Abstract
The rhizospheres of five different potato cultivars (including a genetically modified cultivar) obtained from a loamy sand soil and two from a sandy peat soil, next to corresponding bulk soils, were studied with respect to their community structures and potential function. For the former analyses, we performed bacterial 16S ribosomal RNA gene-based PCR denaturing gradient gel electrophoresis (PCR-DGGE) on the basis of soil DNA; for the latter, we extracted microbial communities and subjected these to analyses in phenotype arrays (PM1, PM2, and PM4, Biolog), with a focus on the use of different carbon, sulfur and phosphorus sources. In addition, we performed bacterial PCR-DGGE on selected wells to assess the structures of these substrate-responsive communities. Effects of soil type, the rhizosphere, and cultivar on the microbial community structures were clearly observed. Soil type was the most determinative parameter shaping the functional communities, whereas the rhizosphere and cultivar type also exerted an influence. However, no genetically modified plant effect was observed. The effects were imminent based on general community analysis and also single-compound analysis. Utilization of some of the carbon and sulfur sources was specific per cultivar, and different microbial communities were found as defined by cultivar. Thus, both soil and cultivar type shaped the potato root-associated bacterial communities that were responsive to some of the substrates in phenotype arrays.
Introduction
Plants are known to produce and secrete (via their root systems) a range of chemical compounds, under which sugars, amino acids, organic acids, and a plethora of signaling molecules of diverse nature [22]. The biological outcome of the secretion of such compounds by plant roots is thought to be (among other effects): (1) defense against harmful organisms and (2) attraction of organisms that are either beneficial or detrimental (pathogens). In total, plants may release up to 30% of their photosynthate via secretion from roots into the adjacent rhizosphere soil. This active release process establishes an environment in which plant–microorganism interactions are bountiful. Such interactions are known to ultimately affect plant growth and health in a plethora of direct and indirect ways, with final beneficial or detrimental outcomes as sketched above [20].
Several studies have indicated that plant roots often exert strong effects on their associated (rhizospheric) bacterial communities [4, 19, 29]. The nature and extent of such effects are likely related to the composition of the root exudates. In other words, different compounds in the exudates may stimulate different root-responsive organisms [22]. Soil type and plant species and age [9, 10, 21, 27] have all been shown to exert clear effects on root exudate composition. On the other hand, the type of soil determines the availability of nutrients, which can have a strong effect on plant physiology, reflected in root exudation patterns and, consequently, the microbial communities in the rhizosphere. For instance, soil pH, aeration, and physicochemical characteristics all result in distinct conditions in the soil and thus impact the rhizosphere microbial communities [3, 12, 23]. Moreover, effects of plant species type on these communities in the rhizosphere have been shown [4], next to effects of genetic modifications [24]. The modification-dependent effects were generally described as being minor compared to the impact of soil type, plant variety, and plant developmental stage. However, we still lack knowledge as to how cultivars of the same plant species, in interaction with soil, affect the community make-up of rhizosphere inhabitants and where a genetically modified (GM) plant is situated in this picture.
Biolog community-level physiological profiling has been previously used [2, 8, 14, 15, 29, 31] to study specific aspects of soil microbial function. For instance, in a key study, no soil effect, but a clear plant type effect, was shown [15]. Another study claimed that soil type is a major driver of the community composition [1]. The number of carbon sources that were available in the previous studies has varied from 31 to 95, depending on the Biolog system used [7, 11, 25]. The novel so-called phenotype array, in which the number of C sources has been doubled to 190 and a range of other resources such as sources of N, P, and S were introduced, now offers a method that allows the medium-throughput analysis of potential soil function. Its use with rhizosphere communities of different cultivars of the same plant species grown in parallel in the same soil is thought to generate information about the function-based selection processes exerted on microorganisms by the roots of such cultivars.
In this study, the effects of selected potato cultivars, including a GM cultivar, growing in two different soils on the functional community of the root-associated microbial communities were investigated, using, for the first time, the novel Biolog phenotype array (C, S, and P). We provide evidence for the contention that potato root exudate compounds shape the potential function of rhizospheric microbial communities, next to their structures. Moreover, we assessed how microbial community structures change per compound and how this relates to soil type.
Materials and Methods
Soil Sampling and DNA Extraction
Two experimental fields—Buinen (B, 52°55′ N–6°49 E) and Valthermond (V, 52°50′ N–6°55 E), located in Drenthe, The Netherlands—were selected for the experiments. These fields contained divergent soil types, i.e., B, loamy sand, 5% OM (organic matter - pH 5.0) and V, sandy peat, 25% OM (pH 5.0). The fields were under agricultural rotation. In a previous growth season, spring barley had been grown in both fields. Five different potato cultivars [Aveka (A), Aventra (Av), Karnico (K), Modena (M; modified from Karnico for low amylose content in the tubers) [6], and Désirée (D)] from B and cultivars A and D from V soil were used in the senescence stage. Cultivars A, Av, K, and M produced tubers with high starch contents and had a low and/or medium growth rate, whereas cultivar D yielded tubers with relatively low starch contents and had high growth rate. For each potato cultivar, four replicate plots randomly distributed over the fields were used. At the start of the growth season, these plots were cropped with 20 plants (tubers) each. The fields were under standard agricultural practice. Samples were taken at senescence stage (EC99) [16].
Sampling was as described previously [18]. Bulk and rhizosphere soils were used directly for DNA extraction. For the extractions, the Powersoil DNA extraction kit (Mo Bio Laboratories Inc., New York, USA) was used as described previously [19].
PCR Amplification and DGGE Community Fingerprintings
On the basis of DNA extracted from the rhizosphere and bulk soils, PCR amplifications targeting the 16S ribosomal RNA (rRNA) genes of general bacteria were run. The reaction contained 1 μl (5 ng) of extracted DNA, which was amplified with DGGE primers GC-341 and 518R (30 cycles) as described previously [19]. All DGGE profiles were generated in the Ingeny Phor-U system (Ingeny International, Goes, The Netherlands) as described previously [19].
