Abstract
Triacylglycerols (TAG) and steryl esters (SE) are the principal storage lipids in all eukaryotic cells. In yeasts, these storage lipids accumulate within special organelles known as lipid bodies (LB). In the lipid accumulation-oriented metabolism of the oleaginous yeast Yarrowia lipolytica, storage lipids are mostly found in the form of TAG, and only small amounts of SE accumulate. We report here the identification of a new DAG acyltransferase gene, DGA2, homologous to the ARE genes of Saccharomyces cerevisiae. This gene encodes a member of the type 1 acyl-CoA:diacylglycerol acyltransferase family (DGAT1), which has not previously been identified in yeasts, but is commonly found in mammals and plants. Unlike the Are proteins in S. cerevisiae, Dga2p makes a major contribution to TAG synthesis via an acyl-CoA-dependent mechanism and is not involved in SE synthesis. This enzyme appears to affect the size and morphology of LB, suggesting a direct role of storage lipid proteins in LB formation. We report that the Are1p of Y. lipolytica was essential for sterol esterification, as deletion of the encoding gene (ARE1) completely abolished SE synthesis. Unlike its homologs in yeasts, YlARE1 has no DAG acyltransferase activity. We also reconsider the role and function of all four acyltransferase enzymes involved in the final step of neutral lipid synthesis in this oleaginous yeast.
Keywords: Oleaginous yeast, Yarrowia lipolytica, Metabolism, Biotechnology, Acyltransferases, Lipids
Introduction
Triacylglycerols (TAG) are the principal form of stored energy in eukaryotic cells. Together with steryl esters (SE), they comprise the neutral lipid fraction of the cell, commonly found within special organelles known as lipid bodies (LB). In yeast, several lipid-metabolizing enzymes localize to the lipid bodies, endowing these organelles with a complex role in lipid biosynthesis, degradation, and trafficking. In oleaginous species, lipids may accumulate to levels exceeding 70% of total biomass (Ratledge 1994), and Yarrowia lipolytica is able to store up to 40% of its dry weight in lipid form (Ratledge and Wynn 2002). However, regardless of lipid accumulation levels, the proportions of free fatty acids (FFA), TAG, and SE appear to differ between species. For instance, Saccharomyces cerevisiae, a non-oleaginous yeast, produces similar amounts of TAG and SE, which accumulate to levels not exceeding 15% of dry weight, whereas FFA are barely detectable (Daum et al. 1998, 2007). By contrast, Y. lipolytica, which specializes in the use of hydrophobic substrates, accumulates more than 90% of storage lipids in the form of TAG and is simultaneously capable of maintaining significant quantities of FFA (Beopoulos et al. 2008, 2009).
Two major metabolic pathways for TAG biosynthesis are known: the glycerol-3-phosphate (G-3-P) pathway and the monoacylglycerol (MAG) pathway, although little evidence of the latter exists in yeasts (Heier et al. 2010). In both pathways, diacylglycerol (DAG), the immediate precursor of TAG, is formed at a branch point. The DAG formed by the MAG pathway is then oriented to the G-3-P pathway for the final esterification step. In yeasts, TAG can be synthesized either by an acyl-CoA-dependent reaction catalyzed by acyl-CoA:diacylglycerol acyltransferase (DGAT) or an acyl-CoA-independent pathway catalyzed by phospholipid:diacylglycerol acyltransferase (PDAT) (Czabany et al. 2007; Daum et al. 2007; Beopoulos et al. 2008). The protein expressing PDAT activity is Lro1p and shares 27% identity with the human lecithin:cholesterol acyltransferase. Lro1p has both phospholipase and acyltransferase functions and mediates the esterification of DAG, using the sn-2 group of phospholipids as an acyl donor (Dahlqvist et al. 2000). However, unlike its human ortholog, yeast Lro1p cannot synthesize steryl esters (Oelkers et al. 2000).
DGAT (EC 2.3.1.20) catalyzes the acyl-CoA-dependent acylation of sn-1,2-diacylglycerol to form TAG. There are two main membrane-bound DGAT families, designated DGAT1 and DGAT2. Saha et al. (2006) isolated a soluble acyltransferase from the cytosolic fraction of developing peanut cotyledons: It shares some identity with DGAT1 and DGAT2 families and is the founder member of a novel, soluble, acyltransferase family named DGAT3. The same team suggested that a soluble acyltransferase identified in the oleaginous yeast Rhodotorula glutinis belongs to this DGAT3 family.
The available data suggest that enzymes of the DGAT2 family are more potent and have higher affinity for their substrates than those of the DGAT1 family (Yen et al. 2008). The DGAT2 family member in many yeasts is a DGA1 homolog, which in S. cerevisiae accounts for 87% of DAG esterification activity (Sandager et al. 2002).
The DGAT1 family shares substantial similarity with acyl-CoA:cholesterol acyltransferases (ACAT), catalyzing sterol esterification in mammals. The first genes to be classified as DGAT1 family members (coding for murine and human proteins) were identified by their sequence similarity to ACAT enzymes, but were shown to encode proteins that possess DGAT activity (Yen et al. 2008). In the yeast S. cerevisiae, two members of the ACAT family, Are1p and Are2p, are responsible for the whole process of sterol esterification and also appear to display low levels of DGAT activity. Sandager et al. (2002) attributed this activity to weak inherent DGAT activity, based on a comparison of ARE orthologs with both functions from plants and mammals. This implies that an ancestral ACAT/DGAT1 family gene evolved to yield enzymes capable of utilizing different substrates such as fatty acyl-CoA acceptors, resulting in the acquisition of DGAT1 activity for long-chain acyl moieties and the abolition of sterol activity. To our knowledge, no enzyme with no sterol esterification (ACAT) activity but full activity with DAG has been identified in yeasts. Hence, no yeast enzyme from the DGAT1 family has been identified.
We show here that the ARE1 and ARE2 homologs in the oleaginous yeast Y. lipolytica appear to have different roles in neutral lipid synthesis and metabolism. Our data clearly indicate that Are1p is the only enzyme with sterol esterification activity, this enzyme thus belonging to the ACAT family, and that the protein encoded by the ARE2 homolog has no affinity for sterol substrates despite its major contribution to DAG esterification. Thus, the enzyme encoded by the ARE2 homolog of Y. lipolytica should be reclassified to the DGAT1 family, under the gene name DGA2, as it and DGA1 are the only genes encoding enzymes capable of catalyzing an acyl-CoA-dependent DAG esterification reaction. We also investigated the role of the enzymes involved in neutral lipid synthesis, providing new insight into the process of lipid metabolism in oleaginous yeasts.
