Abstract
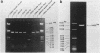
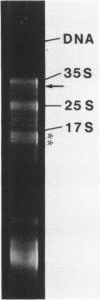
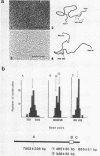
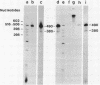
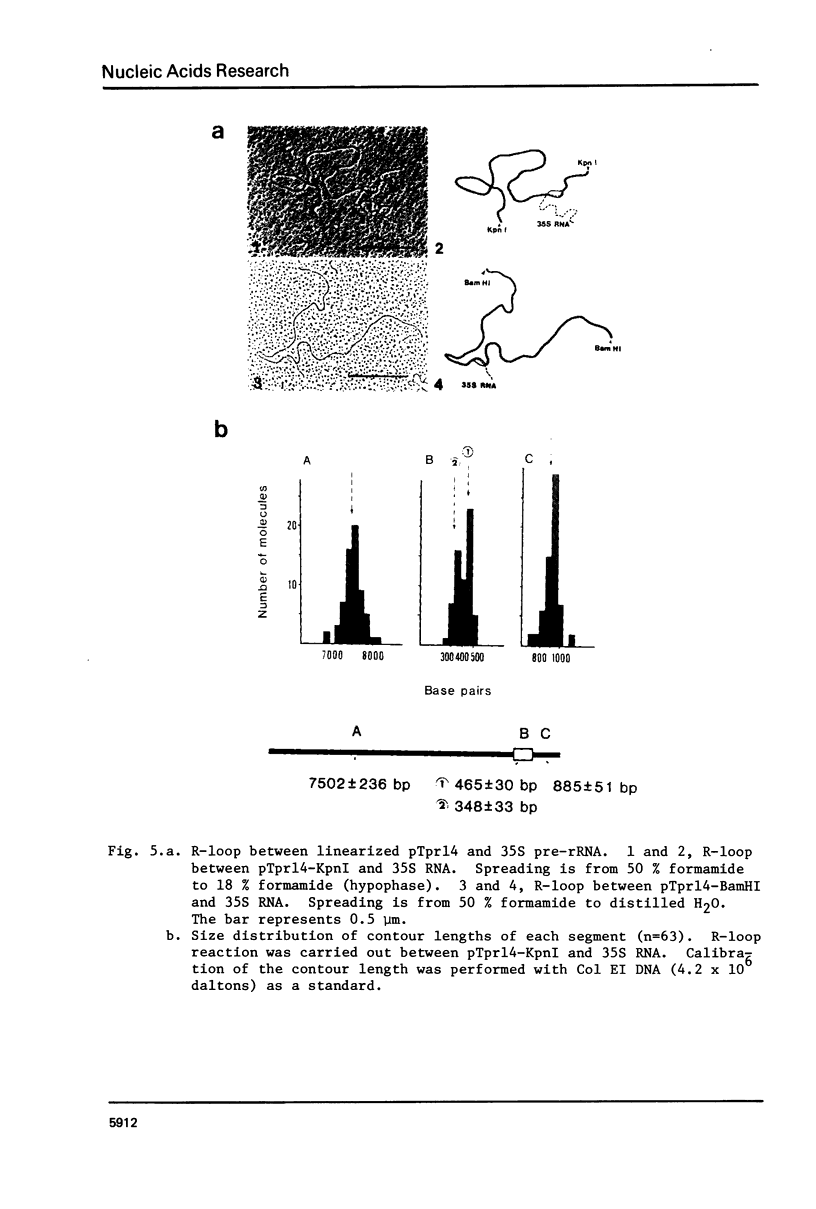
A DNA fragment (1.4 Kb) which codes for 5' region of 35S ribosomal precursor RNA (pre-rRNA) in Tetrahymena pyriformis was cloned with pBR322. The fragment was cleaved from the central part of the palindromic rDNA with restriction endonuclease KpnI and HindIII, and ligated to the larger moiety of pBR322 DNA-HindIII-BamHI fragment together with lambda DNA-KpnI-BamHI fragment through trimolecular ligation. The analysis of R-loop formed between KpnI-linearized recombinant plasmid and 35S pre-rRNA revealed a DNA:RNA hybrid region of 465 +/- 30 base pairs in length. Considering the contraction of DNA:RNA hybrids relative to DNA duplexes (Philippsen et al., J. Mol. Biol., 123, 387-404, 1978), the size of the hybrid region was corrected to about 490 base pairs. Alternatively, the size of DNA which was protected against nuclease S1 due to hybrid formation with 35S pre-rRNA was estimated to be 490 nucleotides long. These data indicate that the transcription initiation site is localized at about 490 base pairs from the HindIII site of the cloned rDNA fragment.