The DGGE profiles in the different gels were digitized and stored as TIFF files. Images were normalized using the markers and the patterns subsequently compared using clustering methods. Similarity matrices consisting of defined numbers within each gel were generated using Pearson’s correlation coefficient (r). Subsequently, the patterns were clustered using the unweighted pair group method with arithmetic averages (UPGMA) with GelCompar II software (Applied Maths, Sint-Martens-Latem, Belgium). Besides, the matrices representing band distribution were used for multivariate analysis using CANOCO (Microcomputer Power, Ithaca, NY, USA).
Metabolic Profiling Using Biolog Phenotype Arrays
Rhizosphere samples (roots with adhering soil; approximately 3 g) at senescence stage were placed in sterile Falcon tubes and treated twice at high speed (260 rpm) with 20 ml of a solution containing phosphate-buffered saline and 0.3 g of Chelex-100 (Bio-Rad, Hercules, CA, USA) and 10 ml of 0.5% polyethylene glycol using a modified protocol [17, 29]. Bacterial cells from the sample were recovered by differential centrifugation as described previously [29]. The resulting cell pellet was resuspended in 40 ml of IF-0 buffer (inoculating fluid, supplied by Biolog). The optical density at 650 nm of the IF-0 solution was adjusted to 0.2 using sterile saline. For PM4 plates, sodium succinate and ferric citrate were added to the solution as is recommended by Biolog. Each bacterial cell suspension was used to inoculate replicate PM1, PM2, and PM4 plates (Biolog, Hayward, CA, USA; 100 ml per well), which were then incubated at room temperature for 48 h. The OD510 values were measured with a microtiter plate reader each 6 h of incubation, and values at 48 h were used for further analysis. Values were normalized, and the created matrix data were used for redundancy analysis (RDA) using CANOCO (Microcomputer Power, Ithaca, NY, USA) [19].
PCR-DGGE Analysis of Communities in Phenotype Array Wells
Wells with developed growth were selected. The liquid in the well (about 40 μl) was sampled, treated for 20 min at 95°C and directly frozen at −20°C for 20 min. One microliter cell lysate was then used as a template for PCR as described above. PCR products were checked on gel for quality and quantity, and DGGE was run of positive products as explained before. The gel images were digitized. Matrices based on intensity of bands were generated by Gelcompar and used to create dendrograms (Pearson correlation). Then, RDA was performed, followed by analysis of similarity (ANOSIM) using Euclidean distance. ANOSIM was carried out using PRIMER 6 (Primer v6, Plymounth), based on transformed data (fourth square). Samples were also grouped per soil and cultivar type. Two-way crossed analyses with replicates were done with 5,000 permutations. The global R value, varying between −1 and 1, was used. R = 0 indicated completely random grouping, while R = 1 indicated that samples within a soil were more similar to each other than to any samples from the other soil. A significant global R indicated that there are differences between soil types somewhere in the analysis.
Results
Bacterial Community Structures in Bulk and Rhizosphere Soils as Assessed by PCR-DGGE
For both bulk and rhizosphere soil samples, bacterial PCR-DGGE patterns were generated. Overall, these patterns revealed 60–70 bands per lane in all samples. Patterns derived from rhizosphere soil samples from the B soil generally revealed higher band numbers and stronger bands in comparison to those from the V soil as well as all bulk soil samples (data not shown).
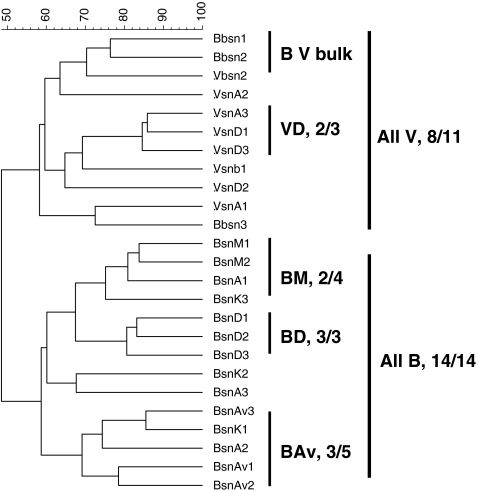
Analyses of all patterns (using UPGMA with Pearson correlation) revealed two major clusters, one encompassing the patterns derived from all B soil derived samples; the other one encompassing those from all V soil ones, next to three B soil samples. The bulk soil samples clustered together inside the V soil cluster. Per soil, the rhizosphere-derived patterns all grouped apart from the corresponding bulk soil ones, thus indicating clear effects of the plant roots on the structures of the local bacterial communities. The rhizosphere effects were more evident for the B than for the V soil. That the effect of soil type was indeed stronger than that of cultivar was also apparent from the fact that the same cultivars grown in B versus V soil yielded different patterns, which grouped them in clusters defined by soil rather than cultivar type (Fig. 1).
Figure 1.
Dendrogram representing the similarity of PCR-DGGE profiles generated with 16S rRNA gene based bacterial DGGE for B and V soil at senescence stage. B Buinen, A Aveka, Av Aventra, K Karnico, M Modena, D Désierée, V Valthermond, sn senescence, b bulk
At the finer level, clustering into two main clusters was observed for the rhizosphere patterns in the B soil (cultivars M and D on the one hand and cultivars A, Av, and K on the other hand). In the V soil, a main cluster encompassed the patterns from cultivars A and D, which were quite similar. These data confirmed the clustering of bacterial PCR-DGGE patterns related to cultivars obtained in a previous study [19].