Materials and methods
Yeast strains, growth, and culture conditions
The Y. lipolytica strains used here were derived from the wild-type strain W29 (ATCC 20460) (see Table 1 and Fig. 1). The auxotrophic strain Po1d (Leu−, Ura−) has been described elsewhere by Barth and Gaillardin (1996). The strains used are presented in Table 1; their construction is depicted in Fig. 1 and described in detail below. The medium and growth conditions for Escherichia coli were as described by Sambrook et al. (1989), and those for Y. lipolytica were as described by Barth and Gaillardin (1996). Rich medium (YPD), minimal glucose medium (YNB), and minimal medium with casamino acids (YNBcas) were prepared as previously described (Mlickova et al. 2004). Minimal medium (YNB) contained 0.17% (w/v) yeast nitrogen base (without amino acids and ammonium sulfate, YNBww; Difco, Paris, France), 0.5% (w/v) NH4Cl, 0.1% (w/v) yeast extract (Bacto-DB), and 50 mM phosphate buffer (pH 6.8). The following carbon sources were added: YNBD glucose medium (2% (w/v) glucose, Merck, Fontenay-sous-Bois Cedex, France) and YNBO3 oleic acid medium (3% oleic acid from Merck −60% purity). Oleic acid was emulsified by sonication in the presence of 0.02% Tween 40. Uracil (0.1 g/l) was added when appropriate. Typically, yeast was cultured as follows: An initial preculture was set up by inoculating YPD medium with a colony from an YPD plate (15 ml in 50 ml baffled Erlenmeyer flasks, shaken at 170 rpm, 28°C, 6 h). The resulting cell suspension was used to inoculate a preculture in YNB medium (50 ml in 500 ml baffled Erlenmeyer flasks, shaken at 170 rpm, 28°C, overnight). Exponentially growing cells from the overnight culture were then harvested by centrifugation and resuspended in fresh YNBO3 medium to an optical density (OD600) of 0.5. Cell growth was assessed as follows: Cultures were centrifuged at 10,000×g for 10 min, the resulting cell pellet was washed three times with equal volumes of SB solution (9 g/l NaCl in 0.5% BSA), and biomass production was determined by measuring OD600 and estimating cell dry weight (after freeze-drying).
Table 1.
E. coli and Y. lipolytica strains and plasmids used in this study
| Strain (host strain) | Plasmid, genotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5a | Φ80dlacZΔm15, recA1, endA1, gyrA96, thi-1, hsdR17 (rk−, mk+), supE44, relA1, deoR, Δ(lacZYA-argF)U169 | Promega |
| JME459 (DH5a) | pBluescript II KS+ (ColE1 ori, LacZ, bla) | Stratagene |
| JME461 (DH5a) | pRRQ2 (Cre ARS68 LEU2 in) | Fickers et al. 2003 |
| JME507 (DH5a) | 1.3 kb IsceI fragment containing URA3 marker, MU cassette | Fickers et al. 2003 |
| JME803 | JMP62-pTEF-URA3ex | Muller et al. 1998; Nicaud et al. 2002 |
| JME735 (DH5a) | 1.5 kb PCR fragment containing ylDGA1PT cassette in pCR4®Blunt-TOPO | This work |
| JME737 (DH5a) | 1.9 kb PCR fragment containing ylLRO1PT cassette in pCR4®Blunt-TOPO | This work |
| JME984 (DH5a) | 1.6 kb PCR fragment containing ylARE1PT cassette in pCR4®Blunt-TOPO | This work |
| JME1035 (DH5a) | 1.6 kb PCR fragment containing ylDGA2PT cassette in pCR4®Blunt-TOPO | This work |
| JME736 (DH5a) | MU cassette in JME735, ylDGA1 PUT cassette | This work |
| JME738 (DH5a) | MU cassette in JME737, ylLRO1PUT cassette | This work |
| JME996 (DH5a) | MU cassette in JME984, ylARE1 PUT cassette | This work |
| JME1040 (DH5a) | ML cassette in JME984, ylDGA2 PLT cassette | This work |
| JME1042 (DH5a) | MU cassette in JME984, ylDGA2 PUT cassette | This work |
| JME1112 (DH5a) | JMP62-pTEF-DGA1-URA3ex | This work |
| JME1114 (DH5a) | JMP62-pTEF-LRO1-URA3ex | This work |
| JME1132 (DH5a) | JMP62-pTEF-DGA2-URA3ex | This work |
| JME1215 (DH5a) | JMP62-pTEF-ARE1-URA3ex | This work |
| Y. lipolytica strains | ||
| W29 | MATA, wild-type | Barth and Gaillardin 1996 |
| Po1d | MATA ura3-302 leu2-270 xpr2-322 | |
| JMY1201 | MATA leu2-270 ura3-302 Δlro1::URA3 | This work |
| JMY1204 | MATA leu2-270 ura3-302 Δdga1::URA3 | This work |
| JMY1217 | MATA leu2-270 ura3-302 Δdga1 | This work |
| JMY1613 | MATA leu2-270 ura3-302 Δare1::URA3 | This work |
| JMY1636 | MATA leu2-270 ura3-302 Δdga2::URA3 | This work |
| JMY1281 | MATA leu2-270 ura3-302 Δdga1Δlro1::URA3 | This work |
| JMY1327 | MATA leu2-270 ura3-302 Δdga1Δlro1 | This work |
| JMY1610 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1 ::URA3 | This work |
| JMY1631 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1 ::URA3 Δdga2 ::LEU2 | This work |
| JMY1877 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1Δdga2 | This work |
| JMY1882 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1Δdga2 pTEF-LRO1-URA3ex | This work |
| JMY1884 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1Δdga2 pTEF-DGA2-URA3ex | This work |
| JMY1892 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1Δdga2 pTEF-DGA1-URA3ex | This work |
| JMY1988 | MATA leu2-270 ura3-302 Δdga1Δlro1Δare1Δdga2 pTEF-ARE1-URA3ex | This work |
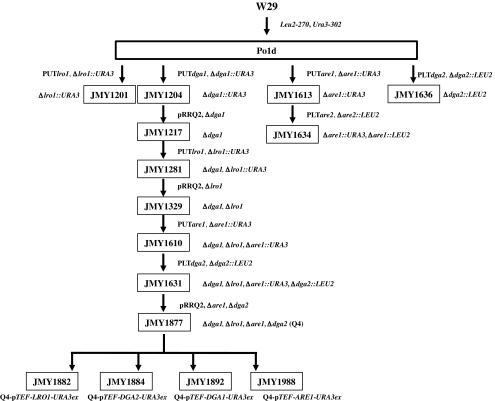
Fig. 1.
Schematic representation of strain constructions. The auxotrophic strain Po1d (Leu− Ura−) was derived from wild-type strain W29. Strain JMY1201, carrying a disrupted LRO1 gene (lro1::URA3), was obtained by introducing the LRO1-PUT cassette into Po1d. Strains JMY1204 and JMY1613, carrying disruptions of DGA1 (dga1::URA3) and ARE1 (are1::URA3), respectively, were constructed similarly. The JMY1636 strain, carrying a DGA2 gene disruption (dga2::LEU2), was then obtained by transforming Po1d with the DGA2-PLT disruption cassette. Additional deletions of genes encoding acyltransferases were obtained by successive gene disruption and marker rescue as described by Fickers et al. (2003): (a) The JMY1204 marker was rescued after transformation with the replicative plasmid pRRQ2, followed by plasmid curing, yielding the strain JMY1217; (b) the LRO1 gene was deleted with the LRO1-PUT disruption cassette, yielding the strain JMY1281; (c) the marker was rescued as above, giving rise to strain JMY 1329; (d) the ARE1 gene was deleted with the ARE1-PUT disruption cassette, giving rise to strain JMY1610; and (e) the DGA2 gene was deleted with the DGA2-PLT disruption cassette, giving rise to the prototrophic JMY1631 quadruple mutant strain. Marker excision gave rise to the auxotrophic strain JMY1877 carrying quadruple deletions (Q4). Each of the acyltransferase-coding genes under the pTEF constitutive promoter was introduced into strain JMY1877 (Q4) giving JMY1882 (Q4-pTEF-LRO1-URA3ex), JMY1884 (Q4-pTEF-DGA2-URA3ex), JMY1892 (Q4-pTEF-DGA1-URA3ex), and JMY1988 (Q4-pTEF-ARE1-URA3ex) overexpression mutants. Similarly, the prototrophic ∆are1∆dga2 double mutant strain JMY1634 was constructed by deleting the DGA2 gene from strain JMY1613 with the DGA2-PLT disruption cassette
General genetic techniques
Standard molecular genetics techniques were used. Restriction enzymes were obtained from Eurogentec S.A. (Liège, Belgium). Genomic DNA from yeast transformants was obtained as described by Querol et al. (1992). An Eppendorf 2720 thermal cycler with both Taq (Promega, Madison, WI, USA) and Pfu (Stratagene, La Jolla, CA, USA) DNA polymerases was used for PCR amplification. Amplified fragments were purified with the QIAgen Purification Kit (Qiagen, Hilden, Germany), and DNA fragments were recovered from agarose gels with the QIAquick Gel Extraction Kit (Qiagen). The STADEN package of programs (Dear and Staden 1991) was used for sequence analysis. Yeast cells were transformed by the lithium acetate method (Gaillardin et al. 1985).
