In a prospective study, human immunodeficiency virus (HIV)–infected patients with pneumococcal pneumonia had nasopharyngeal colonization densities 5 log10 higher than those in concurrently identified HIV-infected asymptomatic controls, as measured by real-time polymerase chain reaction (rtPCR). A nasopharyngeal lytA density of ≥8000 copies/mL at rtPCR may be a useful diagnostic marker for pneumococcal pneumonia.
Abstract
Background. There is major need for a more sensitive assay for the diagnosis of pneumococcal community-acquired pneumonia (CAP). We hypothesized that pneumococcal nasopharyngeal (NP) proliferation may lead to microaspiration followed by pneumonia. We therefore tested a quantitative lytA real-time polymerase chain reaction (rtPCR) on NP swab samples from patients with pneumonia and controls.
Methods. In the absence of a sensitive reference standard, a composite diagnostic standard for pneumococcal pneumonia was considered positive in South African human immunodeficiency virus (HIV)–infected adults hospitalized with radiographically confirmed CAP, if blood culture, induced good-quality sputum culture, Gram stain, or urinary Binax demonstrated pneumococci. Results of quantitative lytA rtPCR in NP swab samples were compared with quantitative colony counts in patients with CAP and 300 HIV-infected asymptomatic controls.
Results. Pneumococci were the leading pathogen identified in 76 of 280 patients with CAP (27.1%) using the composite diagnostic standard. NP colonization density measured by lytA rtPCR correlated with quantitative cultures (r = 0.67; P < .001). The mean lytA rtPCR copy number in patients with pneumococcal pneumonia was 6.0 log10 copies/mL, compared with patients with CAP outside the composite standard (2.7 log10 copies/mL; P < .001) and asymptomatic controls (0.8 log10 copies/mL; P < .001). A lytA rtPCR density ≥8000 copies/mL had a sensitivity of 82.2% and a specificity of 92.0% for distinguishing pneumococcal CAP from asymptomatic colonization. The proportion of CAP cases attributable to pneumococcus increased from 27.1% to 52.5% using that cutoff.
Conclusions. A rapid molecular assay of NP pneumococcal density performed on an easily available specimen may significantly increase pneumococcal pneumonia diagnoses in adults.
Pneumococcal disease remains a major contributor to morbidity and mortality, particularly in those with a high prevalence of human immunodeficiency virus (HIV) infection [1]. Current diagnostic standards for bacterial pneumonia are limited by a lack of emphasis on appropriate specimen collection [2], and reliance on assays with poor sensitivity and specificity [3]. Therefore, the role of Streptococcus pneumoniae in community-acquired pneumonia (CAP) is underestimated, even though S. pneumoniae is reported as the most frequently identified pathogen in both HIV-infected and HIV-uninfected adults in developed and developing countries [1, 4–8]. Easily obtainable specimens and assays with increased sensitivity to identify pneumococcal CAP could improve management and strategies for better prevention of pneumococcal CAP. Nasopharyngeal (NP) swab samples are appealing specimens compared with sputum specimens, which is frequently of poor quality and which are affected by specificity problems [9]. Quantitative real-time polymerase chain reaction (rtPCR) is an attractive tool to diagnose pneumococcal disease [3], particularly with more specific gene targets, such as the main pneumococcal autolysin lytA [10, 11]. Based on the growing appreciation of the association between pneumococcal load and clinical disease, there has been recent interest in quantitative pneumococcal assays [12–14]. This study explored the predictive value of density of pneumococcal NP colonization in relation to diagnosis of pneumococcal pneumonia in HIV-infected South African adults.