Phenotype Array Assays
Carbon Sources
Overall, 173 of the 190 carbon sources were consumed, by any of the bulk- or rhizosphere-derived communities, leaving 17 of the 190 substrates unconsumed. The latter compounds were sec-butylamine, 3-0-β-d galactopyranosyl-d-arabinose, 2-hydroxy benzoic acid, oxalic acid, l-glucose, acetamide, putrescine, 3-hydoxy-2-butane, sedoheptulosan, d,l-octopamine, tyramine, d-threonine, acetoacetic acid, m-hydroxy phenyl acetic acid, glucuronamide, 1,2-propanediol, and d-psicose.
Overall, N-acetyl-d-glucosamine, l-proline, γ-amino butyric acid, and d-ribono-1,4-lactone were the most consumed carbon sources. With the exception of N-acetyl-d-glucosamine and d-ribono-1,4-lactone, the C sources consumed by the B and V rhizosphere soil communities were either different, or the utilization of C sources was faster, than observed for the corresponding bulk soil communities. One peculiarity was that phenylethylamine was only consumed by the B bulk soil communities.
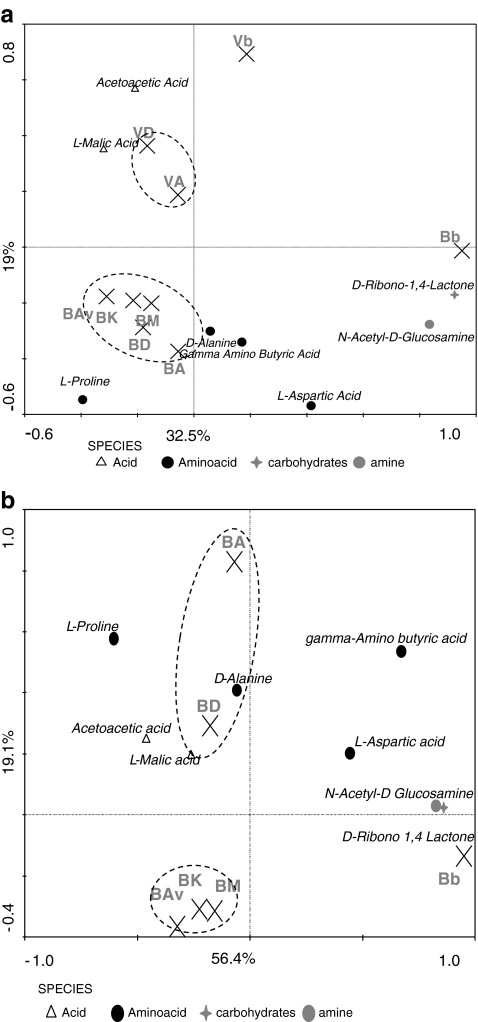
RDA (based on the complete set of carbon sources) revealed a significant effect of soil type on the bulk soil communities’ ability to utilize different carbon sources (P < 0.05; Fig. 2a). Specifically, the communities from the two soils, both from bulk and rhizospheres, were functionally quite different. For the B soil, this analysis further revealed a significant (P < 0.05) effect of the rhizosphere, irrespective of cultivar type, on the potential to consume the phenotype array carbon sources. However, no such effect was observed for the V soil. Furthermore, no significant effect of cultivar type on C substrate consumption was found in any of the two soils. However, the communities derived from the B soil grown cultivars A and D revealed similar substrate utilization profiles, while those from B soil grown cultivars Av, K, and M also grouped together (Fig. 2b). The utilization patterns by the communities from the V soil grown cultivars A and D also grouped together. The analyses further showed that the communities from cultivars A and D grown in different soils had different preferences for the carbon sources of the phenotype array (Fig. 2a).
Figure 2.
Ordination biplots generated by redundancy analysis (RDA) of consumption of carbon sources for a soil B and V and b soil B (B Buinen, V Valthermond, A Aveka, Av Aventra, K Karnico, M Modena, D Désierée, b bulk). The eigenvalues displayed on the diagram axes refer to the percentage variation of carbon consumption; environment correlation accounted for at the respective axis. Compounds shown on the diagrams are analyzed by PCR-DGGE
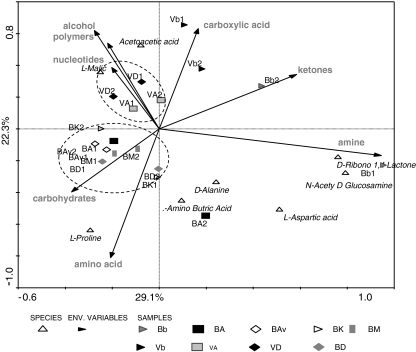
To analyze the putative effects of carbon source classes, all carbon sources of the phenotype array were grouped into eight classes, i.e., amino acids, carboxylic acids, nucleotides, carbohydrates, ketones, alcohols, amines, and polymers. These were subsequently used as environmental data in the analyses. PCA analyses (CANOCO) were then performed to investigate the possible correlation between cultivars and carbon sources (Microcomputer Power, Ithaca, NY, USA). The communities from B bulk soil mainly showed a preference for ketones and amines, whereas those from V bulk soil preferred carboxylic acids. The communities from the rhizospheres also revealed different preferences per soil. Those from B soil grown cultivars had affinity for carbohydrates and amino acids, whereas those from V soil grown ones consumed mainly alcohols, polymers, and nucleotide-based carbon sources (Fig. 3).
Figure 3.