Construction of the LRO1, ARE1, DGA1, and DGA2 quadruple-disrupted strain followed by marker excision
The deletion cassettes were generated by PCR amplification as described by Fickers et al. (2003) using the primer pairs described in Table 2. The PT cassettes were then inserted into the PCR4 RBlunt-TOPO vector to generate the corresponding JME (PT) constructs (Table 1). The auxotrophic marker (either URA3 or LEU2) was then inserted into the vectors IsceI sites to generate the corresponding JME (PUT) vectors (Table 1). The PUT and PLT disruption cassettes were introduced to Y. lipolytica by transformation with the lithium acetate method (Barth et al. 2003). Transformants were selected on YNBcasa and YNBura, respectively. The corresponding ver1 and ver2 primers (Table 2) were used to check gene disruption by PCR amplification of the genomic loci. Marker rescue was performed after transformation with the replicative plasmid pRRQ2 as described by Fickers et al. (2003).
Table 2.
Primers used in this study
| Primers | Sequence (5′→3′)a | Restriction site(s), introduced* |
|---|---|---|
| LRO1-ver1 | CCACGGAGACTGGAACAGATGTCGG | |
| LRO1-P1 | CGATCCCAGTGCTCTAGACTGTC | |
| LRO1-P2 | GCTAGGGATAACAGGGTAATGCGGGGTAGCTGAGACATGTCGCGTG | IsceI |
| LRO1-T1 | GCATTACCCTGTTATCCCTAGCGCGTTCGTCCTCTCATGATTCC | IsceI |
| LRO1-T2 | CCAAACATAGTCATTTGCAGATCC | |
| LRO1-ver2 | CCAAGGCGACCGTCTGGCCAAAC | |
| DGA1-ver1 | CGGACACCTCTTTTATGCTGCGGGC | |
| DGA1-P1 | GGCGGATCCTGGTGCATTTTTGC | |
| DGA1-P2 | GCTAGGGATAACAGGGTAATGCGCAAACTCATCTGGGGGAGATCC | IsceI |
| DGA1-T1 | GCATTACCCTGTTATCCCTAGCGAGCTTATCAGTCACGGTCCACG | IsceI |
| DGA1-T2 | CCATGGAGGTGTCCCCAAACG | |
| DGA1-ver2 | CCCCCAAGCATACCGACCGTCGC | |
| ARE1-ver1 | CCAACCGGTACATACTATGATAGGAATGAGGC | |
| ARE1-P1 | TGGCCTGAAGGCTCCAATGGCACCATTCTC | |
| ARE1-P2 | CGATTACCCTGTTATCCCTACCTGTGTGTGCGGAGAGTGTCTTGTAAG | IsceI |
| ARE1-T1 | GGTAGGGATAACAGGGTAATCGGCACAGTCGCTTCACCACTTGTGCCTATAG | IsceI |
| ARE1-T2 | AGCATGTGCACGTAGGCCACCGCTAGTTC | |
| ARE1-ver2 | AAACAATGACAAATACAACTCTAGACTCCAAAAC | |
| DGA2-ver1 | GCCTGGACCCCGACTACCGCTAG | |
| DGA2-P1 | CCAGAACCCTTGTACATCAGCCTATATGC | |
| DGA2-P2 | CGATTACCCTGTTATCCCTACCTATGGTGTGGGGATGTTATG | IsceI |
| DGA2-T1 | GGTAGGGATAACAGGGTAATCGGCATAACACTCATCAGTAGC | IsceI |
| DGA2-T2 | GTATAGTGGGGCATATTGTAATTAAAAGCACTG | |
| DGA2-ver2 | GCCTATGGGATCTTTCATTGCTCTTG | |
| ACT1_F | TCCAGGCCGTCCTCTCCC | |
| ACT1_R | GGCCAGCCATATCGAGTCGCA | |
| DGA1_F | TGTACCGATTCCAGCAGT | |
| DGA1_R | GGTGTGGGAGATAAGGCAA | |
| LRO1_F | CTCCGCCGACTTCTTTATG | |
| LRO1_R | GAAGTATCCGTCTCGGTG | |
| DGA2_F | TTCTCATCTTCCAGTACGCCTA | |
| DGA2_R | GGCAATAAGATTGAGACCGTT | |
| ARE1_F | TCCTCAAGCGACACGTCTA | |
| ARE1_R | CAGCAACAGCAGGTATCC |
Primers P1, P2, T1, and T2 were used for the construction of the disruption cassettes. Primers ver1 and ver2 were used to check for gene disruption by PCR amplification of the genomic loci. Primers P2 and T1 contained the IsceI endonuclease site (underlined). Primers F and R were used for qPCR amplification
*Underlined sequences correspond to introduced restriction sites
Cloning and expression of the individual Y. lipolytica acyltransferases under control of the TEF constitutive promoter in the quadruple mutant
For the construction of the single acyltransferase overexpression mutants (LRO1, ARE1, DGA1, DGA2), the genes were placed under the control of the yeast TEF constitutive promoter (Muller et al. 1998). Acyltransferase genes were amplified by PCR using Y. lipolytica W29 genomic DNA as the template and the corresponding primer pairs (Table 2) as described in the previous section. Acyltransferase-coding genes were then inserted between the BamHI-AvrII sites of the expression vector pTEF, JMP62 (JME 803) containing the selective marker URA3 (Nicaud et al. 2002). Plasmids were digested with NotI and used to transform the quadruple mutant Y. lipolytica strain JMY1877 by the lithium acetate method (Barth et al. 2003). Transformants were selected on YNBcasa medium. A schematic representation of the strains constructed is depicted in Fig. 1.
RNA isolation and transcript quantification
Biological materials were frozen in liquid nitrogen and stored at −80°C. Samples were crushed in liquid nitrogen, and total RNA was extracted with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. For RT-qPCR experiments, the total RNA of each sample was treated with DNAseI (Ambion). We checked that there was no contaminating genomic DNA by PCR with the DNAseI-treated total RNA as a template and Taq (Promega, Madison, WI, USA). The amount of total RNA was checked with an Agilent 2100 Bioanalyzer, Agilent RNA 6000 Nano reagents, and RNA Chips. Total DNAseI-treated RNA (1 μg) was treated with Thermoscript RT (Invitrogen, Carlsbad, USA) as described by manufacturer. Quantitative PCR was then performed in a LightCycler 1.5 machine (Roche, Meylan, France), with LightCycler Fast Start DNA Master Sybr Green (Roche, Meylan, France), according to the manufacturer’s instructions. Relative quantification was based on the 2ΔCT method, using YlACT, encoding actin, as the reference for calibration. The amplification reaction program was set as follows: 95°C for 8 min, 95°C for 10 s, 60°C for 6 s, and 72°C for 10 s (45 cycles), 95°C for 15 s, 60°C for 15 s, 95°C for 15 s, and 40°C for 5 min. Three independent replicates, prepared from independent biological samples, were analyzed. The primers used for qPCR are shown in Table 2.