METHODS
Adults (≥18 years old) admitted to Chris Hani Baragwanath Hospital in Soweto, South Africa, with acute pneumonia (symptoms for <14 days) were enrolled from December 2005 until September 2007. Pneumonia was defined as the presence of ≥2 of the following: cough, dyspnea, pleuritic chest pain, or fever (>38.0°) plus crackles or bronchial breathing at auscultation. Patients were excluded if they had a previous diagnosis of tuberculosis and were on antituberculous treatment, or if only antituberculous treatment was initiated on admission. Radiologically confirmed pneumonia (referred to as CAP) was defined as any new infiltrate in association with a compatible clinical syndrome. The main investigation group for this analysis was HIV-infected patients with CAP. As controls, we simultaneously enrolled a convenience sample of HIV-infected adults from March 2006 until October 2007 attending the Chris Hani Baragwanath Hospital outpatient HIV clinic who were afebrile, asymptomatic for respiratory illness, and had no active tuberculosis.
In eligible patients with pneumonia, we obtained 10 mL of blood for aerobic blood cultures and induced sputum using hypertonic (5%) saline within 12 hours of admission. Blood and sputum were processed using standard microbiological culture and staining techniques (BacTAlert for blood cultures; BioMérieux). A pneumococcal latex agglutination test (Wellcogen Bacterial Antigen Kit; Wellcogen) and a nested PCR assay for pneumococcus [15], were performed if growth in blood cultures was detected but failed to identify an organism.
In both patients with pneumonia and controls, NP swab samples were collected from a single nostril with dacron swabs (Medical Wire and Equipment), placed in 1 mL of skim milk, tryptone, glucose, and glycerin (STGG) medium and stored at 4°C for ≤12 hours before processing [16]. Quantitative counts of pneumococci were obtained by plating out serial dilutions of STGG medium onto blood agar plates with 5 mg/mL gentamicin. S. pneumoniae was identified based on colony morphology, optochin susceptibility, and bile solubility.
An immunochromatographic membrane test (ICT) (Binax Now Streptococcus pneumoniae; Binax) was performed on unconcentrated urine specimens from patients with pneumonia and controls, according to the manufacturer’s recommendations. Urine was tested for the presence of antimicrobial substances by using a pansensitive Bacillus subtilis strain. Duplex quantitative rtPCR targeting ply and lytA genes on NP swab samples was performed by blinded study personnel (supplemental material). PCR results were considered positive only if the lytA gene was amplified (cycle threshold [CT], <45) [10].
Statistical Analysis
For patients with CAP, a composite diagnostic standard for pneumococcal pneumonia was defined as positive if pneumococci were demonstrated by blood culture, urinary ICT, or good-quality (>25 neutrophils and <10 epithelial cells per high-power field) sputum Gram stain or culture [17]. Bacterial colony counts were log-transformed. For mathematical reasons, we assigned a value of 1 colony-forming unit (CFU)/mL to patients in whom pneumococcus was not identified in NP cultures or with lytA rtPCR. Correlations between colony counts identified by culture and lytA rtPCR were performed using Pearson’s correlation coefficient. Optimal cutoff values for quantitative NP colonization cultures and quantitative lytA rtPCR from NP swab samples were identified by calculating receiver operating characteristic (ROC) curves to distinguish pneumococcal CAP from colonization. Continuous variables were compared with 2-sided pooled t tests or the Wilcoxon rank sum test, and proportions with the χ2 test or Fisher’s exact test, as appropriate. Kappa statistics were applied to calculate the agreement between reported antibiotic use and measured antimicrobial activity in urine specimens. The Cochran-Armitage χ2 test was calculated for trend. Differences were considered statistically significant at P ≤ .05 (2-sided P values). Analyses were performed with SAS software (version 9.1; SAS Institute) and Epi Info 2002 software (Centers for Disease Control and Prevention). The study was approved by the ethics committees of the University of the Witwatersrand and Emory University. All patients and controls provided written informed consent.
RESULTS
Demographic Characteristics
Of 514 patients with clinical pneumonia, 92.4% had a chest radiograph available on admission. Of these 475, 370 (77.9%) had radiologically confirmed CAP. Of patients with CAP, 50 (13.5%) refused HIV testing, 40 (10.8%) were HIV uninfected, and 280 (75.7%) were HIV infected (Table 1). No patient or control had received a pneumococcal (polysaccharide or conjugate) vaccine.