Ordination biplot generated by principal components analysis (PCA) on the basis of the consumption of carbon sources (B Buinen, V Valthermond, A Aveka, Av Aventra, K Karnico, M Modena, D Désierée, b bulk). The eigenvalues displayed on the diagram axes refer to the percentage variation of the sulfur consumption rate; environment correlation accounted for at the respective axis. The carbon sources of plates PM1 and PM2 were grouped into eight classes, i.e., amino acids, organic acids, heterocyclic compounds, sugars, lipids, alcohols, amines, and polymers, and introduced as environmental data. Compounds shown on the diagram are analyzed by PCR-DGGE
Sulfur and Phosphorus Sources
The utilization patterns of different phosphorus (59) and sulfur (35) sources were also analyzed across the samples, using the Biolog PM4 plate. Overall, most (57/59) of the phosphorus compounds were consumed by any of the bulk and rhizosphere soil communities, leaving only two compounds (tripolyphospate and d-mannose-6-phosphate), which were not consumed by any community. Overall, six out of the 59 phosphorus sources were highly consumed; these were the heterocyclic compounds adenosine-3′-monophosphate, adenosine-2′-monophosphate, uridine-2′,3′-cyclic monophosphate, guanosine-2′,3′-cyclic monophosphate, cytidine-2′,3′ cyclic monophosphate, and guanosine-5′-monophosphate. The remaining phosphorus sources (51 of 59) were differentially used.
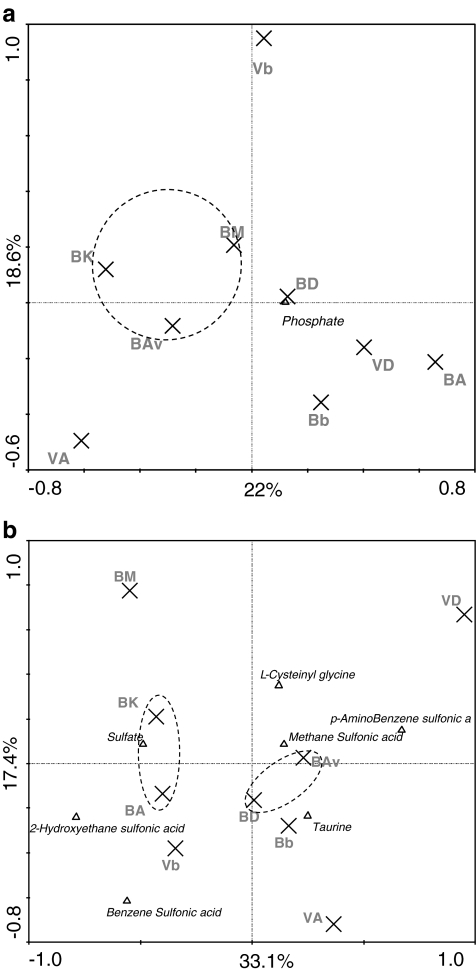
RDA analysis of the data from all 59 phosphorus sources revealed a rhizosphere effect, but this was not significant in any of the two soils. Furthermore, the utilization patterns by communities from the B soil were similar between cultivars Av, K, and M, whereas those by communities from cultivars D and A clustered apart from the former (Fig. 4a). In the V soil, no significant effect of cultivar was observed.
Figure 4.
Ordination biplot generated by redundancy analysis (RDA) of consumption of (a) phosphorus and (b) sulfur sources. ([B] Buinen, [V] Valthermond, [A] Aveka, [Av] Aventra, [K] Karnico, [M] Modena, [D] Désierée, [b] bulk). The eigenvalues displayed on the diagram axes refer to the percentage variation of sulfur consumption rate; environment correlation accounted for at the respective axis. Compounds shown on the diagrams are analyzed by PCR-DGGE
Of the 35 sulfur sources analyzed, 31 were utilized by any of the analyzed communities. Thus, four (tetramethylene sulfone, l-methionine, S-methyl-l-cystine and d-methionine) remained unconsumed. Another four (taurine, 2-hydroxy ethane sulfonic acid, sulfate, and benzene sulfonic acid) were highly consumed by all communities analyzed. The remaining sulfur sources (27/35) were differentially used.
RDA analysis performed on the sulfur utilization assays revealed no clear rhizosphere effects for the communities in both soils, except for the cultivar D community in the V soil. However, in the B soil, the communities from cultivars Av and D on the one hand and A and K on the other revealed similar utilization patterns, whereas those from cultivar M grouped apart from the other cultivars (Fig. 4b).
PCR-DGGE Analysis of Communities in Selected Wells of the Phenotype Arrays
To analyze the structures of the communities that contributed to the observed consumption patterns in the phenotype arrays and to find key indicator carbon/sulfur/phosphorus sources that might best indicate community structure changes due to soil type, rhizosphere, or cultivar type effects, the communities of selected wells were analyzed by PCR-DGGE. In the selection, the resources that (1) were consumed to the greatest extent and (2) were presumably representative of potato root exudates (carbon sources) were selected. Thus, N-acetyl-d-glucosamine, l-proline, d-alanine, γ-amino butyric acid, acetic acid, l-malic acid, glycyl-l-proline, l-aspartic acid, and d-ribono-1,4-lactone were selected as the carbon sources. As to the phosphorus and sulfur sources, phosphate and sulfate, l-cysteinyl-glycine, benzene sulfonic acid, methane sulfonic acid, taurine, p-aminobenzene sulfonic acid and 2-hydroxyethane sulfonic acid were selected. Several of the nine carbonaceous resources have indeed previously been suggested to represent characteristic C compounds in potato root exudates [5].
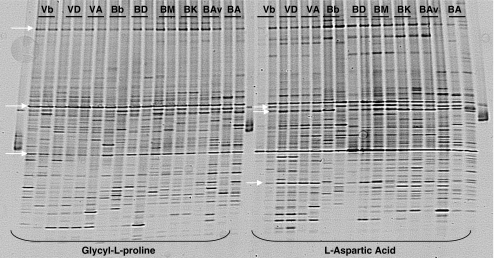
For all substrates, the PCR-DGGE patterns revealed the presence of 20–30 bands per lane. Several bands were shared per analyzed carbon, phosphorus and/or sulfur source, but fluctuating bands (variably present or absent) were also observed. Per gel, two carbon sources could be analyzed. To avoid the complication posed by gel-to-gel differences, the commonality of microbial community structure occurring with different substrates was compared on the same gel. Thus, the patterns derived from γ-amino butyric acid and N-acetyl-d-glucosamine, next to d-alanine and l-proline, shared three very dominant bands, which appeared in all samples regardless of the origin of the communities. Furthermore, the patterns from glycyl-l-proline and l-aspartic acid shared three dominant bands, which also appeared for all samples (Fig. 5). In the glycyl-l-proline wells, one extra common band was observed, whereas in the l-aspartic acid ones, three extra common bands were present (Fig. 5). The patterns from d-ribono-1,4-lactone revealed four common bands across all communities (from different cultivars and corresponding bulk soils)
Figure 5.