Light microscopy
For light microscopy, we fixed 10 ml of a growing yeast culture by adding 1.34 ml of formaldehyde stock solution (50 mM K-Pi buffer, pH 6.8; 0.5 mM MgCl2; 4.8% formaldehyde) and incubating for a further hour at 28°C, with shaking at 250 rpm. Fixed cells were harvested, resuspended to an OD600 of 2.5 in the formaldehyde stock solution, and incubated for 5 h at room temperature. The cells were then washed twice with 50 mM K-Pi buffer (pH 6.8) and stored at 4°C in 0.1 M K-Pi buffer (pH 7.5) at an OD600 of 2.5 until observation under the microscope.
Fluorescence microscopy
For the visualization of LB, Nile red (1 mg/ml solution in acetone; Molecular Bioprobe, Montluçon France) was added to the cell suspension (1/10, v/v), which was then incubated for 15 min at room temperature. Alternatively, LipidTOX™ Green neutral lipid stain (2.5 mg/ml in ethanol; Invitrogen) was added to the cell suspension (OD600 of 5) and incubated for 10 min at room temperature. An AXIO Imager.M1 fluorescence microscope (Zeiss, Le Pecq, France) with a ×100 oil immersion objective and the AxioVision Rel. 4.6 software were used for observation and recording the images.
Lipid analysis
Lipids from the equivalent of 10 OD units of freeze-dried culture cells were extracted by the procedure described by Folch et al. (1957) for TLC analysis and then converted into their methyl esters, as described by Browse et al. (1986), for GC analysis. GC analysis of fatty acid methyl esters (FAME) was carried out on a Varian 3900 gas chromatograph equipped with a flame ionization detector and a Varian FactorFour vf-23ms column, with a bleed specification at 260°C of 3pA (30 m, 0.25 mm, 0.25 μm). FA were identified by comparison with commercial fatty acid methyl ester standards (FAME32, Supelco) and quantified by the internal standard method with the addition of 50 μg of commercial C17:0 (Sigma).
Lipid fractionation (SPE)
Total lipids were fractionated into TAG and FFA for lipid class quantification, using an Isolute SPE Aminopropyl column (IST, France) as described elsewhere (Laffargue et al. 2007). The column was equilibrated three times with 3 ml of hexane at a normal flow rate. Total lipids extracted by the Folch method (as described in the previous section) in CHCl3 (1 ml) were loaded onto the column, and the neutral lipid fraction was collected. The total elution of neutral lipids was achieved by washing the column three times with 3 ml of CHCl3/isopropanol (2/1) each time. The FFA fraction was collected by washing the column three times with 3 ml of diethylether containing 2% acetic acid, with a normal flow rate. The solvent fraction was evaporated under N2 flux. The samples were then subjected to direct transmethylation followed by GC analysis. TLC plates were used to check the extraction. A comparison of the GC profiles of the fractionated and the unfractionated samples confirmed the efficiency of the procedure.
TLC lipid analysis
Precoated TLC plates (silica G60, 20 × 20 cm, 0.25 mm thick) from Merck (Germany) were used. Lipid classes were separated with a dual-solvent development system. System A (half-plate migration) is petroleum ether/ethyl ether/acetic acid, 20/20/0.8 (v/v/v). System B (whole-plate migration) is petroleum ether/Et2O, 49/1 (v/v). We sprayed the plates with 1% vanillin in ethanol and incubated them at 105°C. The lipid bands were apparent after 10 min of incubation.
Results
In silico genomic study of the enzymes involved in storage lipid synthesis in Y. lipolytica
Several reviews of the genes involved in storage lipid synthesis in S. cerevisiae have recently been published (Sandager et al. 2002; Czabany et al. 2007). We compared these genes in S. cerevisiae (a non-oleaginous yeast) and Y. lipolytica (an oleaginous yeast), using the protein amino acid sequences deduced from the S. cerevisiae gene sequences available from the SGD site (http://www.yeastgenome.org/), the Genolevures site (http://cbi.labri.fr/Genolevures/), and the results of a proteomic study of Y. lipolytica LBs (Athenstaedt et al. 2006). The results are summarized in Table 3.
Table 3.
Genes involved in storage lipid synthesis in Y. lipolytica and S. cerevisiae. S. cerevisiae gene names and Enzyme Commission numbers, Y. lipolytica orthologs (gene name), and corresponding functions
| Gene | SC name | YL name | EC number | Function |
|---|---|---|---|---|
| SCT1 | YBL011w | YALI0C00209g | EC 2.3.1.15 | Glycerol-3-phosphate acyltransferase |
| SLC1 | YDL052c | YALI0E18964g | EC 2.3.1.51 | 1-acyl-sn-glycerol-3-phosphate acyltransferase |
| PAP | YMR165C | EC 3.1.3.4 | Phosphatidate phosphatase | |
| DGA1 | YOR245c | YALI0E32769g | EC 2.3.1.20 | Diacylglycerol acyltransferase (DGAT2) |
| LRO1 | YNR008w | YALI0E16797g | EC 2.3.1.158 | Phospholipid:diacylglycerol acyltransferase |
| TGL3 | YMR313c | YALI0D17534g | EC 3.1.1.3 | Triacylglycerol lipase |
| TGL4 | YKR089c | YALI0F10010g | EC 3.1.1.3 | Triacylglycerol lipase |
| TGL5 | YOR081c | EC 3.1.1.3 | Triacylglycerol lipase | |
| ARE1 | YCR048w | YALI0F06578g | EC 2.3.1.26 | Acyl-CoA:sterol acyltransferase |
| ARE2 | YNR019w | EC 2.3.1.26 | Acyl-CoA:sterol acyltransferase | |
| DGA2 | YALI0D07986g | Diacylglycerol acyltransferase (DGAT1) | ||
| TGL1 | YKL140w | YALI0E32035g | EC 3.1.1.13 | Cholesterol esterase |
Bioinformatic data were obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/) and the Genolevures database (http://cbi.labri.fr/Genolevures/)
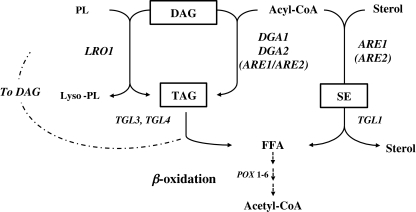
For the acyl-CoA-independent reaction, the Y. lipolytica homolog of the phospholipid:diacylglycerol acyltransferase gene (LRO1) is encoded by YALI0E16797g. For the acyl-CoA-dependent reaction, a diacylglycerol acyltransferase (DGA1) gene homolog, YALI0E32769g, was identified in Y. lipolytica. Two steryl ester synthases, encoded by the ARE1 and ARE2 genes, are involved in DAG acylation, via the acyl-CoA-dependent mechanism, in S. cerevisiae (Fig. 2). The sequences of Are1p and Are2p are 46% identical in S. cerevisiae. In Y. lipolytica, there is only one ARE1/ARE2 ortholog, YALI0F06578g (encoded by YlARE1), with an amino acid sequence about 30% identical to that of the corresponding protein in S. cerevisiae. However, a new homolog was identified in Y. lipolytica, YALI0D07986g (subsequently renamed YlDGA2), with a lower level of sequence identity to the protein encoded by YlARE1 (17%), but greater identity to a DGAT from the plant Perilla frutescens (22%), human DGAT1 (28%), and the DGAT1 from the plant Arabidopsis thaliana (25%). No gene with greater similarity to ScARE2 than YlARE1 was found in Y. lipolytica. We therefore focused on characterizing the “close hit” YALI0D07986g.
