells per mm of PE width. F Percentage of ERG positive cells within the individual EC populations (LB-EC, SV-EC and PE-EC). White dashed line demarks the border of the heart. Yellow dashed line demarks the border of the LB. Pink dashed line demarks the SV. Yellow arrows indicate examples of LB-EC, pink arrow indicate examples of SV-EC and white arrows indicate examples of PE-EC. Scale bars = 50μm * p value ≤ 0.05 # p value ≤ 0.1
Comparison of the distribution of ERG positive EC in the different populations over time reveals a change in temporal distribution of ERG positive cells within the individual populations similar to the change seen with PECAM-1. At E9.0 there are more ERG positive SV-EC (69.54% ± 11.16) compared to the other two populations (LB-EC 20.83% ± 18.04; p value = 0.016, and PE-EC 9.64% ± 9.58; p value = 0.002). A similar trend was observed at E9.5 with more ERG positive cells in the SV-EC population (56.35% ± 7.29) compared to the LB-EC (28.96% ± 2.83; p value = 0.004), or the PE-EC (14.68% ± 4.46; p value = 0.001) (Figure 3F). In contrast, by E9.75 there were more ERG positive cells in the LB-EC population (62.40% ± 4.66) compared to the SV-EC (22.60% ± 10.79; p-value = 0.004) or PE-EC (15.00% ± 11.24; p-value = 0.003) (Figure 3F). These data indicate that there is an increase in the total number of EC during PE development from E9.0 through E9.75. Our data also suggest a shift of EC from being primarily associated with SV at E9.0 and E9.5 to being primarily associated with LB at E9.75.
The PE contains lineage precursor for various types of cardiac associated cells. To address whether the ERG positive cells represent a PE cell population that is distinct from the WT1 population, we examined the co-expression of ERG with the nuclear PE marker WT1. Although WT1 is a commonly used marker for the PE it is not expressed in all cells within the PE (Supplemental Figure 2) (Moore et al., 1999; Perez-Pomares et al., 2002b). In E9.5 embryos we observed 16.03% ± 6.38 of total ERG cells co-expressed WT1 suggesting that the majority of EC in the PE represent a distinct non-WT1 population. Numerous studies indicate that the PE contains lineage precursors for coronary smooth muscle cells (Mikawa and Gourdie, 1996; Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Vrancken Peeters et al., 1999; Guadix et al., 2006). To examine whether smooth muscle cells are present in the PE, we examine embryos at E9.75 for the expression of smooth muscle myosin heavy chain (SM-MHC). At E9.75 SM-MHC is not expressed in the PE (Supplemental Figure 3A). SM-MHC expression is first visible in the heart at E14.5 (Supplemental Figure 3B).
Early and late endothelial cell markers are expressed in the PE
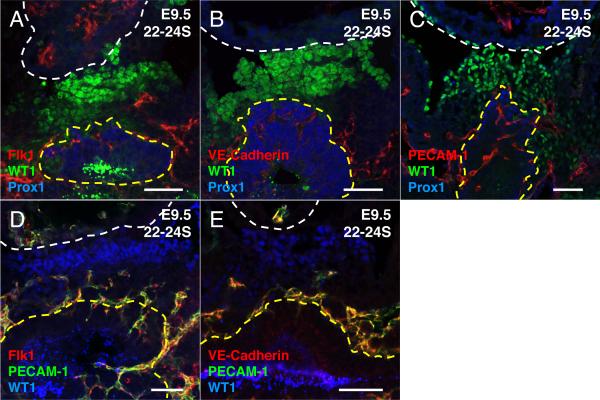
The initial appearance of various EC markers reflects the status of EC differentiation, maturation and vessel morphogenesis (Vittet et al., 1996; Drake and Fleming, 2000; Ferguson et al., 2005). Early markers such as vascular endothelial growth factor receptor 2 (FLK1) are initially expressed in hematopoietic endothelial progenitor cells (angioblasts), and its expression is maintained in EC upon differentiation (Millauer et al., 1993; Yamaguchi et al., 1993). Vascular endothelial cadherin (VE-Cadherin) is a cell adhesion molecule that is not expressed in endothelial progenitor cells but is expressed in differentiated EC (Lampugnani et al., 1992). Similarly, PECAM-1 is initially expressed in differentiated EC, although its expression may precede that of VECadherin (Drake and Fleming, 2000). Therefore FLK1, PECAM-1 and VECadherin expression were used to assess the differentiation status of the various EC populations in E9.5 mouse PE. All three EC markers, FLK1, PECAM-1 and VE-Cadherin, were observed in the PE of E9.5 embryos (n=3) (Figure 4A–C). To determine if these observations represent different stages of EC differentiation and maturation, we conducted co-expression analyses with PECAM-1 in combination with FLK1 or VE-Cadherin (n=3). We observed 100% co-localization of PECAM-1 expression with FLK1, as well as with VE-Cadherin in the LB-EC, SV-EC and PE-EC populations (Figure 4D–E). These data suggest that the various EC populations present in the E9.5 mouse PE are at the same level of EC maturation.