Table 1.
Demographics of Patients with Community-Acquired Pneumonia and HIV-Infected Asymptomatic Controls
| Patients With Radiologically Confirmed Pneumonia |
|||||||||
| Characteristic | HIV-Infected (n = 280; 75.7%) | HIV-Uninfected (n = 40; 10.8%) | HIV Status Unknown (n = 50; 13.5%) | Total (n = 370) | Pa | Pb | HIV-Infected Controls (n = 300) | Pc | Pd |
| Age, mean ± SD, years | 36.5 ± 9.7 | 41.9 ± 15.1 | 46.1 ± 17.9 | 38.4 ± 12.2 | .035 | <.001 | 38.7 ± 8.1 | .04 | .7 |
| Female sex | 176/280 (62.9) | 19/40 (47.5) | 20/50 (40.0) | 215/370 (58.1) | .06 | .002 | 249/300 (83.3) | <.001 | <.001 |
| Smoker (current) | 36/276 (11.4) | 13/40 (4.1) | 10/49 (20.1) | 59/365 (16.2) | .004 | .19 | 23/298 (7.7) | .04 | <.001 |
| New HIV diagnosis | 134/280 (47.9) | NA | NA | 134/370 (36.2) | … | … | … | … | … |
| Receiving HAART at admission | 27/280(9.6) | NA | NA | … | … | … | 286/300 (95.3) | <.001 | <.001 |
| CD4 cell count, mean ± SD, cells/mm3 | 132.5 ± 153.8 | NA | NA | … | … | … | 320 ± 187.2 | <.001 | … |
| Receiving cotrimoxazole at admission | 30/280 (10.7) | 0 | 0 | 30/320 (9.4) | … | … | 145/300 (48.3) | <.001 | <.001 |
| Measured urinary antimicrobial activity | 136/251 (54.2) | 17/38 (44.7) | 21/45 (46.7) | 174/334 (52.1) | .29 | .35 | 144/299 (48.2) | .16 | .32 |
| CURB-65 | |||||||||
| 0 | 69/275 (25.1) | 12/40 (30.0) | 10/48 (20.8) | 91/363 (25.1) | .51 | .53 | NA | … | … |
| 1 | 109/275 (39.6) | 16/40 (40.0) | 18/48 (37.5) | 143/363 (39.4) | .97 | .78 | NA | … | … |
| 2 | 70/275 (25.5) | 8/40 (20.0) | 11/48 (22.9) | 89/363 (24.5) | .46 | .71 | NA | … | … |
| 3 | 24/275 (8.7) | 3/40 (7.5) | 8/48 (16.7) | 35/363 (9.6) | .99 | .11 | NA | … | … |
| 4 | 3/275 (1.1) | 1/40 (2.5) | 1/48 (2.1) | 5/363 (1.4) | .42 | .48 | NA | … | … |
| Hospital death | 36/271 (13.3) | 6/39 (15.4) | 10/47 (21.3) | 52/357 (14.6) | .76 | .15 | NA | … | … |
Unless otherwise specified, data represent No. (%) of patients. Among patients with unknown human immunodeficiency virus (HIV) serostatus, voluntary informed counseling and testing for HIV was performed using the HIV enzyme-linked immunosorbent assay (ELISA) test (Axsym system; HIV1/2 Abbott); reactive tests were confirmed by a second ELISA (ElexSys 2010; Roche).
Abbreviations: CURB-65, confusion, urea, respiratory rate, blood pressure, age ≥65 years [18] (the number reflects the sum of criteria present); HAART, highly active antiretroviral therapy; HIV, human immunodeficiency virus; NA, not applicable; SD, standard deviation.
P value for comparison between HIV-infected and HIV-uninfected patients with pneumonia.