Fig. 2.
Biosynthesis of triacylglycerols and steryl esters in Y. lipolytica. G-3-P glycerol-3- phosphate, DHAP dihydroxyacetone phosphate, LPA lysophosphatidic acid, PA phosphatidic acid, DAG diacylglycerol, TAG triacylglycerol, PL phospholipid SE steryl ester, FFA free fatty acids
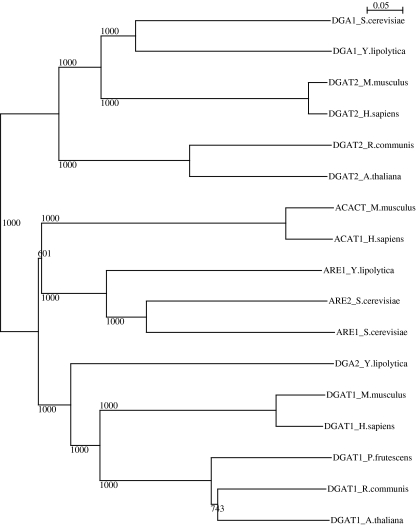
The full-length YALI0D07986g cDNA contained an open reading frame of 1,581 bp encoding a 526-amino acid protein with four potential transmembrane domains. Blast analyses were used to find close orthologs among the DGAT1 and ACAT families of species representative of three different kingdoms (see Fig. 3 for the resulting phylogenetic tree). The dendrogram has two main branches corresponding to entirely ACAT-related or entirely DGAT2-related sequences. The ACAT-related family itself branches into the DGAT1 (lower branch) and sterol acyltransferase-related enzymes (middle branch). YlDGA2 clusters with the DGAT1 family, even though DGAT1-type enzymes have not been described in other yeast species. Furthermore, the protein sequences of the representative families clustered according to the kingdom to which the species concerned belonged.
Fig. 3.
Dendrogram of the DGAT2 and ACAT membrane-bound families. The protein sequences deduced from the various full-length cDNAs of species belonging to three different kingdoms were aligned and bootstrapped with the ClustalX program (www.clustal.org). The tree was then generated using the NJplot program (Perrière and Gouy 1996). The proteins sequences used were DGAT1 (NP03611.2), DGAT2 (NP115953.1), and ACAT1 (NP003092.4) sequences from humans (Homo sapiens); DGAT1 (NP034176.1), DGAT2 (NP080660.1), and ACACT (NP033256.2) from the mouse (Mus musculus); the DGAT1 (XP002514132.1) and DGAT2 (XP002528531.1) sequences from the castor bean (Ricinus communis); the DGAT1 (NP179535.1) and DGAT2 (NM115011.3) sequences from Arabidopsis (Arabidopsis thaliana); the DGAT1 (AAG23696.1) sequences from the plant P. frutescens; the DGA1 (NP014888.1), ARE1 (CAY78257.1), and ARE2 (NP014416.1) sequences from the baker’s yeast (S. cerevisiae); and the DGA1 (XP504700.1), DGA2 (XP502557.1), and ARE1 (XP505086.1) sequences from Y. lipolytica. GenBank accession numbers are provided in parentheses. Note that sterol-related ACAT plant proteins were omitted due insufficient annotation
In vivo contribution of acyltransferases to storage lipid synthesis
We estimated the relative contributions of the individual genes to TAG and SE synthesis, by constructing two sets of yeast strains (Fig. 1): The first set was obtained by disrupting one or more genes in the wild-type Po1d strain to generate JMY1201 (∆lro1), JMY1204 (∆dga1), JMY1613 (∆are1), JMY1636 (∆dga2), JMY1634 (∆dga2∆are1), JMY1610 (∆dga1∆lro1∆are1), and the quadruple (Q4)-disrupted strain JMY1877 (∆dga1∆dga2∆lro1∆are1). The single deletions were used to determine the consequences of the loss of a single acyltransferase gene for TAG and/or SE accumulation. The double and triple deletion strains were used to analyze the functions of Are1p and Dga2p, and the quadruple mutant was used to investigate the possible contribution of other genes to storage lipid synthesis. The second set of yeast strains each overexpressed a single gene under the control of the constitutive TEF promoter, in the quadruple mutant background. These strains were JMY1882 (Q4-pTEF-LRO1), JMY1884 (Q4-pTEF-DGA2), JMY1892 (Q4-pTEF-DGA1), and JMY1988 (Q4-pTEF-ARE1) and were used to examine the roles of the individual genes.
All the strains constructed were viable and had growth rates similar to that of the parental Po1d strain (data not shown). The YNB(YE)O3 medium used in this study has a high C/N ratio, inducing lipid accumulation (Beopoulos et al. 2009). As storage lipids accumulate mostly during the late exponential and stationary growth phases, we analyzed lipid content at the beginning and end of the stationary phase and 37 h thereafter (11 and 48 h of culture, respectively). Lipid accumulation should be maximal after 11 h of culture in these conditions and should be affected by lipid mobilization due to β-oxidation after 48 h (Beopoulos et al. 2008).
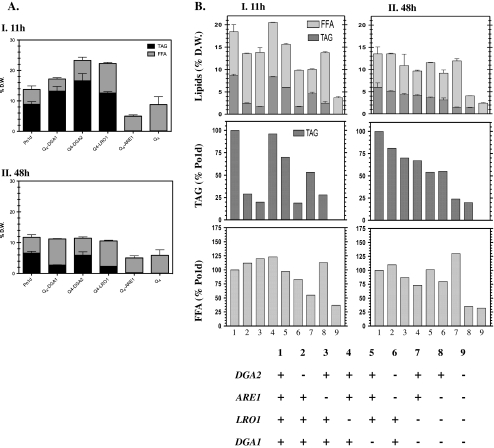
The patterns of storage lipid accumulation differed between strains. The ∆dga1∆dga2∆lro1∆are1 strain (the Q4 strain) contained no storage lipids and lacked LBs at all growth phases (Figs. 4 and 7b, respectively), confirming that the four acyltransferase-coding genes were essential for storage lipid synthesis. However, the Q4 strain produced significant amounts of FFA, accounting for about 5% of dry weight at all time points, although these amounts were less than these produced by the wild-type strain.
Fig. 4.
A TAG and FFA lipid fractions of wild-type (Po1d) and single acyltransferase gene overexpression strains in the Q4 background: I beginning of the stationary phase (11 h); II late stationary phase (48 h). B TAG and FFA lipid fractions from wild-type, single, double, triple, and quadruple mutant strains in I beginning of the stationary phase (11 h) and II late stationary phase (48 h). The presence or absence of a particular gene in each strain is indicated in the table at the bottom of the figure. Light gray bars indicate the amount of FFA; dark gray bars indicate the amount of TAG. The first row indicates the amount of lipids (FFA and TAG) as a percentage of yeast dry weight. The second row shows the amount of TAG found in mutant strains as a percentage of the TAG found in the wild-type strain. The third row shows the amount of FFA found in mutant strains as a percentage of the FFA found in the wild-type strain. Lipid quantification data in A and B represent mean values from three independent experiments
Fig. 7.