Figure 4.
Early and late EC marker positive cells are found in E9.5 mouse PE. A–E Immunofluorescence analysis of sagittal sections from E9.5 22–24 somite (22–24S) mouse embryos. A–C Sections were stained with the PE marker WT1 (green), LB marker Prox1 (blue), EC markers A Flk1 (red) B VE-Cadherin (red) C PECAM-1 (red). D–E Flk1 and VE-Cadherin positive cells also co-express PECAM-1 in the E9.5 mouse PE. Sections were stained with WT1 (blue), PECAM-1 (green) and D FLK1 (red) E VE-Cadherin (red). White dashed line demarks the border of the heart. Yellow dashed line demarks the border of the LB. Scale bars = 50μm
Discussion
The precise origin of the coronary EC still remains unclear. The coronary vasculature was initially thought to arise from invaginations of the endocardium into the myocardium (Viragh and Challice, 1981). An extra cardiac origin for the vasculature was subsequently proposed when it was observed that the appearance of coronary EC closely followed the appearance of the epicardium (Mikawa and Fischman, 1992; Poelmann et al., 1993). Using quail/chick chimeras to perform lineage-tracing experiments, coronary EC were shown to originate from an extra cardiac source termed the PE, which was also known to contribute to the formation of the epicardium (Viragh et al., 1993; Mikawa and Gourdie, 1996; Dettman et al., 1998; Perez-Pomares et al., 1998b; Perez-Pomares et al., 1998a; Perez-Pomares et al., 2004; Guadix et al., 2006; Watanabe et al., 2006). Additional evidence that the PE and PE-derived epicardial tissues are required for normal coronary vascular formation came from genetic knock-out studies, studies that ablated the PE, and studies that inhibited PE migration. When chick PE was ablated or PE migration was mechanically blocked, there was a reduction or delay in epicardial formation followed by a defect in the formation of the coronary vasculature (Gittenberger-de Groot et al., 2000; Manner et al., 2005). In α4-integrin global null mice, PE cells were able to migrate to the heart, however there was a defect in formation of the epicardium as well as a defective coronary endothelial vascular formation at E12.5 (Yang et al., 1995). Furthermore, in Fog2 global null mice, the epicardium was able to form, however there was a loss in coronary vascular formation due to an inhibition of epicardial epithelial-mesenchymal transition (Tevosian et al., 2000). Together these studies reinforce a model in which the epicardium and the coronary vasculature are derived from a common PE origin, and that the PE is required for formation of the coronary vasculature through its role in epicardial development.
Since the PE forms adjacent to the highly vascularized LB, it has also been proposed that avian coronary EC may originate from the underlying LB (Poelmann et al., 1993). Consistent with this model, LB derived EC are observed migrating to the heart along with the cells of the avian PE (Poelmann et al., 1993; Vrancken Peeters et al., 1997; Lie-Venema et al., 2005). A more recent study, using mouse in vivo cre recombinase based lineage-tracing approaches, has challenged the PE origin of coronary EC (Red-Horse et al., 2010). It was observed that coronary EC originated primarily from the SV with some coronary EC also being derived from endocardial invaginations. In contrast to other studies the authors concluded, based on a Tbx18-cre lineage reporting system, that there was no significant coronary EC contribution from the PE. Tbx18 is expressed in the PE, although it is unclear whether the cre reporter used in these studies is expressed in all cells of the PE or in a subset of cells that may not contribute coronary EC (Kraus et al., 2001; Cai et al., 2008).