16S rRNA DGGE profile for the compounds glycyl-L-proline and aspartic acid. Indicated bands are common throughout all the samples of both analyzed compunds. Arrows indicate the common bands in the analyzed samples. Samples are also indicated above the lanes. B Buinen, A Aveka, Av Aventra, K Karnico, M Modena, D Désierée, V Valthermond, b bulk
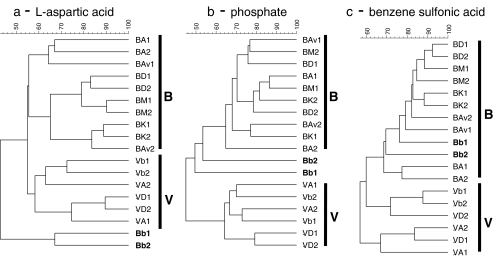
Cluster (UPGMA-based) analysis of the PCR-DGGE data obtained for each substrate showed clear effects of the soil origin of the analyzed community on the clustering of bulk and rhizosphere patterns for four (γ-amino butyric acid, l-malic acid, glycyl-l-proline, and l-aspartic acid) of the nine selected carbon sources. Moreover, the effect of the rhizosphere was related to the type of carbon source analyzed (not shown). Figure 6a shows the clustering obtained for l-aspartic acid, as a representative of clusterings of analyzed carbon sources. Further analyses revealed that the rhizosphere effects observed for the communities (obtained from all cultivars) in the l-aspartic acid wells (Fig. 6a) from plants grown in both the B and the V soils were all significant (P < 0.05). The structures of the communities that were consuming l-aspartic acid were found to be soil type and rhizosphere specific. Moreover, those for the l-proline, γ-amino butyric acid, and l-malic acid wells from plants grown in the B soil also showed significant differences (P < 0.05). In this respect, the rhizosphere effects in the B soil were found to be stronger than those in the V soil. Strikingly, the patterns obtained from the l-aspartic acid responsive communities derived from B soil grown cultivars A and Av on the one hand and K, M, and D on the other hand, grouped together. The same was true for the communities in the l-proline, γ-amino butyric acid, l-malic acid and N-acetyl-d-glucosamine wells. Together, this indicated dichotomous selection of substrate-responsive microbial communities by the two cultivar clusters (A/Av versus K/M/D) in the B soil.
Figure 6.
Dendrograms representing the similarity of PCR-DGGE profiles generated with 16S rRNA gene based bacterial DGGE for carbon, phosphorus and sulfur sources: a l-aspartic acid, b phosphate, c benzene sulfonic acid, for B and V soil at senescence stage. B Buinen, A Aveka, Av Aventra, K Karnico, M Modena, D Désierée, V Valthermond, b bulk
Given their high consumption rates, the communities in the wells with either phosphate or the seven selected sulfur sources were analyzed with PCR-DGGE. Effects of soil type on the resulting patterns were observed for phosphate and six of the seven sulfur sources (Table 1). Moreover, a comparison of the patterns obtained from B bulk and rhizosphere soil communities for phosphate consumption revealed community structures to be different between the rhizosphere and bulk soils (Fig. 6b). On the other hand, in the V soil, the only effect of the presence of roots (i.e., a rhizosphere effect) was observed for cultivar D. In this analysis, there were only three common bands across all cultivars and bulk soils. Moreover, the responsive communities were similar across the cultivars they were derived from, and hence, no significant cultivar effect was observed.
Table 1.
ANOSIM analysis for soil and cultivar effect (Primer 6)
| Test for difference between soil (across all cultivars and bulk soil) | Test for difference between cultivars (across all soils) | ||||||
|---|---|---|---|---|---|---|---|
| Type | Compounds | Global R | Significance | Type | Compounds | Global R | Significance |
| Carbon | d-Alanine | 0.258 | 0.259 | Carbon | d-Alanine | 0 | 0.835 |
| Carbon | Glycyl-l-proline | 0.667 | 0.05 | Carbon | Glycyl-l-proline | 0.512 | 0.003 |
| Carbon | l-Proline | 0.408 | 0.037 | Carbon | l-Proline | 0.735 | 0.0004 |
| Carbon | d-Ribono-1,4-lactone | 0.333 | 0.037 | Carbon | d-Ribono-1,4-lactone | 0 | 0.5 |
| Carbon | N-Acetyl-d-glucosamine | 0.917 | 0.037 | Carbon | N-Acetyl-d-glucosamine | 0.445 | 0.006 |
| Carbon | Amino butyric acid | 0.803 | 0.037 | Carbon | Amino butyric acid | 0.332 | 0.056 |
| Carbon | l-Malic acid | 0.333 | 0.222 | Carbon | l-Malic acid | 0.32 | 0.011 |
| Carbon | Acetoacetic acid | 0.667 | 0.074 | Carbon | Acetoacetic acid | 0.439 | 0.002 |
| Carbon | l-Aspartic acid | 1 | 0.037 | Carbon | l-Aspartic acid | 0.584 | 0.002 |
| Phosphorus | Phosphate | 0.583 | 0.054 | Phosphorus | Phosphate | 0.494 | 0.003 |
| Sulfur | Sulfate | 0.75 | 0.037 | Sulfur | Sulfate | 0.044 | 0.414 |
| Sulfur | p-Amino benzene sulfonic acid | 0.667 | 0.037 | Sulfur | p-Amino benzene sulfonic acid | 0.603 | 0.0002 |
| Sulfur | Benzene sulfonic acid | 0.668 | 0.037 | Sulfur | Benzene sulfonic acid | 0.316 | 0.0031 |
| Sulfur | 2-Hydroxyethane sulfonic acid | 0.833 | 0.037 | Sulfur | 2-Hydroxyethane sulfonic acid | 0.614 | 0.0002 |
| Sulfur | Taurine | 0.833 | 0.037 | Sulfur | Taurine | 0.683 | 0.0004 |
| Sulfur | Methane sulfonic acid | 0.5 | 0.037 | Sulfur | Methane sulfonic acid | 0.36 | 0.007 |
| Sulfur | l-Cysteinyl-glycine | 0.667 | 0.074 | Sulfur | L-Cysteinyl-glycine | 0.382 | 0.02 |
Global R value is between −1 and 1. R = 0 indicating completely random grouping, while R = 1 indicates that communities within a soil are more similar to each other than to any samples from other soil. A significant global R indicates that there are differences between the soil types somewhere in the analysis
Significant effects of (bulk) soil type on the community structures that had established in wells with the six different sulfur sources (Fig. 6c, shown is benzene sulfonic acid) were found. In addition, effects of rhizosphere derivation were seen for the communities obtained from all B soil grown cultivars (with benzene sulfonic acid, taurine, and p-amino benzene sulfonic acid) and from cultivars A, Av, and K (with sulfate, l-cysteinyl-glycine, and 2-hydroxyethane sulfonic acid).