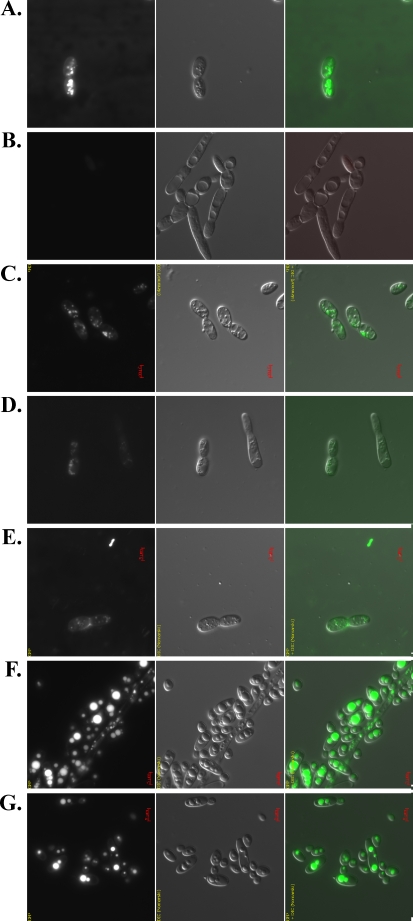
Observation of the phenotypes of the disrupted mutants after 24 h of culture in media containing 3% oleic acid. Fluorescence microscopy (left) with BodiPy neutral lipid staining, visual microscopy (middle), and combined photomicrographs (right) of A Po1d, B quadruple mutant, C ∆are1, D ∆dga1∆lro1, E ∆dga1∆lro1∆are1, F ∆dga2, and G ∆are1∆dga2 disrupted strains
Contribution of the genes to TAG synthesis
We analyzed the lipid content of the disrupted mutants and of the Q4 strains overexpressing each of the acyltransferases individually. We separated aliquots of total lipids into TAG and FFA fractions on a solid-phase extraction column. Total lipid content, TAG, and FFA fractions were then analyzed individually by gas chromatography.
We found that the TAG fraction of the Q4 strains expressing the DGA1, DGA2, and LRO1 genes individually under the control of the TEF promoter accounted for 76%, 70%, and 57% of total lipids at 11 h of culture, respectively, whereas wild-type TAG levels accounted for 64% of total lipids (Fig. 4a). The TAG fraction accounted for 28%, 55%, and 35% of total lipids at 48 h of culture for these strains and for 58% of total lipids for the wild-type strain. Surprisingly, the strain overexpressing only the ARE1 gene was completely devoid of TAG at all time points, like the Q4 strain, demonstrating a complete lack of DAG acyltransferase activity for Are1p. The strain producing Dga2p contained more TAG, at all time points, than the other strains. These results demonstrate the acyltransferase capacity of Lro1p, Dga1p, and Dga2p as, the TEF promoter drove expression of these genes.
The Q4 strain was the only disrupted strain with no TAG at all, in any of the growth phases tested (Fig. 4b). This was confirmed by TLC analysis (see Fig. 5, Q 4). However, the patterns of TAG synthesis differed significantly between strains during culture. The LRO1 and DGA1 single-deletion strains (Fig. 4b columns 4 and 5, respectively) contained 95% and 70% the amount of TAG in the wild type (Po1d, column 1), respectively, at 11 h. At the end of culture (48 h), they contained 65% and 55% the wild-type amount of TAG, due to the increasing activity of these two enzymes over time in the wild type. Similarly, the TAG content of the Δlro1Δdga1 double-disrupted strain increased from 45% to 75% of wild-type levels between 11 and 48 h of culture. The ARE1 and DGA2 single deletion strains contained very little TAG at 11 h (26% and 20% of wild-type levels, respectively) but TAG abundance increased thereafter, approaching wild-type levels at 48 h (80% and 70% of wild-type levels, respectively). ARE1 does not participate in TAG synthesis, but this observation suggests that acyltransferase genes act in synergy in DAG esterification. TAG levels in the Δlro1Δdga1Δare1 triple mutant remained at about 20% those in the wild-type throughout culture, demonstrating the relative contribution of the DGA2 gene to TAG synthesis.
Fig. 5.
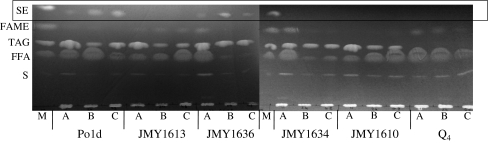
Neutral lipids separated by TLC. Plates were overloaded for visualization of the minor SE fraction, which accounts for less than 5% of total lipids in Y. lipolytica. Lipids were extracted using the Folch method from the following strains: Po1d; JMY1613 (Δare1); JMY1636 (Δdga2); JMY1634 (Δare1Δdga2); JMY1610 (Δdga1Δlro1Δare1). A, B, and C deposits correspond to the lipid extracts at 11, 24, and 48 h of culture, respectively. M TLC marker containing S sterols, FFA free fatty acids, TAG triacylglycerols, FAME fatty acid methyl esters, and SE steryl esters. FAME are artifacts produced during the Folch extraction procedure. The box is used to highlight the SE fraction
Contribution of the genes to SE synthesis
Y. lipolytica produces only minute amounts of steryl esters (<5% of total lipids), so our determinations of the SE fraction were qualitative. We overloaded TLC plates, for estimation of the relative amounts of SE in the various samples. However, these overloaded plates gave blurry results. Nevertheless, steryl esters were undetectable in all strains lacking ARE1 (Fig. 5; strains JMY1613, JMY1634, JMY1610, and Q4), whereas they were produced by all other strains. The dga2∆ mutant strain (JMY1636) was perfectly capable of synthesizing steryl esters, but the quantities of SE produced were below wild-type levels (Po1d), suggesting possible regulatory mechanisms operating between the ACAT genes.
FFA accumulation and the contribution of acyltransferases to total lipid accumulation
We assessed the extent to which the absence of the various acyltransferases affected overall lipid composition, by analyzing the amounts of FFA during the various growth phases. Total lipid levels were high at the start of the stationary growth phase (11 h) and were lower after 48 h of culture, due to β-oxidation (Beopoulos et al. 2008). The total lipid content of the Q4 mutant was about 25% that in the wild-type strain throughout the culture, and the Q4 mutant was unable to accumulate FFA to levels exceeding 5% of dry weight (Fig. 4b). Similarly, the FFA content of the triple mutant after 48 h was also very low. By contrast, all the other strains retained high FFA levels until the end of the time course. FFA level fluctuations depended on the strain genotype, consistent with the regulation of TAG synthesis and lipid accumulation.
Acyltransferase gene expression in the Po1d wild-type and derived strains
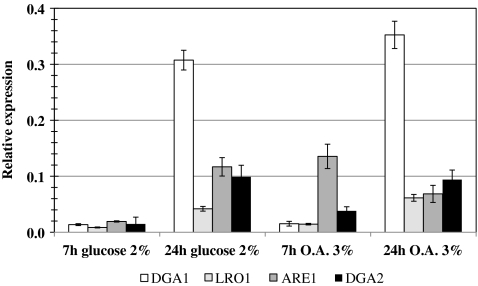
We analyzed expression of the acyltransferase genes in the Po1d wild-type strain, by RT-qPCR. Cells were cultured in YNB(YE)O3 accumulation medium and in YNB(YE)D2 medium (containing 2% glucose), favoring FA synthesis. Gene expression was assessed by determining mRNA abundance at the end of the exponential growth phase and during stationary phase (7 and 24 h, respectively).