In this study we used the expression WT1 to demark the boundaries of the PE. WT1 is commonly used as a PE marker (Moore et al., 1999; Perez-Pomares et al., 2002b). Various inducible and non-inducible WT1-CRE mouse models have been recently generated (Zhou et al., 2008; Zhou et al., 2010; del Monte et al., 2011). WT1-CRE lineage analysis has identified lineage positive cells that co-express with EC markers however the rate of co-expression is very low indicating that the WT1-CRE lineage cells are likely not the primary source of coronary EC (Zhou et al., 2008; Zhou et al., 2010; S. Cossette unpublished observations). During our analysis we observed that WT1 it is not uniformly expressed throughout the PE, consistent with the idea that the PE contains a heterogeneous populations of cells (Supplemental Figure 2). Furthermore, WT1 was found to be minimally co-expressed with EC in the PE (16.03% of the total EC population in the PE). These observations, along with the WT1-CRE lineage analysis of Zhou et al., allow us to hypothesize that the PE may contain coronary EC precursors that are originate from non-WT1 population of PE cells.
Based on the above studies, we believe it is likely that there are multiple possible origins of coronary EC (PE, LB, SV and/or endocardium). While the precise contribution of the PE to the coronary vasculature is still unclear, many studies suggest the PE contains a heterogeneous population of cells that are specified to differentiate into various cell types such as epicardial cells, smooth muscle cells, perivascular fibroblasts, cardiomyocytes and EC (Mikawa and Gourdie, 1996; Dettman et al., 1998; Gittenberger-de Groot et al., 1998; Vrancken Peeters et al., 1999; Guadix et al., 2006; Zhou et al., 2010). To address further the origin of EC in the PE, we have in the current study systematically examined their appearance in the developing mouse PE. Our study has identified three apparently different populations of EC within the PE that appear to have distinct origins: a population associated with the developing LB (LB-EC), a population associated with SV (SV-EC) and a population of that is not associated with either of these two regions (PE-EC) as illustrated in Figure 1B. Further, the relative contribution of these populations to the PE appears to dynamically change during PE development.
Similar to what was observed in the chick (Vrancken Peeters et al., 1997; Lie-Venema et al., 2005), we observed mouse EC migrating into the PE from the adjacent SV and the LB. In contrast to what is seen in the chick, we did not detect any EC that may have migrated with PE cells onto the surface of the heart at E9.5 or E9.75 (Figure 2B–C, 3B–C and 4A–E). Although we were unable to detect EC migration to the heart, it is possible that EC migration from the PE mainly occurs at later times during PE development. By E9.75 PE migration has begun and is not complete until E10.0 to E10.5. Analysis at these later embryonic stages may result in the identification of migrating EC with the PE cells. Another hypothesis is that undifferentiated angioblasts, which are not detected by our analysis, are able to migrate to the heart.
In our analysis we also observed EC that are not associated with the SV or the LB. The precise origin of these cells, designated PE-EC, remains to be determined. One possibility is that the PE-EC may have initially migrated from the SV and the LB and subsequently lost contact with these structures. Alternatively they may reflect a population that has undergone differentiation from EC progenitors (angioblasts) that reside within the PE. Previous studies in avian systems suggest the presence of angioblasts within the PE (Perez-Pomares et al., 1998a; Perez-Pomares et al., 2004; Guadix et al., 2006; Watanabe et al., 2006). However, the presence of resident angioblasts in the mouse PE remains uncertain. EC progenitor analysis using FLK1 expression in conjunction with PECAM-1 and VE-Cadherin expression at E9.5 has so far failed to identify any undifferentiated angioblasts at this stage and also suggest that the EC present in the PE represent the same level of EC maturation (Figure 4A–E). However, it remains possible that there exists a population of EC progenitors in the PE that express low levels of FLK1 that is undetectable by immunofluorescence. Further analysis using additional early angioblast markers, more sensitive expression analyses such as flow cytometry of dissociated PE cells and lineage tracing experiments are needed to further address whether there are resident EC progenitor cells in the PE at these embryonic stages.