On another notice, the patterns from aminobenzene sulfonic acid and 2-hydroxyethane sulfonic acid had five common bands across all cultivars and bulk soils, whereas three bands were common in the patterns from sulfate, methane sulfonic acid, and l-cysteinyl glycine per compound. There were four common bands in the community analysis of benzene sulfonic acid across all samples. Hence, it can be assumed that, across our samples, a limited number of similar bacterial types responded to these diverse sources of S.
Discussion
The aim of the current study was to investigate the effects of different potato cultivars growing in two different soil types on the consumption of different carbon, sulfur, and phosphorus sources by root-associated and bulk soil bacterial communities. We used functional analyses generated with the novel medium-throughput Biolog phenotype arrays and 16S rRNA gene profiles generated by PCR-DGGE, both directly and indirectly (from selected Biolog wells). This combined approach was chosen because functionality in the rhizosphere is thought to depend on (1) the microbial communities that are locally present in the soil, (2) the spectrum of potential activity of this local microbiota, and (3) the selective effect that plant roots exert on these local communities and functionalities.
The main finding in this study, which confirms previous studies, was that soil type exerts the most profound influence on the structural and functional bacterial communities in the potato rhizosphere. First, the communities, when analyzed by direct PCR-DGGE, clearly differed between the two soils. Second, the communities in the rhizospheres of the very same cultivars were often also quite divergent between the two soil types. Besides, tendencies for consumption of carbon sources showed differences based on soil type. In previous studies, it was also shown that soil type is a key factor that determines the bacterial community composition in the rhizosphere of plants grown across soils [1, 13]. In the current study, evidence supporting this contention came from the fact that the bacterial communities from the same cultivar types grouped per soil, instead of per genotype, in the direct PCR-DGGE-based analyses. However, the analyses also revealed effects of the rhizosphere, which were strongest in the B soil. Thus, the roots of the same potato cultivars, when grown in different soils, modulate the local communities to a certain extent, but do not establish similar communities. Importantly, the same cultivars, when grown in the two different soils, were also shown to yield different microbial communities involved in the consumption of particular carbon, sulfur, and phosphorus sources, as shown in the RDA analyses (Figs. 3 and 4). Considering the rhizosphere effects, this indicated that the same cultivars, upon growth in the two different soils, had attracted rather different communities with different functional capabilities.
Strikingly, the selective effects of the rhizosphere were more clearly shown in the B soil than in the V soil. This might be explained by the higher organic matter content of the latter soil compared to the former, which was thought to diminish the effects of growing roots in it. In a previous study [19], it was also shown that, due to its higher nutritional status, the rhizospheric bacterial community structure in the V soil is affected to a lower extent by root exudates than that in the B soil. In support of this contention, in a previous study, the rhizosphere effect along wheat roots was also shown to be positive and strong in soil low in organic matter but not in high organic matter soil [28].
The communities from the B and V rhizosphere soils either consumed different C sources than those from the bulk soils or the utilization rates were simply higher than those in the latter. One should bear in mind that, in the current study, potential in situ utilization was measured, which may only remotely reflect the processes that play a role in situ. For instance, the carbon sources l-proline, N-acetyl-d-glucosamine, and d-alanine, which were utilized to the highest degree by communities from the rhizosphere soils (as well as from the bulk soils), may have been present as a result of exudation or degradation of organic matter.