Acyltransferase gene expression was weak throughout the exponential growth phase in medium containing glucose (Fig. 6, 7 h glucose), where concentration of synthesized FA is low to promote neutral lipid formation. However, after 24 h, when FA concentration was high, the DGA1 gene was strongly expressed, consistent with the major contribution of its product to TAG formation. Similar results were obtained after 24 h of culture in medium containing oleic acid, suggesting that DGA1 is expressed later than the ARE1 and DGA2 genes during neutral lipid synthesis. LRO1 gene expression followed a similar pattern. The ARE1 gene was strongly expressed in all conditions tested, except exponential growth in glucose medium. This is not particularly surprising, as ARE1 is the only gene encoding a sterol acyltransferase in Y. lipolytica.
Fig. 6.
Expression of the acyltransferase genes at 7 and 24 h of culture in media containing 2% glucose or 3% oleic acid (O.A.). Total RNA was isolated from the Po1d wild-type strain. Reverse transcription-quantitative PCR was carried out, with gene-specific primers (Table 2), for expression analysis. Three independent cDNA preparations obtained in independent experiments were used for the analysis
Observation of the phenotype of mutant strains
As Y. lipolytica is able to accumulate FFA in addition to TAG, we investigated whether this yeast formed LBs in the absence of storage lipids. We stained the cells in late stationary phase cultures with BodiPy dye, such that neutral lipids could be visualized under UV light. The wild-type strain (Po1d) formed about 15 medium-sized LBs per cell (Fig. 7A), whereas lipid bodies were completely absent from the Q4 strain (Fig. 7B). LBs were fewer in number and slightly smaller in the ARE1 single mutant (Fig. 7C), the LRO1 and DGA1 single mutants and the Δlro1Δdga1 double mutant (Fig. 7D; only the Δlro1Δdga1 mutant is shown), and in the Δlro1Δdga1Δare1 triple mutant (Fig. 7E) than in the parental wild-type strain. Surprisingly, the strains from which the DGA2 gene was deleted formed large LBs (the Δdga2 and Δare1Δdga2 strains, Fig. 7F and G, respectively). These strains had a FA composition similar to that of the other strains tested. The differences in LB morphology were therefore due to the loss of Dga2p, which therefore seems to be important for LB formation or the prevention of LB coalescence.
Discussion
The production of lipids by microorganisms for chemical, pharmaceutical, nutritional additive, or fuel use is increasingly attracting the interest of biotech companies, which are seeking renewable sources of products able to displace petrol-derived composites and to maintain ecological sustainability. Yeasts are good candidate microorganisms for such functions because they are easy to culture and to manipulate genetically and they have a high accumulation potential. Studies identifying and characterizing the genes involved in their storage lipid metabolism are paving the way to manipulation of the accumulation ratio and lipid composition of these species. S. cerevisiae was one of the first yeasts in which lipid metabolism was studied (Oelkers et al. 2002; Sandager et al. 2002; Sorger et al. 2004), but it is important to explore the lipid-synthesizing pathways of oleaginous yeasts, which are of greater potential utility for lipid-producing applications.
We show here that, in the oleaginous yeast Y. lipolytica, the DGA2 gene, which displays sequence similarity to the ARE1 gene of Y. lipolytica and the ARE genes of S. cerevisiae, plays a role different from that of the common ACAT-related genes identified in yeasts. Phylogenetic analysis of the genes contributing to the acyl-CoA dependent acylation of DAG from various organisms revealed that the YlDGA2 gene clustered with the DGAT1 family. This family forms a branch separate from the ACAT-related enzymes with sterol acyltransferase activity. However, DGAT1 and sterol acyltransferase-related enzymes clearly belong to different, but functionally convergent protein classes. All these families have highly similar domains in common, suggesting that these genes may all be derived from a common ancestor. Nevertheless, the DGA2 gene of Y. lipolytica is clearly only very distantly related to plant and mammalian enzymes.
As we were unable to identify any other gene homologous to S. cerevisiae ARE genes, we confirmed that the four Y. lipolytica acyltransferases were the only genes involved in DAG acylation: The quadruple-deleted strain (Q4) had no storage lipids in any of the growth phases studied. However, this strain nonetheless maintained significant amounts of FFA. Y. lipolytica is unusual in being able to accumulate large amounts of FFA without toxic effects on the cell. This residual lipid fraction may be responsible for the sustained viability of the Q4 strain during stationary phase. We investigated the substrate specificity of the product encoded by the DGA2 gene by studying the expression of this gene, under the control of the constitutive TEF promoter, in the quadruple mutant background. The same expression protocol was used for each of the four acyltransferases individually. Dga2p had strong TAG acyltransferase activity, whereas no SE was produced at any growth stage. SE were identified by TLC in all strains lacking the DGA2 gene. We therefore conclude that the Y. lipolytica DGA2 gene belongs to the DGAT1 family and has no sterol esterification activity. No other gene of the ACAT family and DGAT1 subfamily has, to our knowledge, been identified in yeasts.
We investigated the role of the other Y. lipolytica ACAT enzyme, encoded by the ARE1 gene, and found that the strain overexpressing the ARE1 gene only was completely devoid of TAG. By contrast, all other Q4 strains in which a single gene was overexpressed were devoid of SE. TLC analysis of deletion strains confirmed these findings because the SE fraction was undetectable in all strains lacking ARE1. Our results indicate that the Are1p of Y. lipolytica has no TAG acyltransferase activity and contributes to storage lipid synthesis as a sterol acyltransferase. Thus, each member of the ACAT family in Y. lipolytica has a different activity, and these activities are different from those of the ARE-encoded enzymes of S. cerevisiae, which participate in the synthesis of both SE and TAG.
However, the low TAG content of the strains with single or double deletions of ARE1 and DGA2 at 11 h of culture, despite the lack of participation of Are1p in TAG synthesis, may indicate that acyltransferase genes act in synergy in DAG esterification. Indeed, the ∆are1 strain had lower levels of TAG levels than the wild-type strain, and the Δare1Δdga2 double-disrupted strain contained less TAG than the DGA2 single-deletion strain at the end of culture. The situation may be similar for SE synthesis, as the SE levels of the ∆dga2 strain were below wild-type levels. However, we can draw no firm conclusions about the quantitative contribution of each gene to any such regulation.
One interesting finding of this study is the relative contributions of the four acyltransferase genes to neutral lipid synthesis. Our results strongly suggest that the DGAT2 enzyme, encoded by the DGA1 gene, is the major TAG-synthesizing enzyme in Y. lipolytica. However, the acyltransferase activity potential of its counterpart from the DGAT1 family, encoded by the DGA2 gene, was greater when the gene was expressed in the quadruple mutant strain under the control of a strong constitutive promoter. This suggests that there may be a mechanism regulating gene expression. In addition, the acyltransferase gene context of a strain seems to affect the activities of the individual genes, suggesting that there may be a mechanism for regulating overall lipid synthesis. During culture, the TAG levels of the LRO1 and DGA1 single-deletion strains gradually decreased over time, indicating an increase in the activity of these two enzymes. By contrast, the DGA2 and ARE1 single-deletion strains displayed only very low levels of TAG synthesis at the start of culture, suggesting that these enzymes were strongly expressed during the exponential growth phase. These results suggest that DGA1 is expressed after the ARE1 and DGA2 genes during neutral lipid synthesis. LRO1 gene expression followed a similar pattern, consistent with the decrease in TAG levels in the ∆dga1∆lro1 double mutant strain between 11 and 48 h of culture. Oelkers et al. (2002) demonstrated that transcription of the DGAT2 gene of S. cerevisiae (ScDGA1) was upregulated by various culture conditions, such as growth to saturation and nitrogen deprivation. In general, DGAT2 activity is considered to make a more marked contribution to TAG formation during stationary phase (Yen et al. 2008; Kohlwein 2010). However, this is not the case for the genes of the ACAT family. DGA2 gene expression levels were intermediate, with the expression of this gene stronger than that of LRO1, consistent with its “second-string” role in TAG synthesis. This finding is consistent with TAG levels in the ∆dga1∆lro1 mutant, in which only Dga2p contributes to TAG formation.