Analysis of the temporal expression of PECAM-1 and ERG in PE identified a temporal increase in the overall number of EC in the PE from E9.0 through to E9.75 (Figure 2D and 3D–E) as well as a change in the pattern and number of EC in each population (LB-EC, SV-EC and PE-EC) during PE development (Figure 2E and 3F). Based on these observations, we propose a model in which the SV at E9.0 provide the majority EC to the PE with some EC being derived from de novo differentiation and migration of resident angioblasts surrounding the LB and within the PE itself (Figure 5A). At E9.5 the SV remains the largest contributor of EC, however with the expansion of the LB there is also an increase of EC in the LB-EC population (Figure 5B). By E9.75 the LB has extensively invaded into the PE and is now the largest contributor of EC either from direct migration or the merging with resident SV-EC and PE-EC (Figure 5C). Our model is consistent with observations in avian systems of EC migration into the PE from the SV and LB (Lie-Venema et al., 2005). Our results have elucidated the dynamics of PE development, however, to understand the molecular mechanisms underlying EC differentiation and migration during these earliest steps of coronary vasculogenesis a more careful examination of the expression of pro-angiogenic and vasculogenic factors is needed.
Figure 5.
Schematic model of EC involvement in the developing mouse. Individual panels depict sagittal sections of A E9.0, B E9.5 and C E9.75 mouse PEs. See Figure 1B for description of symbols and colors. A In E9.0 PE, angioblasts within the PE and on the surface of the LB begin to differentiate into EC. EC from the SV migrate into the PE and provide the majority of EC to the PE. B In E9.5 PE, the LB is expanding and is now fully surrounded by EC and there is an increase in LB-EC however SV-EC is still the largest population of EC in the PE. WT1 positive PE cells begin to migrate to the surface of the heart. C In E9.75, the LB has greatly expanded into the PE. The LB is now the largest contributor of EC within the PE.
An open question that remains to be addressed is the exact origin of the EC within the different populations. Do PE-associated EC originate from de novo differentiation of angioblasts, or do they originate by a migration of EC from the SV or the LB? To address whether increased proliferation may account for the increase in the number of EC in the PE we examined PH3 co-expression (data not shown). We found no significant PH3 expression in EC, which is consistent with longstanding observation by Mikawa and colleagues. Our studies support a model in which the increase in EC number in the PE is mainly due to migration of EC from LB and SV in E9.5 PE (data not shown). Another question that remains to be addressed is whether the subpopulations of EC in the PE identified in our study contribute to the coronary vasculature, or whether there might be an as yet to be determined PE population of undifferentiated EC progenitors (angioblasts) that is able to migrate to the heart and differentiate into coronary EC. To properly address this issue a more careful inducible cre based lineage tracing analysis will be required.
In summary, we have identified three apparently distinct EC populations within the developing PE that display a change in developmental pattern between E9.0 and E9.75. While these studies have documented the appearance of EC in the PE, it remains to be determined by other more robust lineage tracking approaches whether these EC are able to migrate to the surface of the heart along with the rest of the cells of the PE as well as the extent to which these EC contribute to the actual coronary vasculature.
Experimental Procedures
Mice
The mice were housed in the Medical College of Wisconsin Biological Resource Center, and all experiments were performed in accordance with an IACUC approved animal procedure protocol. Wild-type mixed background mice were mated and the presence of a vaginal plug the following morning was assessed. If a vaginal plug was observed, the females were determined to be pregnant and the resulting embryos were designated E0.5.
Embryo Harvest
Embryos were collected at the following stages: E9.0 (16–18 somites), E9.5 (22–24 somites) and E9.75 (26–28 somites). Staged embryos were fixed in 4% paraformaldehyde for 30–45 minutes and immediately subjected to cryo-embedding.
