Abstract
We have determined histone stoichiometries in nuclei from several sources by a direct chemical method, with the particular aim of quantitating histone H1 and, in chicken erythrocytes, H5, and of distinguishing between one and two molecules per nucleosome. The four histones H3, H4, H2A and H2B are found in equimolar amounts, as expected for the core histone octamer. The molar ratio of H1 in lymphocyte and glial nuclei is 1.0 per octamer, and in liver nuclei from three species 0.8 per octamer. These results suggest that each nucleosome has one H1 molecule; nucleosomes could acquire two molecules of H1 only at the expense of others containing none. The stoichiometry of H5 in chicken erythrocyte nuclei is similar to that of H1 in other nuclei, being about 0.9 molecules per nucleosome; the H1 also present in these nuclei amounts to 0.4 molecules per nucleosome.
Full text
PDF



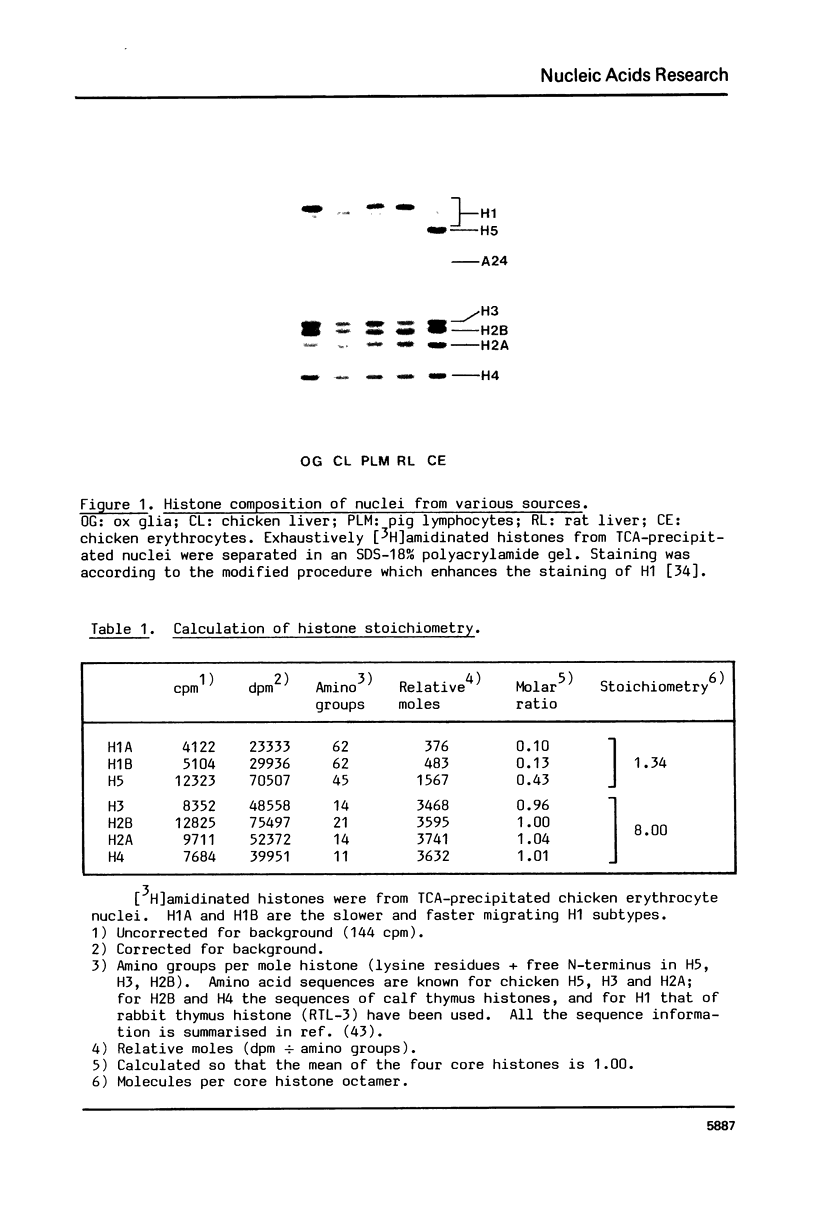
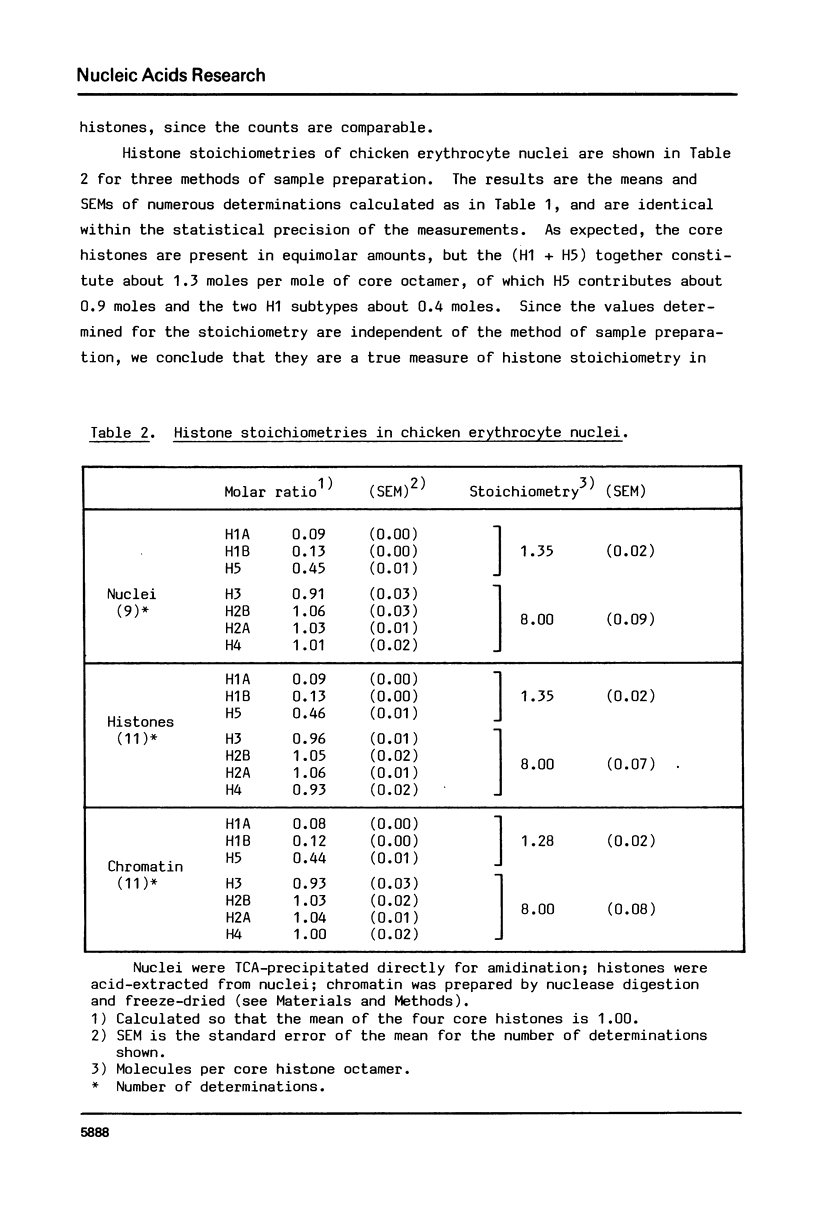

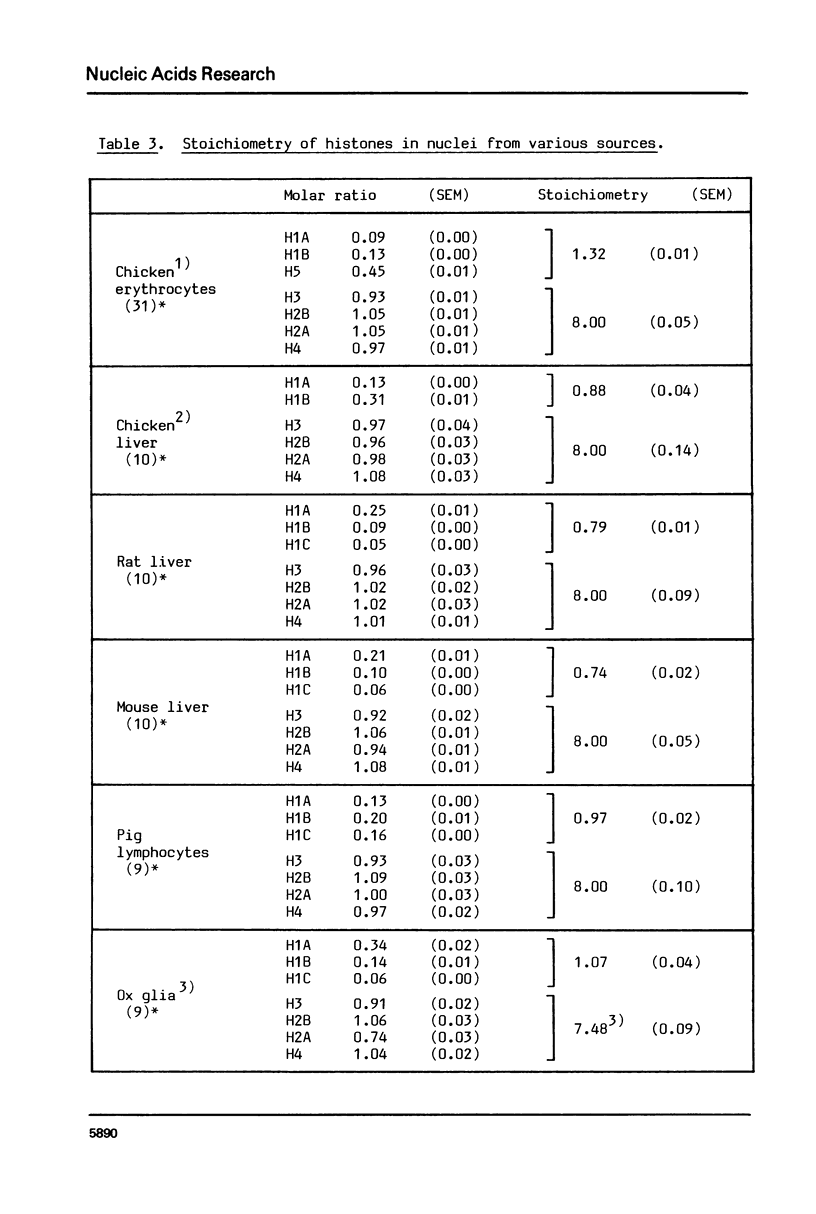




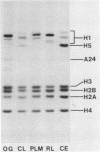
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright S. C., Nelson P. P., Garrard W. T. Histone molar ratios among different electrophoretic forms of mono- and dinucleosomes. J Biol Chem. 1979 Feb 25;254(4):1065–1073. [PubMed] [Google Scholar]
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J., Leadlay P. F., Perham R. N. Synthesis of methyl[3H]acetimidate of high specific radioactivity, a reagent for radiolabeling proteins. Anal Biochem. 1980 Dec;109(2):410–413. doi: 10.1016/0003-2697(80)90669-7. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Danson M. J., Hale G., Hooper E. A., Perham R. N. Self-assembly and catalytic activity of the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Nature. 1977 Jul 28;268(5618):313–316. doi: 10.1038/268313a0. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Harrison R. A., Perham R. N. The stoichiometry of polypeptide chains in the pyruvate dehydrogenase multienzyme complex of E. coli determined by a simple novel method. FEBS Lett. 1975 Dec 15;60(2):427–430. doi: 10.1016/0014-5793(75)80764-2. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Perham R. N., Coggins J. R. Methods for obtaining peptide maps of proteins on a subnanomole scale. Anal Biochem. 1975 Sep;68(1):175–184. doi: 10.1016/0003-2697(75)90692-2. [DOI] [PubMed] [Google Scholar]
- Bode J., Henco K., Wingender E. Modulation of the nucleosome structure by histone acetylation. Eur J Biochem. 1980 Sep;110(1):143–152. doi: 10.1111/j.1432-1033.1980.tb04849.x. [DOI] [PubMed] [Google Scholar]
- Butler P. J., Thomas J. O. Changes in chromatin folding in solution. J Mol Biol. 1980 Jul 15;140(4):505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- Caron F., Thomas J. O. Exchange of histone H1 between segments of chromatin. J Mol Biol. 1981 Mar 15;146(4):513–537. doi: 10.1016/0022-2836(81)90045-0. [DOI] [PubMed] [Google Scholar]
- Farmbrough D. M., Fujimura F., Bonner J. Quantitative distribution of histone components in the pea plant. Biochemistry. 1968 Feb;7(2):575–585. doi: 10.1021/bi00842a010. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Garrard W. T., Pearson W. R., Wake S. K., Bonner J. Stoichiometry of chromatin proteins. Biochem Biophys Res Commun. 1974 May 7;58(1):50–57. doi: 10.1016/0006-291x(74)90889-4. [DOI] [PubMed] [Google Scholar]
- Goldknopf I. L., Busch H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc Natl Acad Sci U S A. 1977 Mar;74(3):864–868. doi: 10.1073/pnas.74.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin G. H., Nicolas R. H., Johns E. W. A quantitative analysis of histone H1 in rabbit thymus nuclei. Biochem J. 1977 Nov 1;167(2):485–488. doi: 10.1042/bj1670485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. C., Hooper E. A., Perham R. N. Subunit stoichiometry of tobacco ribulose 1.5-bisphosphate carboxylase. FEBS Lett. 1980 Jun 2;114(2):237–239. doi: 10.1016/0014-5793(80)81123-9. [DOI] [PubMed] [Google Scholar]