P value for comparison between HIV-infected patients with pneumonia and patients with pneumonia of unknown HIV serostatus;
P value for comparison between HIV-infected patients with pneumonia and HIV-infected asymptomatic controls.
P value for comparison between all patients with pneumonia and HIV-infected asymptomatic controls.
Antimicrobial activity in admission urine specimens was detected in 54.2% of HIV-infected patients with CAP; there was a poor correlation (κ = 0.07) with patients’ reports of antibiotic use, because only 16.2% patients with urinary antimicrobial activity reported taking an antibiotic in the previous 48 hours. For further analysis, we thus used only measured antimicrobial activity as the indicator of recent antibiotic use.
Compared with HIV-infected adults with radiologically confirmed CAP, the asymptomatic HIV-infected control subjects (n = 300) had a higher proportion of females, higher CD4 cell counts, and were more likely to be receiving highly active antiretroviral therapy (HAART) and cotrimoxazole prophylaxis (Table 1). There was no difference in urinary antimicrobial activity between case patients and controls.
Diagnostic Utility of Different Pneumococcal Assays
S. pneumoniae was identified in 27.1% of patients with CAP based on the composite diagnostic standard (15.4% based on the composite diagnostic standard without urinary ICT). The performance of individual assays is summarized in Table 2 and Supplementary Table 1. Urinary ICT results were positive in 66.7% of patients with bacteremic and in 72.7% of patients with nonbacteremic pneumococcal CAP (P = .62) and in 0.3% asymptomatic controls. In the subgroup of 48 patients with CAP and good-quality sputum specimens, pneumococcus was identified in 62.5%, compared with 46 of 232 (19.8%) without good-quality sputum specimens (P < .001).
Table 2.
Diagnostic Yield of Colonization Density for Streptococcus pneumoniae in HIV-Infected Patients With Community-Acquired Pneumonia
| Diagnostic Standard | Pneumococcal Etiology Identified Without Assaya | Pneumococcal Etiology Identified With Assayb | Additional Yield (1 – Specificity) From Assayc | Sensitivityd | PPV |
| Composite diagnostic standard: blood culture, good-quality sputum culture, Gram stain, and urine ICT | |||||
| Density at NP swab culture, ≥15 000 CFU/mL | 75/274 (27.4) | 117/274 (42.7) | 42/199 (21.1) | 54/75 (72.0) | 54/96 (56.3) |
| Previous antimicrobial therapy | 32/135 (23.7) | 49/135 (36.3) | 17/103 (16.5) | 22/32 (68.8) | 22/39 (56.4) |
| No previous antimicrobial therapy | 39/111 (35.1) | 59/111 (53.1) | 20/72 (27.8) | 28/39 (71.8) | 28/48 (58.3) |
| Density at NP lytA rtPCR, ≥8000 copies/mL | 73/265 (27.5) | 139/265 (52.5) | 66/192 (34.4) | 60/73 (82.2) | 60/126 (47.6) |
| Previous antimicrobial therapy | 30/130 (23.1) | 64/130 (49.2) | 34/100 (34.0) | 25/30 (83.3) | 25/59 (42.4) |
| No previous antimicrobial therapy | 39/107 (36.4) | 62/107 (57.9) | 23/68 (33.8) | 32/39 (82.1) | 32/55 (58.2) |
| Composite diagnostic standard: blood culture, good-quality sputum culture, and Gram stain | |||||
| Density at NP swab culture, ≥15 000 CFU/mL | 42/274 (15.3) | 111/274 (40.5) | 69/232 (29.7) | 27/42 (64.3) | 27/96 (28.1) |
| Previous antimicrobial therapy | 15/135 (11.1) | 45/135 (33.3) | 30/120 (25.0) | 9/15 (60.0) | 9/39 (23.1) |
| No previous antimicrobial therapy | 23/111 (20.7) | 57/111 (51.4) | 34/88 (38.6) | 14/23 (60.9) | 14/48 (29.2) |
| Density at NP lytA rtPCR, ≥8000 copies/mL | 42/265 (15.8) | 135/265 (50.9) | 93/223 (41.7) | 33/42 (78.6) | 33/126 (26.2) |
| Previous antimicrobial therapy | 14/130 (10.8) | 63/130 (48.5) | 49/116 (42.2) | 10/14 (71.4) | 10/59 (16.9) |
| No previous antimicrobial therapy | 24/107 (22.4) | 59/107 (55.1) | 35/83 (42.2) | 20/24 (83.3) | 20/55 (36.4) |
Data represent No. (%) of patients.