Sectioning
Whole embryos were embedded in a 1:1 (20% sucrose in PBS:OCT) solution. An entire embryo was subjected to thick sectioning (20–30 μm) and placed on a single slide for analysis. The embryo sections were subjected to immunofluorescence analysis as previously described (Battle et al., 2006; Holtz and Misra, 2008). The following reagents were used: EC surface markers anti-platelet endothelial cell adhesion molecule-1 (PECAM-1; BD Bioscience, 550274 and R&D Systems, AF3628), anti-vascular endothelial-cadherin (VE-Cadherin; BD Bioscience, 550548), and anti-vascular endothelial growth factor receptor 2 (FLK1; BD Bioscience, 550549), EC nuclear marker anti-ETS related gene (ERG; Santa Cruz, sc-353), PE nuclear marker anti-Wilm's Tumor 1 (WT1; DAKO, m3561), LB nuclear markers anti-prospero homebox 1 (Prox1; Abcam, ab11941 and ab37128), and anti-hepatic nuclear factor 4 (HNF4; Santa Cruz, sc-8987), as well as the smooth muscle cell marker smooth muscle myosin heavy chain (SM-MHC; Biomedical Technologies BT-562).
Images were captured using a Nikon Eclipse 90i microscope equipped with a Nikon C-1 Confocal and EZ C1 imaging software. Z-series were obtained for each 20–30μm section. Image processing was performed in Adobe Photoshop CS4.
Cytometric Analysis
To determine whether the SV or LB contribute EC to the PE, EC within the PE area were assigned to 3 different populations: 1) EC that are associated with the LB (LB-EC), 2) EC associated with the SV (SV-EC), and 3) any EC that were in the PE and not associated with SV or LB (PE-EC) (Figure 1B). The boundaries of the PE was defined by the expression of the PE marker gene WT1 and the LB markers Prox1 and HNF4 (Moore et al., 1999; Li et al., 2000; Burke and Oliver, 2002; Perez-Pomares et al., 2002b; Tremblay and Zaret, 2005). PECAM-1 is a commonly used EC surface marker (Baldwin et al., 1994; Drake and Fleming, 2000). For this reason PECAM-1 expression is ideal for determining the pattern of EC streaming/migration as well as examining the pattern of EC differentiation. To establish the expression pattern of PECAM-1 within PE, whole embryos at E9.0 (n=3), E9.5 (n=5) and E9.75 (n=3) were sectioned to provide complete examination of the PE and surrounding structures. Sections were subjected to confocal z-series analysis in 2μm increments, with an average of 45 steps per E9.0 embryo and 70–75 steps per E9.5 and E9.75 embryo. Individual steps containing PECAM-1 positive cells within PE were scored. Scoring entailed the detection of PECAM-1 positive cells and assignment to one or more population. The percentage of PE steps containing PECAM-1 positive cells was calculated using the number of steps scored to contain PECAM-1 positive cells within the PE divided by the total number of PE steps as defined by WT1 positive staining. The extent of PECAM-1 positive steps within the three different populations (LBEC, SV-EC and PE-EC) was calculated using the number of steps containing PECAM-1 positive cells in a particular population divided by the total number of steps containing PECAM-1 positive cells in one or more of the three populations. See supplemental information for details (Supplemental Figure 1A).
PECAM-1 exhibits cell surface expression, and it is therefore not an optimal marker to quantitate individual ECs. Consequently, individual cells were quantitated using the nuclear EC marker ERG (Vlaeminck-Guillem et al., 2000; McLaughlin et al., 2001; Mohamed et al., 2010). Since the pattern of PECAM-1 positive EC may differ from the actual number of EC, we used ERG in conjunction with PECAM-1 to quantitate the number of cells within the developing PE and within each population. For analysis of ERG positive PEs, whole embryos at stages E9.0 (n=3), E9.5 (n=3) and E9.75 (n=3) were imaged and positive cells were counted in either 2 or 6μm increment confocal z-series steps. Careful attention was paid to ensure that a single cell/nuclei was counted only once. The percentage of PE steps that contained ERG positive cells was calculated in the same manner as described for PECAM-1. The relative extent of ERG positive expression between each of the three individual populations (LB-EC, SV-EC, and PE-EC) was determined by calculating the number of individual ERG positive cells in a particular population divided by the total number of individual ERG positive cells in all three populations. To determine the average number of cells per micron (μm) of PE, the total number of ERG positive cells in the PE was divided by the total width of the PE analyzed. The percentage of ERG positive cells that co-express WT1 was determined by calculating the number of ERG and WT1 co-expressing cells divided by the total number of individual ERG positive cells. See supplemental information for more details (Supplemental Figure 1B).