- Hale G., Hooper E. A., Perham R. N. Amidination of pyruvate dehydrogenase complex of Escherichia coli under denaturing conditions. Biochem J. 1979 Jan 1;177(1):136–137. doi: 10.1042/bj1770136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Hofstaetter T., Yakuwa N. Asymmetry of chromatin subunits probed with histone H1 in an H1-DNA complex. Biochemistry. 1978 May 16;17(10):1880–1883. doi: 10.1021/bi00603a012. [DOI] [PubMed] [Google Scholar]
- Hesketh T. R., Smith G. A., Houslay M. D., Warren G. B., Metcalfe J. C. Is an early calcium flux necessary to stimulate lymphocytes? Nature. 1977 Jun 9;267(5611):490–494. doi: 10.1038/267490a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Joffe J., Keene M., Weintraub H. Histones H2a, H2b, H3, and H4 are present in equimolar amounts in chick erythroblasts. Biochemistry. 1977 Mar 22;16(6):1236–1238. doi: 10.1021/bi00625a032. [DOI] [PubMed] [Google Scholar]
- Johns E. W. The electrophoresis of histones in polyacrylamide gel and their quantitative determination. Biochem J. 1967 Jul;104(1):78–82. doi: 10.1042/bj1040078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Rhodes D., Smith J., Finch J. T., Thomas J. O. A low resolution structure for the histone core of the nucleosome. Nature. 1980 Oct 9;287(5782):509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Carlson R. D., Wright E. B., Olins D. E. Chromatin nu bodies: isolation, subfractionation and physical characterization. Nucleic Acids Res. 1976 Dec;3(12):3271–3291. doi: 10.1093/nar/3.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D., Chalkley R. An electrophoretic analysis of Drosophila histones. I. Isolation and identification. Exp Cell Res. 1972 Aug;73(2):295–302. doi: 10.1016/0014-4827(72)90051-1. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. The heterogeneity of histones. I. A quantitative analysis of calf histones in very long polyacrylamide gels. Biochemistry. 1969 Oct;8(10):3972–3979. doi: 10.1021/bi00838a013. [DOI] [PubMed] [Google Scholar]