Abbreviations: CFU, colony-forming units; ICT, immunochromatographic membrane test; NP, nasopharyngeal; PPV, positive predictive value; rtPCR, real-time polymerase chain reaction.
Pneumococcal etiology according to composite diagnostic standard.
Pneumococcal etiology according to composite diagnostic standard or assay in question.
Number of patients in whom the assay in question was the only test with positive results and no assay of the composite diagnostic standard had positive results.
In reference to composite diagnostic standard.
Quantitative Colonization Density
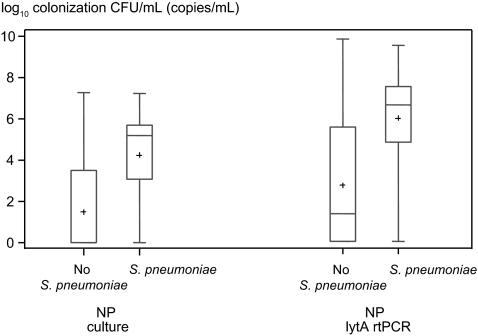
Patients with CAP were more frequently colonized with pneumococcus than controls (Table 3). The density of pneumococcal colonization was higher in HIV-infected patients with CAP attributed to pneumococcus (n = 76) based on the composite diagnostic standard than in those in whom pneumococcus was not identified (n = 204) on NP swab samples (4.2 vs 1.5 log10 CFU/mL; P < .001) (Figure 1, Supplementary Figure 1). Among HIV-infected patients with CAP, the proportion categorized as pneumococcal based on the composite diagnostic standard was higher with increasing NP colonization densities (P < .001).
Table 3.
Pneumococcal Nasopharyngeal Carriage in HIV-Infected Patients With Community-Acquired Pneumonia and Asymptomatic Controls
| Asymptomatic Controls | Patients With CAP | P | |
| NP carriage of Streptococcus pneumoniae, No. (%) of subjects | |||
| Culture | 35/298 (11.7) | 125/278 (44.9) | <.001 |
| lytA rtPCR | 57/288 (19.8) | 167/266 (62.8) | <.001 |
| Quantitative lytA rtPCR results, mean ± SD, log10 copies/mL | |||
| Previous antimicrobial therapy | 0.8 ± 1.7 | 5.9 ± 2.3 | <.001 |
| No previous antimicrobial therapy | 0.8 ± 1.8 | 6.0 ± 2.3 | <.001 |
| Current HAART | 0.8 ± 1.7 | 6.4 ± 4.3 | <.001 |
| No current HAART | 1.1 ± 2.2 | 5.9 ± 2.3 | <.001 |
| Current smoking | 0.8 ± 1.6 | 5.1 ± 2.6 | <.001 |
| No current smoking | 0.8 ± 1.8 | 6.2 ± 2.2 | <.001 |
| CD4 cell count, cells/mm3 | |||
| ≤200 | 0.6 ± 1.7 | 5.8 ± 2.5 | <.001 |
| >200 to ≤500 | 1.0 ± 1.8 | 6.6 ± 1.6 | <.001 |
| >500 | 0.5 ± 1.5 | 4.0 ± 2.2 | <.001 |
Abbreviations: CAP, community-acquired pneumonia; HAART, highly active antiretroviral therapy; NP, nasopharyngeal; rtPCR, real-time polymerase chain reaction; SD, standard deviation.