Supplementary Material
Acknowledgements
We would like to thank Drs. Mary Holtz, Ramani Ramchandran and Jong-In Park for critical reading of the manuscript; Dr. Stephen Duncan and Kurt Kolander for useful discussions; Dr. John Corbett for use of confocal imagery equipment. This work was supported by a predoctoral training grant to S.M.C. from the American Heart Association Heartland Division, and grants from the National Institutes of Health (HL084636) and the Sophia Wolf Quadracci Memorial Fund for Stem Cell Research to R.P.M.
References
- Baldwin H, Shen H, Yan H, DeLisser H, Chung A, Mickanin C, Trask T, Kirschbaum N, Newman P, Albelda S, et al. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–2553. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- Battle M, Konopka G, Parviz F, Gaggl A, Yang C, Sladek F, Duncan S. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci U S A. 2006;103:8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- Cai C, Martin J, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup W, Denton C, McCulloch A, Chen J, Evans S. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Monte G, Casanova J, Guadix J, MacGrogan D, Burch J, Perez-Pomares J, de la Pompa J. Differential Notch signaling in the epicardium is required for cardiac inflow development and coronary vessel morphogenesis. Circ Res. 2011;108:824–836. doi: 10.1161/CIRCRESAHA.110.229062. [DOI] [PubMed] [Google Scholar]
- Dettman R, Denetclaw WJ, Ordahl C, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- Drake C, Fleming P. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- Ferguson, Kelley R, Patterson C. Mechanisms of endothelial differentiation in embryonic vasculogenesis. Arterioscler Thromb Vasc Biol. 2005;25:2246–2254. doi: 10.1161/01.ATV.0000183609.55154.44. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot A, Vrancken Peeters M, Mentink M, Gourdie R, Poelmann R. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, Poelmann RE. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circ Res. 2000;87:969–971. doi: 10.1161/01.res.87.11.969. [DOI] [PubMed] [Google Scholar]
- Guadix J, Carmona R, Munoz-Chapuli R, Perez-Pomares J. In vivo and in vitro analysis of the vasculogenic potential of avian proepicardial and epicardial cells. Dev Dyn. 2006;235:1014–1026. doi: 10.1002/dvdy.20685. [DOI] [PubMed] [Google Scholar]
- Holtz ML, Misra RP. Endothelial-specific ablation of Serum Response Factor causes hemorrhaging, yolk sac vascular failure, and embryonic lethality. BMC Dev Biol. 2008;8:65. doi: 10.1186/1471-213X-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- Lampugnani M, Resnati M, Raiteri M, Pigott R, Pisacane A, Houen G, Ruco L, Dejana E. A novel endothelial-specific membrane protein is a marker of cell-cell contacts. J Cell Biol. 1992;118:1511–1522. doi: 10.1083/jcb.118.6.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ning G, Duncan S. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- Lie-Venema H, Eralp I, Maas S, Gittenberger-De Groot A, Poelmann R, DeRuiter M. Myocardial heterogeneity in permissiveness for epicardium-derived cells and endothelial precursor cells along the developing heart tube at the onset of coronary vascularization. Anat Rec A Discov Mol Cell Evol Biol. 2005;282:120–129. doi: 10.1002/ar.a.20154. [DOI] [PubMed] [Google Scholar]
- Manner J, Schlueter J, Brand T. Experimental analyses of the function of the proepicardium using a new microsurgical procedure to induce loss-ofproepicardial-function in chick embryos. Dev Dyn. 2005;233:1454–1463. doi: 10.1002/dvdy.20487. [DOI] [PubMed] [Google Scholar]
- McLaughlin F, Ludbrook V, Cox J, von Carlowitz I, Brown S, Randi A. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. Blood. 2001;98:3332–3339. doi: 10.1182/blood.v98.12.3332. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Fischman D. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci U S A. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Millauer B, Wizigmann-Voos S, Schnurch H, Martinez R, Moller N, Risau W, Ullrich A. High affinity VEGF binding and developmental expression suggest Flk-1 as a major regulator of vasculogenesis and angiogenesis. Cell. 1993;72:835–846. doi: 10.1016/0092-8674(93)90573-9. [DOI] [PubMed] [Google Scholar]