Acyltransferase gene expression, as assessed by RT-qPCR, was weak during the exponential growth phase in medium containing glucose, in which the concentration of FA synthesized is too low for neutral lipid formation. However, after 24 h, when FA concentration was high, the DGA1 gene was strongly expressed, consistent, again, with its major contribution to TAG formation. The ARE1 gene was strongly expressed in all conditions tested, except exponential growth in glucose medium. This is not particularly surprising, as ARE1 is the only gene encoding a sterol acyltransferase in Y. lipolytica. As the genes encoding TAG acyltransferases appear to have overlapping functions, they may contribute to TAG synthesis to different extents during culture.
Another striking observation was the formation of large LB in all strains lacking Dga2p. We interpret this as indicating that Dga2p either contributes to LB formation or prevents LB coalescence. In support of a role in preventing coalescence, Dga2p contains four transmembrane domains, which may contribute to repulsion between LBs. No proteins involved in LB stabilization have yet been identified in yeasts, by contrast to the situation in plants and mammals. However, the most plausible hypothesis suggests a direct role of Dga2p in LB biogenesis. The most widely accepted model of LB formation involves the biogenesis of lipid droplets in the ER membrane due to the accumulation of neutral lipids within its bilayer. This accumulation is due to the high concentration of neutral lipid-synthesizing enzymes at this site (Walther and Farese 2009). If Dga2p participates abundantly in the initiation of neutral lipid synthesis, then its absence may reduce the number of ER sites involved, resulting in fewer, but larger LBs.
In conclusion, we describe here the roles of the four acyltransferases involved in the final acylation step of TAG assembly in the oleaginous yeast Y. lipolytica. It is clear from the enzyme specificity data for this yeast that TAG synthesis is favored over SE synthesis. Dga2p, an enzyme of the DGAT1 acyltransferase family, participates in TAG synthesis, rather than sterol esterification, by contrast to the functions reported for common ACAT-related enzymes in yeasts. Our findings provide insight into the lipid synthesis metabolism of oleaginous yeasts, which are good candidates for applications in lipid production. Nevertheless, from a biotechnological point of view, the quadruple mutant is also a good candidate for heterologous acyltransferase expression, particularly for modification of the TAG fatty acid profile.
Acknowledgments
A. Beopoulos was supported by INRA. This work was financially supported in part by Aerospace Valley, by the Lipicaero French national research program (ANR-PNRB) No. 0701C0089, and by the European Commission in the framework of the LipoYeasts project (contract number 213068). We thank Julie Sappa of Alex Edelman and Associates for her excellent help in correcting the English version of the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Athanasios Beopoulos, Email: abeopoulos@grignon.inra.fr.
Ramdane Haddouche, Email: rhaddouche@grignon.inra.fr.
Thierry Chardot, Email: thierry.chardot@versailles.inra.fr.
Jean-Marc Nicaud, Email: jean-marc.nicaud@grignon.inra.fr.
References
- Athenstaedt K, Jolivet P, Boulard C, Zivy M, Negroni L, Nicaud JM, Chardot T. Lipid particle composition of the yeast Yarrowia lipolytica depends on the carbon source. Proteomics. 2006;6:1450–1459. doi: 10.1002/pmic.200500339. [DOI] [PubMed] [Google Scholar]
- Barth G, Gaillardin C. Yarrowia lipolytica. In: Wolf K, editor. Non conventional yeasts in biotechnology, a handbook. Berlin: Springer; 1996. pp. 313–388. [Google Scholar]
- Barth G, JM B, Dominguez A, Kerscher S, Ogrydziak D, Titorenko V, Gaillardin C. Functional genetics of Yarrowia lipolytica. In: dW JH, editor. Functional genetics of industrial yeasts, vol 1. Topics in current genetics. Berlin: Springer; 2003. pp. 227–271. [Google Scholar]
- Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, Chardot T, Nicaud JM. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beopoulos A, Cescut J, Haddouche R, Uribelarrea JL, Molina-Jouve C, Nicaud JM. Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res. 2009;48:375–387. doi: 10.1016/j.plipres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Browse J, McCourt PJ, Somerville CR. Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem. 1986;152:141–145. doi: 10.1016/0003-2697(86)90132-6. [DOI] [PubMed] [Google Scholar]
- Czabany T, Athenstaedt K, Daum G. Synthesis, storage and degradation of neutral lipids in yeast. Biochim Biophys Acta. 2007;1771:299–309. doi: 10.1016/j.bbalip.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Daum G, Wagner A, Czabany T, Athenstaedt K. Dynamics of neutral lipid storage and mobilization in the yeast. Biochimie. 2007;89:243–248. doi: 10.1016/j.biochi.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickers P, Le Dall MT, Gaillardin C, Thonart P, Nicaud JM. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J Microbiol Methods. 2003;55:727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Gaillardin C, Ribet AM, Heslot H. Integrative transformation of the yeast Yarrowia lipolytica. Curr Genet. 1985;10:49–58. doi: 10.1007/BF00418493. [DOI] [Google Scholar]
- Heier C, Taschler U, Rengachari S, Oberer M, Wolinski H, Natter K, Kohlwein SD, Leber R, Zimmermann R. Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1801:1063–1071. doi: 10.1016/j.bbalip.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein SD. Triacylglycerol homeostasis: insights from yeast. J Biol Chem. 2010;285:15663–15667. doi: 10.1074/jbc.R110.118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffargue A, de Kochko A, Dussert S. Development of solid-phase extraction and methylation procedures to analyse free fatty acids in lipid-rich seeds. Plant Physiol Biochem. 2007;45:250–257. doi: 10.1016/j.plaphy.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Mlickova K, Roux E, Athenstaedt K, d’Andrea S, Daum G, Chardot T, Nicaud JM. Lipid accumulation, lipid body formation, and acyl coenzyme a oxidases of the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2004;70:3918–3924. doi: 10.1128/AEM.70.7.3918-3924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Sandal T, Kamp-Hansen P, Dalboge H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast. 1998;14:1267–1283. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1267::AID-YEA327>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Nicaud JM, Madzak C, van den Broek P, Gysler C, Duboc P, Niederberger P, Gaillardin C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002;2:371–379. doi: 10.1016/S1567-1356(02)00082-X. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- Perrière G, Gouy M. www-query: an online retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Querol A, Barrio E, Huerta T, Ramon D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl Environ Microbiol. 1992;58:2948–2953. doi: 10.1128/aem.58.9.2948-2953.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge C. Yeasts, moulds, algae and bacteria as sources of lipids. In: Kamel BS, Kakuda Y, editors. Technological advances in improved and alternative sources of lipids. London: Blackie Academic and Professional; 1994. pp. 235–291. [Google Scholar]
- Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol. 2002;51:1–51. doi: 10.1016/S0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- Saha S, Enugutti B, Rajakumari S, Rajasekharan R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006;141:1533–1543. doi: 10.1104/pp.106.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S. Storage lipid synthesis is non-essential in yeast. J Biol Chem. 2002;277:6478–6482. doi: 10.1074/jbc.M109109200. [DOI] [PubMed] [Google Scholar]
- Sorger D, Athenstaedt K, Hrastnik C, Daum G. A yeast strain lacking lipid particles bears a defect in ergosterol formation. J Biol Chem. 2004;279:31190–31196. doi: 10.1074/jbc.M403251200. [DOI] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]