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach R., Grummt I., Allet B. The nucleotide sequence of the initiation region of the ribosomal transcription unit from mouse. Nucleic Acids Res. 1981 Apr 10;9(7):1559–1569. doi: 10.1093/nar/9.7.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Eckert W. A., Kaffenberger W., Krohne G., Franke W. W. Introduction of hidden breaks during rRNA maturation and ageing in Tetrahymena pyriformis. Eur J Biochem. 1978 Jul 3;87(3):607–616. doi: 10.1111/j.1432-1033.1978.tb12413.x. [DOI] [PubMed] [Google Scholar]
- Eckert W. A., Kaffenberger W. Regulation of rRNA metabolism in Tetrahymena pyriformis. I. Nutritional shift-down. Eur J Cell Biol. 1980 Apr;21(1):53–62. [PubMed] [Google Scholar]
- Engberg J., Andersson P., Leick V., Collins J. Free ribosomal DNA molecules from Tetrahymena pyriformis GL are giant palindromes. J Mol Biol. 1976 Jun 25;104(2):455–470. doi: 10.1016/0022-2836(76)90281-3. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashinakagawa T., Sezaki M., Kondo S. Isolation of nucleoli from Tetrahymena pyriformis. Dev Biol. 1979 Apr;69(2):601–611. doi: 10.1016/0012-1606(79)90314-2. [DOI] [PubMed] [Google Scholar]