- Mohamed A, Tan S, Mikhalkevich N, Ponniah S, Vasioukhin V, Bieberich C, Sesterhenn I, Dobi A, Srivastava S, Sreenath T. Ets family protein, erg expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer. 2010;1:197–208. doi: 10.7150/jca.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares J, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002a;46:1005–1013. [PubMed] [Google Scholar]
- Perez-Pomares J, Carmona R, Gonzalez-Iriarte M, Macias D, Guadix J, Munoz-Chapuli R. Contribution of mesothelium-derived cells to liver sinusoids in avian embryos. Dev Dyn. 2004;229:465–474. doi: 10.1002/dvdy.10455. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. Immunolocalization of the vascular endothelial growth factor receptor-2 in the subepicardial mesenchyme of hamster embryos: identification of the coronary vessel precursors. Histochem J. 1998a;30:627–634. doi: 10.1023/a:1003446105182. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. The origin of the subepicardial mesenchyme in the avian embryo: an immunohistochemical and quail-chick chimera study. Dev Biol. 1998b;200:57–68. doi: 10.1006/dbio.1998.8949. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev Biol. 2002b;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- Poelmann R, Gittenberger-de Groot A, Mentink M, Bokenkamp R, Hogers B. Development of the cardiac coronary vascular endothelium, studied with antiendothelial antibodies, in chicken-quail chimeras. Circ Res. 1993;73:559–568. doi: 10.1161/01.res.73.3.559. [DOI] [PubMed] [Google Scholar]
- Red-Horse K, Ueno H, Weissman I, Krasnow M. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian S, Deconinck A, Tanaka M, Schinke M, Litovsky S, Izumo S, Fujiwara Y, Orkin S. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tremblay K, Zaret K. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Viragh S, Challice CE. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- Viragh S, Gittenberger-de Groot AC, Poelmann RE, Kalman F. Early development of quail heart epicardium and associated vascular and glandular structures. Anat Embryol (Berl) 1993;188:381–393. doi: 10.1007/BF00185947. [DOI] [PubMed] [Google Scholar]
- Vittet D, Prandini M, Berthier R, Schweitzer A, Martin-Sisteron H, Uzan G, Dejana E. Embryonic stem cells differentiate in vitro to endothelial cells through successive maturation steps. Blood. 1996;88:3424–3431. [PubMed] [Google Scholar]
- Vlaeminck-Guillem V, Carrere S, Dewitte F, Stehelin D, Desbiens X, Duterque-Coquillaud M. The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech Dev. 2000;91:331–335. doi: 10.1016/s0925-4773(99)00272-5. [DOI] [PubMed] [Google Scholar]
- Vrancken Peeters M, Gittenberger-de Groot A, Mentink M, Hungerford J, Little C, Poelmann R. The development of the coronary vessels and their differentiation into arteries and veins in the embryonic quail heart. Dev Dyn. 1997;208:338–348. doi: 10.1002/(SICI)1097-0177(199703)208:3<338::AID-AJA5>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Vrancken Peeters M, Gittenberger-de Groot A, Mentink M, Poelmann R. Smooth muscle cells and fibroblasts of the coronary arteries derive from epithelial-mesenchymal transformation of the epicardium. Anat Embryol (Berl) 1999;199:367–378. doi: 10.1007/s004290050235. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Nakagawa M, Hanato T, Takeuchi Y, Hara M, Yoshida T, Imanaka-Yoshida K. In vitro model for mouse coronary vasculogenesis. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:714–722. doi: 10.1002/ar.a.20340. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Dumont DJ, Conlon RA, Breitman ML, Rossant J. flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- Yang J, Rayburn H, Hynes R. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development. 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu S, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien K, Pu W. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, von Gise A, Ma Q, Hu Y, Pu W. Genetic fate mapping demonstrates contribution of epicardium-derived cells to the annulus fibrosis of the mammalian heart. Dev Biol. 2010;338:251–261. doi: 10.1016/j.ydbio.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.