Figure 1.
Quantitative colonization densities in human immunodeficiency virus–infected patients with community-acquired pneumonia. Pneumococcal colonization densities are shown as colony counts, obtained with nasopharyngeal (NP) swab cultures and lytA real-time polymerase chain reaction (rtPCR) of NP swab samples. Counts are depicted after logarithmic transformation for those who had Streptococcus pneumoniae identified by the composite diagnostic standard and those who did not. Plus signs represent means; lengths of boxes, interquartile ranges between 25th and 75th percentiles; horizontal lines in boxes, medians; and whiskers, minimum and maximum values (t test, P < .001 for NP counts from cultures and for lytA rtPCR; Wilcoxon rank sum test [used after assignment of value 1 to the colony count for those who were not colonized], P < .001 for NP counts and lytA rtPCR). Abbreviations: CFU, colony-forming units; NP, nasopharyngeal; rtPCR, real-time polymerase chain reaction.
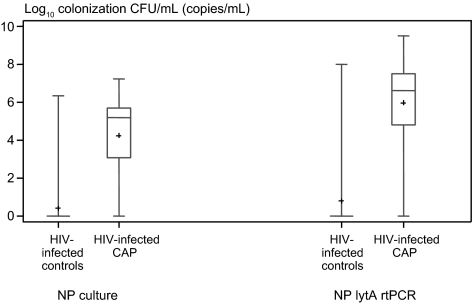
NP colonization densities were also higher in patients with pneumococcal CAP than asymptomatic controls (4.2 vs 0.4 log10 CFU/mL; P < .001) (Figure 2, Supplementary Figure 2). ROC curves showed very good diagnostic accuracy (area under the curve [AUC], 0.85) to distinguish pneumococcal CAP from colonization in HIV-infected persons. A model consisting of NP colonization density, current HAART, and CD4 cell count accurately distinguished between pneumococcal CAP from colonization in HIV-infected persons (AUC, 0.98). The optimal cutoff for colonization density to distinguish between asymptomatic colonization and pneumococcal CAP in HIV-infected persons was 15 000 CFU/mL (sensitivity, 72.0%; specificity, 95.6%) (Supplementary Table 2). In a separate analysis limited to those patients with CAP and controls with pneumococcal colonization, the 15 000 CFU/mL cutoff resulted in higher sensitivity (88.5%) but lower specificity (59.4%). If this cutoff was assumed to represent true-positive cases and was used to diagnose pneumococcal etiology, 42.7% of CAP cases in HIV-infected patients would be attributed to pneumococcus (Supplementary Table 3).
Figure 2.
Nasopharyngeal (NP) colonization densities in human immunodeficiency virus (HIV)–infected patients with pneumococcal community-acquired pneumonia (CAP) and HIV-infected asymptomatic controls. Pneumococcal colonization densities were measured as colony counts, obtained with NP swab cultures and lytA real-time polymerase chain reaction (rtPCR) of NP swab samples. Pluses represent means; lengths of boxes, interquartile ranges between 25th and 75th percentiles; horizontal lines in boxes, medians; and whiskers, minimum and maximum values (t test, P < .001 for NP counts from cultures and for lytA rtPCR; Wilcoxon rank sum test [used after assignment of value 1 to colony count to those who were not colonized], P < .001 for NP counts and lytA rtPCR). Abbreviations: CAP, community-acquired pneumonia; CFU, colony-forming units; HIV, human immunodeficiency virus; NP, nasopharyngeal; rtPCR, real-time polymerase chain reaction.
Results of lytA rtPCR
In HIV-infected patients, lytA rtPCR was more frequently positive in pneumococcal (94.5%) than nonpneumococcal CAP based on the composite diagnostic standard (50.8%; P < .001). There was no difference in lytA PCR positivity in NP swab samples between bacteremic and nonbacteremic pneumococcal CAP (94.7% vs 94.4%; P = .99), and these rates were higher than in asymptomatic HIV-infected persons (19.8%; P < .001).