- Kaback D. B., Angerer L. M., Davidson N. Improved methods for the formation and stabilization of R-loops. Nucleic Acids Res. 1979 Jun 11;6(7):2499–2317. doi: 10.1093/nar/6.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffenberger W., Eckert W. A. Regulation of rRNA metabolism in Tetrahymena pyriformis. II. Nutritional shift-up. Eur J Cell Biol. 1980 Jun;21(2):200–207. [PubMed] [Google Scholar]
- Karrer K. M., Gall J. G. The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J Mol Biol. 1976 Jun 25;104(2):421–453. doi: 10.1016/0022-2836(76)90280-1. [DOI] [PubMed] [Google Scholar]
- Klein R. D., Selsing E., Wells R. D. A rapid microscale technique for isolation of recombinant plasmid DNA suitable for restriction enzyme analysis. Plasmid. 1980 Jan;3(1):88–91. doi: 10.1016/s0147-619x(80)90037-2. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Shimada N., Higashinakagawa T. Effect of cycloheximide on the nucleolar RNA synthesis in rat liver. J Mol Biol. 1970 Oct 14;53(1):91–106. doi: 10.1016/0022-2836(70)90047-1. [DOI] [PubMed] [Google Scholar]
- Niles E. G. Isolation of a high specific activity 35S ribosomal RNA precursor from Tetrahymena pyriformis and identification of its 5' terminus, pppAp. Biochemistry. 1978 Oct 31;17(22):4839–4844. doi: 10.1021/bi00615a035. [DOI] [PubMed] [Google Scholar]
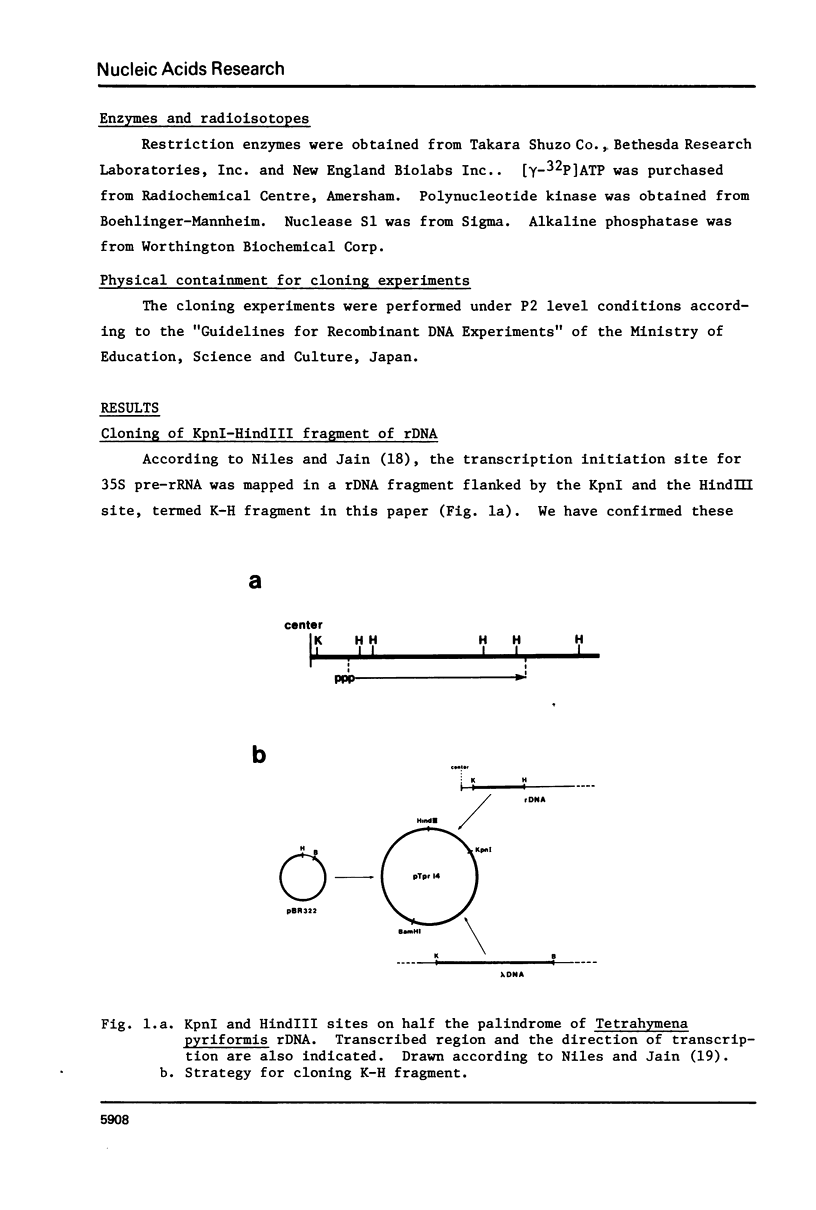
- Niles E. G., Jain R. K. Physical map of the ribosomal ribonucleic acid gene from Tetrahymena pyriformis. Biochemistry. 1981 Feb 17;20(4):905–909. doi: 10.1021/bi00507a039. [DOI] [PubMed] [Google Scholar]