- Rall S. C., Okinaka R. T., Strniste G. F. Histone composition of nucleosomes isolated from cultured Chinese hamster cells. Biochemistry. 1977 Nov 1;16(22):4940–4944. doi: 10.1021/bi00641a031. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P., Jr Chemical evidence that chromatin DNA exists as 160 base pair beads interspersed with 40 base pair bridges. Nucleic Acids Res. 1976 Jan;3(1):117–127. doi: 10.1093/nar/3.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden P. H., Randle P. J. Regulation of pig heart pyruvate dehydrogenase by phosphorylation. Studies on the subunit and phosphorylation stoicheiometries. Biochem J. 1978 Aug 1;173(2):659–668. doi: 10.1042/bj1730659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. The study of histone--histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- Thomas J. O., Thompson R. J. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977 Apr;10(4):633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- Thompson R. J. Studies on RNA synthesis in two populations of nuclei from the mammalian cerebral cortex. J Neurochem. 1973 Jul;21(1):19–40. doi: 10.1111/j.1471-4159.1973.tb04222.x. [DOI] [PubMed] [Google Scholar]
- Urban M. K., Neelin J. M., Betz T. W. Correlation of chromatin composition with metabolic changes in nuclei of primitive erythroid cells from chicken embryos. Can J Biochem. 1980 Sep;58(9):726–731. doi: 10.1139/o80-102. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Holt C., Strickland W. N., Brandt W. F., Strickland M. S. More histone structures. FEBS Lett. 1979 Apr 15;100(2):201–218. doi: 10.1016/0014-5793(79)80337-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978 Apr;5(4):1179–1188. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. B., Olins D. E. Histone stoichiometry in chicken erythrocyte nuclei. Biochem Biophys Res Commun. 1975 Apr 7;63(3):642–650. doi: 10.1016/s0006-291x(75)80432-3. [DOI] [PubMed] [Google Scholar]