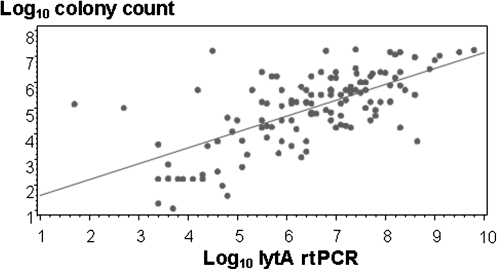
There was good correlation between quantitative culture and quantitative lytA rtPCR on NP swab samples among HIV-infected persons with either CAP or asymptomatic colonization (Figure 3). The quantitative lytA rtPCR detected higher mean copy numbers in pneumococcal (6.0 log10 copies/mL) than in nonpneumococcal (2.7 log10 copies/mL; P < .001) CAP based on the composite diagnostic standard (Figure 1), and higher than in asymptomatic controls (0.8 log10 copies/mL; P < .001). Previous antimicrobial therapy did not affect lytA rtPCR positivity or density (Tables 2 and 3). HIV-infected patients with pneumococcal CAP who died during hospitalization had a higher mean quantitative lytA rtPCR than did survivors (7.7 vs 6.1 log10 copies/mL; P = .02).
Figure 3.
Correlation between quantitative lytA real-time polymerase chain reaction and quantitative colony counts from nasopharyngeal swabs in human immunodeficiency virus–infected persons with community-acquired pneumonia and asymptomatic controls (Pearson’s correlation coefficient r = .67; P < .001). Abbreviation: rtPCR, real-time polymerase chain reaction.
ROC curves for lytA rtPCR showed excellent diagnostic accuracy (AUC, 0.93) to distinguish pneumococcal CAP from colonization in HIV-infected persons and good diagnostic accuracy to distinguish between pneumococcal and nonpneumococcal etiology in patients with CAP (AUC, 0.78). The optimal lytA rtPCR cutoff for NP colonization density to distinguish between asymptomatic colonization and pneumococcal CAP in HIV-infected persons was 8000 copies/mL (sensitivity, 82.2%; specificity, 92.0%) (Supplementary Table 4). Among those HIV-infected patients with CAP and controls with pneumococcal colonization, this cutoff resulted in a higher sensitivity (87.0%) but lower specificity (59.6%). If this cutoff was assumed to represent true-positive cases and therefore was used to diagnose pneumococcal etiology in CAP, 34.4% additional cases of pneumococcal etiology would be identified in HIV-infected patients (total pneumococcal CAP, 52.5%). Its performance was very similar if restricted to patients with CAP without a concomitant new diagnosis of tuberculosis (Supplementary Table 5).
In 14 of 40 (35.0%) HIV-uninfected patients with radiologically confirmed CAP, pneumococci were identified by the composite diagnostic standard. There was no difference in mean colonization densities between HIV-uninfected and HIV-infected patients (6.8 vs 6.3 log10 copies/mL; P = .46). The lytA rtPCR cutoff of 8000 copies/mL yielded 4 (16%) more patients (1 patient had no lytA rtPCR results, resulting in an overall pneumococcal etiology rate of 18/39 (46.2%) in HIV-uninfected patients with available lytA rtPCR results).
The lytA rtPCR copy numbers were significantly higher in patients with CAP than in controls when stratified for CD4 cell counts (Supplementary Figure 3), the presence or absence of urinary antimicrobial activity, smoking status, and current HAART (Table 3). The proportion of pathogenic bacteria identified among the composite pneumococcal group was similar to that among patients with only a positive lytA density assay (Supplementary Table 6).