Soils represent grossly oligotrophic (carbon-limited) habitats [20], whereas the rhizospheres of plants may, to some extent, alleviate carbon starvation stress. Incubating soil cell extracts with high concentrations of readily decomposable organic substrates, like done in the phenotype array, is likely to favor copiotrophic microorganisms that are able to grow explosively (r-strategists), outcompeting oligotrophic ones (K-strategists)[1]. The PCR-DGGE community analyses performed on selected substrates of the phenotype array thus reported on such opportunistic communities, as previously outlined by Smalla and co-workers [30]. Notwithstanding this caveat of the method, our results revealed the rhizosphere soil communities to cluster apart from the corresponding bulk soil ones, indicating that particular bacterial responders to the Biolog phenotype array substrates had been selected by the rhizospheres. Whether this was due to the particular phenotype array compound being present in root exudates is an open question. However, some of the compounds analyzed may be quite important in plant–bacterium interactions [31]. For instance, l-malic acid is secreted by the roots of varied plants and apparently provides an effective nutritive and/or signaling molecule that modulates the establishment of beneficial rhizobacteria. A regulatory role of such root-secreted metabolites may thus be indicated [26]. Moreover, l-malic acid as well as other organic acids may affect the availability of phosphorus [5]. Interestingly, our RDA analysis revealed, for cultivars A and D (in soil V), similar tendencies towards l-malic acid and phosphate, indicating a link between malic acid and phosphate solubilization. These two cultivars might, for instance, secrete more l-malic acid into soil V in comparison to other cultivars, allowing higher phosphate uptake. This might also hint at the available phosphate levels in the two different soils. Thus, l-malic acid, next to compounds such as l-aspartic acid, γ-amino butyric acid, and glycyl-l-proline, might indeed have served as a specific modulator of the root-associated bacterial communities. This was possibly specific per cultivar type, presumably in accordance with root physiology as reflected in, for instance, exudation patterns [1, 8, 29]. The differential availability of such C sources in the rhizospheres of the various potato cultivars may have influenced the proliferation of particular communities of microorganisms. For instance, there were two major cultivar groups, i.e., (1) cultivars A and Av and (2) cultivar D, the only high-starch-tuber cultivar, grouping together with cultivars K and M. The phenotype array-based functional analyses revealed that, in terms of substrates used and bacterial PCR-DGGE patterns in selected wells, the genetically modified cultivar M—derivative from cultivar K—fitted the data obtained with the range of cultivars tested. Thus, cultivar M, compared to the other cultivars (including K), did not exert any specific effect on the utilization patterns of carbon, phosphorus, and sulfur sources by its associated microflora. This finding supports the conclusions from a previous study, which analyzed the abundances and community structures of microbial communities in a different field set-up with potato [19]. However, in another study, cluster analysis of ERIC-PCR fingerprints performed with strains obtained from different alfalfa genotypes revealed differences in the types of bacteria enriched in the different rhizospheres [7]. This indicates differences at the level of cultured bacteria, reflecting an effect of the type of modification on the respective communities.
In contrast to the “cultivar-specific” compounds (l-malic acid, l-aspartic acid, γ-amino butyric acid, and glycyl-l-proline), the compounds d-alanine, N-acetyl-d-glucosamine, acetoacetic acid, phosphate, and hydroxyethane sulfonic acid were apparently cultivar-aspecific, as they showed the same tendencies across cultivars with respect to the communities they selected. These compounds might represent resources that are generally available in potato root exudates to the bacterial consumers.
Overall, our results showed that the roots of five potato cultivars were colonized by bacterial populations that differed between the two soils used. Moreover, in each soil, the communities were different between rhizospheres and corresponding bulk soil, and this effect was strongest in the B soil. Modulation of the rhizosphere colonization patterns presumably occurred through the assimilation, by the local microflora, of different root-secreted compounds, which is known to be governed by the physiologies of the respective roots in interaction with the surrounding soil. Next to the soil type and rhizosphere effects, the microbial communities at the different potato cultivars showed differences in the consumption of carbon, phosphorus, and sulfur sources, which indicated the existence of different functional roles in the communities of the different cultivars. However, no GM plant effect (cultivar M) was observed. Thus, a clear hierarchy in shaping the root-associated communities, i.e., soil > rhizosphere > cultivar, can be envisaged. Since soil ecosystems differ in their buffering capacity against perturbation as a function of biotic and abiotic factors, natural variations within a system owing to environmental effects must be considered [21]. Therefore, the local conditions must be taken into account when the effect of a cultivar needs to be assessed. Last but not least, root exudates normally contain diverse compounds in fluctuating concentrations in an integrated environment (instead of per well like in the phenotype array). Due to this complexity, it is still very difficult to pinpoint the drivers of the in situ microbial populations at the potato root surface.
Acknowledgment
This work was supported by an NWO-ERGO project awarded to JDvE. The set-up of potato fields were laid out by Avebe (Foxhol, NL). We acknowledge Eelco Hoogwout and Emilia Hannula for the help in sampling.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Buyer JS, Roberts DP, Russek-Cohen E. Soil and plant effects on microbial community structure. Can J Microbiol. 2002;48:955. doi: 10.1139/w02-095. [DOI] [PubMed] [Google Scholar]
- 2.Campbell CD, Grayston SJ, Hirst DJ. Use of rhizosphere carbon sources in sole carbon source tests to discriminate soil microbial communities. Journal Microbiol Meth. 1997;30:33. doi: 10.1016/S0167-7012(97)00041-9. [DOI] [Google Scholar]