- Norgard M. V., Keem K., Monahan J. J. Factors affecting the transformation of Escherichia coli strain chi1776 by pBR322 plasmid DNA. Gene. 1978 Jul;3(4):279–292. doi: 10.1016/0378-1119(78)90038-0. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Reeder R. H., Sollner-Webb B., Wahn H. L. Sites of transcription initiation in vivo on Xenopus laevis ribosomal DNA. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5402–5406. doi: 10.1073/pnas.74.12.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M., Blank D., Fahrner K., Hereford L., Ricciardi R., Roberts B., Ruby S., Woolford J. R-looping and structural gene indentification of recombinant DNA. Methods Enzymol. 1979;68:454–469. doi: 10.1016/0076-6879(79)68035-7. [DOI] [PubMed] [Google Scholar]
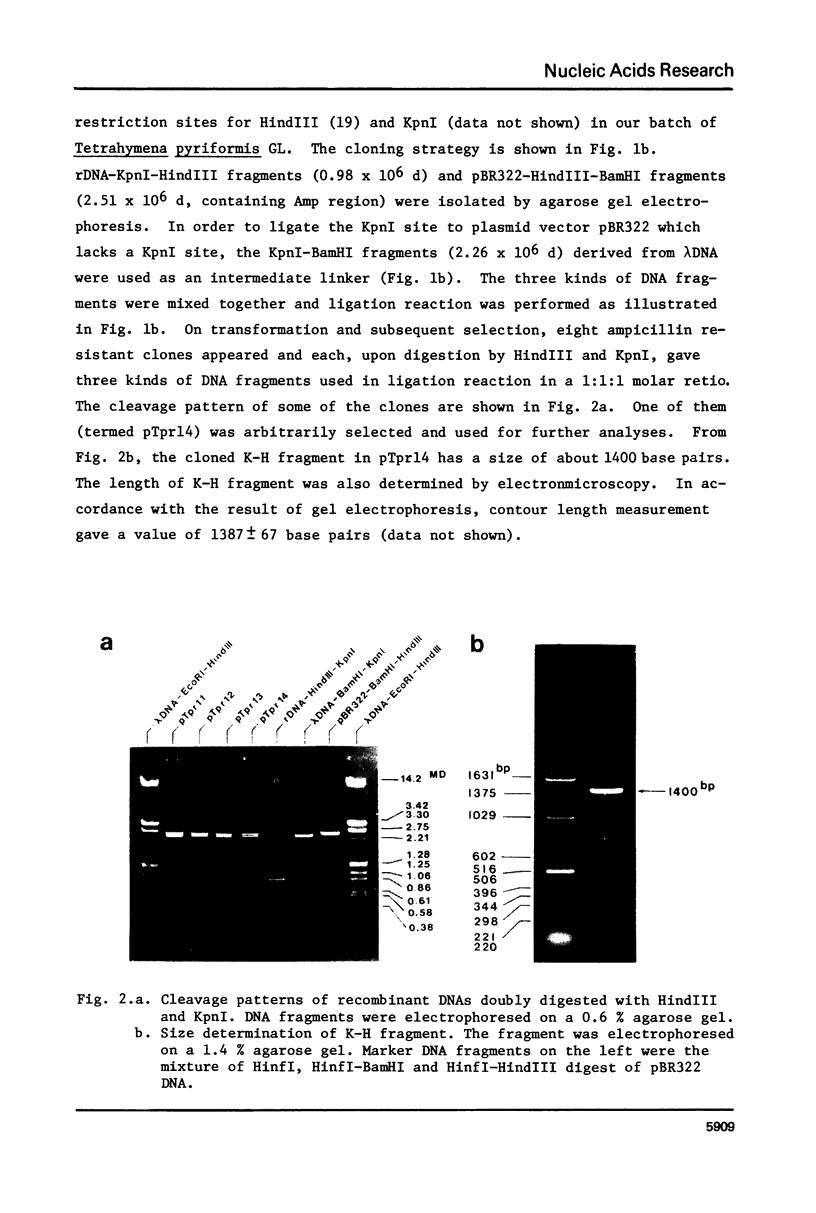
- Weil P. A., Segall J., Harris B., Ng S. Y., Roeder R. G. Faithful transcription of eukaryotic genes by RNA polymerase III in systems reconstituted with purified DNA templates. J Biol Chem. 1979 Jul 10;254(13):6163–6173. [PubMed] [Google Scholar]
- Weinberg R. A., Loening U., Willems M., Penman S. Acrylamide gel electrophoresis of HeLa cell nucleolar RNA. Proc Natl Acad Sci U S A. 1967 Sep;58(3):1088–1095. doi: 10.1073/pnas.58.3.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]