- 3.Cavigelli M, Robertson G, Klug M. Fatty acid methyl ester (FAME) profiles as measures of soil microbial community structure. Plant and Soil. 1995;170:99. doi: 10.1007/BF02183058. [DOI] [Google Scholar]
- 4.Costa R, Götz M, Mrotzek N, Lottmann K, Berg G, Smalla K. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guild. FEMS Mic Eco. 2006;56:236. doi: 10.1111/j.1574-6941.2005.00026.x. [DOI] [PubMed] [Google Scholar]
- 5.Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant and Soil. 2002;245:35. doi: 10.1023/A:1020809400075. [DOI] [Google Scholar]
- 6.de Vetten N, Wolters A, Raemakers K, van der Meer I, ter Stege R, Heeres E, Heeres P, Visser R. A transformation method for obtaining marker-free plants of a cross-pollinating and vegatatively propagated crop. Nature Biotech. 2003;21:439. doi: 10.1038/nbt801. [DOI] [PubMed] [Google Scholar]
- 7.Di Giovanni GD, Watrud LS, Seidler RJ, Widmer F. Comparison of parental and transgenic alfalfa rhizosphere bacterial communities using Biolog GN metabolic fingerprinting and enterobacterial repetitive intergenic consensus sequence-PCR (ERIC-PCR) Microb Ecol. 1999;37:129. doi: 10.1007/s002489900137. [DOI] [PubMed] [Google Scholar]
- 8.Gagliardi JV, Buyer JS, Angle JS, Russek-Cohen E. Structural and functional analysis of whole-soil microbial communities for risk and efficacy testing following microbial inoculation of wheat roots in diverse soils. Soil Biol and Biochem. 2001;33:25. doi: 10.1016/S0038-0717(00)00110-3. [DOI] [Google Scholar]
- 9.Garbeva P, Postma J, van Veen JA, van Elsas JD. Effect of above ground plant species on soil microbial community structure and its impact on supply of Rhizoctonia solani AG3. Env Microb. 2006;8:233. doi: 10.1111/j.1462-2920.2005.00888.x. [DOI] [PubMed] [Google Scholar]
- 10.Garbeva P, van Veen JA, van Elsas JD (2004) Microbial diversity in soil: selection of microbial population by plant and soil type and implications for disease suppressiveness. Ann Rev Phy 42:243–270 [DOI] [PubMed]
- 11.Garland JL. Patterns of potential C source utilization by rhizosphere communities. Soil Biol and Biochem. 1996;28:223. doi: 10.1016/0038-0717(95)00113-1. [DOI] [Google Scholar]
- 12.Gelsomino A, Keijzer-Wolters A, Cacco G, van Elsas JD. Assessment of bacterial community structure in soil by PCR and DGGE. J Mic Met. 1999;38:1. doi: 10.1016/S0167-7012(99)00054-8. [DOI] [PubMed] [Google Scholar]
- 13.Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl Environ Microbiol. 2003;69:1800. doi: 10.1128/AEM.69.3.1800-1809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grayston SJ, Campbell CD, Bardgett RD, Mawdsley JL, Clegg CD, Ritz K, Griffiths BS, Rodwell JS, Edwards SJ, Davies WJ, Elston DJ, Millard P. Assessing shifts in microbial community structure across a range of grasslands of differing management intensity using CLPP, PLFA and community DNA techniques. Appl Soil Ecol. 2004;25:63. doi: 10.1016/S0929-1393(03)00098-2. [DOI] [Google Scholar]
- 15.Grayston SJ, Wang S, Campbell CD, Edwards AC. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol and Biochem. 1998;30:369. doi: 10.1016/S0038-0717(97)00124-7. [DOI] [Google Scholar]
- 16.Hack H, Gall H, Klemke T, Klose R, Meier U, Stauss R, Witzenberger A. Phänologische entwicklungsstadien der Kartoffel (Solanum tuberosum L.) Nachrichtenbl Deut Pflanzenschutzd. 1993;45:11. [Google Scholar]
- 17.Herron P, Wellington E. New method for extraction of streptomycete spores from soil and application to the study of lysogeny in sterile amended and nonsterile soil. App Env Micr. 1990;56:1406. doi: 10.1128/aem.56.5.1406-1412.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inceoglu Ö, Hoogwout EF, Hill P, Van Elsas JD. Effect of DNA extraction method on the apparent microbial diversity of soil. App Env Micr. 2010;76:3378–3382. doi: 10.1128/AEM.02715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inceoglu Ö, Salles JF, Van Overbeek L, Van Elsas JD. Effect of plant genotype and growth stage on the β-proteobacterial community associated with different potato cultivars in two fields. AEM. 2010;76:3675–3684. doi: 10.1128/AEM.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 21.Lupwayi N, Rice W, Clayton G. Soil microbial diversity and community structure under wheat as influenced by tillage and crop rotation. Soil Biol Biochem. 1998;30:1733. doi: 10.1016/S0038-0717(98)00025-X. [DOI] [Google Scholar]
- 22.Lynch J, Whipps J. Substrate flows in the rhizosphere. Plant Soil. 1990;129:1. doi: 10.1007/BF00011685. [DOI] [Google Scholar]
- 23.Marschner P, Yang C, Lieberei R, Crowley D. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem. 2001;33:1437. doi: 10.1016/S0038-0717(01)00052-9. [DOI] [Google Scholar]
- 24.Rasche F, Hödl V, Poll C, Kandeler E, Gerzabek M, van Elsas JD, Sessitsch A (2006) Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effect of soil, wild type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol Ecol 56:219 [DOI] [PubMed]
- 25.Ros M, Goberna M, Pascual JA, Klammer S, Insam H. 16S rDNA analysis reveals low microbial diversity in community level physiological profile assays. J Microbiol Meth. 2008;72:221. doi: 10.1016/j.mimet.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Rudrappa T, Czymmek KJ, Pare PW, Bais HP (2008) Root-Secreted Malic Acid Recruits Beneficial Soil Bacteria. Plant Physiol 148:1547 [DOI] [PMC free article] [PubMed]
- 27.Salles JF, van Elsas JD, van Veen J. Effect of agricultural management regime on Burkholderia community structure in soil. Mic Eco. 2006;52:267. doi: 10.1007/s00248-006-9048-6. [DOI] [PubMed] [Google Scholar]
- 28.Semenov AM, van Bruggen AHC, Zelenev VV. Moving waves of bacterial populations and total organic carbon along roots of wheat. Microb Ecol. 1999;37:116. doi: 10.1007/s002489900136. [DOI] [PubMed] [Google Scholar]
- 29.Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. App Env Micr. 2001;87:4742. doi: 10.1128/AEM.67.10.4742-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smalla K, Wachtendorf U, Heuer H, Liu W, Forney L. Analysis of Biolog GN substrate utilization patterns by microbial communities. App Env Micr. 1998;64:1220. doi: 10.1128/aem.64.4.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vahjen W, Munch J, Tebbe CC. Carbon source utilization of soil extracted microorganisms as a tool to detect the effects of soil supplemented with genetically engineered and non-engineered Corynebacterium glutamicum and a recombinant peptide at the community level. FEMS Microbiol Ecol. 1995;18:317. doi: 10.1111/j.1574-6941.1995.tb00188.x. [DOI] [Google Scholar]