DISCUSSION
This study provides evidence for a new diagnostic assay for S. pneumoniae as a cause of CAP. HIV-infected patients with pneumococcal CAP have lytA rtPCR copy numbers that are 3 log10 and 5 log10 higher than patients with nonpneumococcal CAP and asymptomatic controls, respectively. When a lytA rtPCR density cutoff of 8000 copies/mL was applied, 52.5% of patients with CAP were categorized as having “pneumococcal CAP,” compared with 27.1% for a composite standard of blood, urine, and sputum tests. The NP density assay identified another third of patients with CAP of probable pneumococcal etiology, compared with the use of the composite diagnostic standard.
Studies investigating the burden of pneumococcal pneumonia are hampered by the lack of established reference standards because of the limited sensitivity and specificity of current assays to discriminate between infection and colonization and the difficulties of obtaining good-quality sputum samples. Quantitative lytA rtPCR on NP swab samples demonstrated pneumococci in >50% of our patients with CAP. This proportion of pneumococcal etiology in CAP is similar to the 46% detected using multiple diagnostic assays in both classic analysis and latent class analysis in Kenyan adults including convalescent serum samples [1, 19]. Our classification of “pneumococcal” and “nonpneumococcal” CAP was based on a composite of 4 standard microbiological assays in the absence of a diagnostic reference standard [14, 20].
The rate of pharyngeal pneumococcal colonization—a conditio sine qua non for pneumococcal disease [21, 22] —in our controls (12-13%) was similar to that in Native American (15%) [23], Alaskan (18%) [24], and Kenyan adults (6%) [25], but lower than that found in Gambian adults (51%) using a more sensitive latex typing assay from a sweep of the confluent growth at primary inoculum from the NP that does not rely on detecting individual pneumococcal colonies [26]. However, colonization can be a coincidental finding rendering qualitative colonization an unacceptably nonspecific diagnostic tool.
We therefore used quantitative measures of colonization density. For the first time we show the correlation of pneumococcal colonization density in adults with CAP versus asymptomatic controls based on bacterial cultures and molecular methods. Our study is also the first to evaluate a specific molecular target, lytA, quantitatively against the full range of the current standard pneumococcal diagnostics. Importantly, results of quantitative bacterial cultures and quantitative lytA rtPCR correlated well with those of the molecular method showing higher sensitivity and similar specificity in distinguishing between CAP and asymptomatic carriage. Molecular methods have the advantage of providing timely and accurate results with automation, are independent of the effects of previous antibiotics and are characterized by high specificity for their targets as shown in this and other studies [14, 27]. A cutoff of 104 DNA copies/mL, using the Spn9802 target in NP aspirates, had a specificity of 96.4% for the detection of pneumococcal CAP (defined by NP aspiration culture only) [12]. Correlation was good between semiquantitative cultures (classified as +, ++, or +++) and quantitative Spn9802 rtPCR, and adult patients with pneumonia had lower CT values than asymptomatic controls [12]. This suggests that 103–104/mL is probably a critical colonization density at which there is a transition from carriage to disease in adults. This supports our proposed hypothesis of the pathogenesis of pneumococcal pneumonia based on the concept that carriage and disease serotypes are identical in >75% of patients with invasive pneumococcal disease and that viral respiratory infections that predispose patients to pneumococcal pneumonia [28] may do so after uncontrolled replication of newly acquired pneumococcal serotypes in the nasopharynx, and subsequently the lungs [29].
With a rising burden of organisms in the nasopharynx, probably at critical colonization densities, the risk of microaspiration [30] increases, finally resulting in lobar pneumonia, the typical but not only manifestation of pneumococcal CAP [31]. The observation that the mean colony counts in patients with CAP categorized as nonpneumococcal by the composite diagnostic standard were lower than those in patients with pneumococcal CAP, but higher than in asymptomatic controls, supports the idea that many of these patients had undetected cases of pneumococcal CAP. Conversely, our data imply that NP colonization densities could become an integral part of the diagnosis of pneumococcal CAP in adults. Our observation that mortality increased with higher NP colonization density among patients with pneumococcal CAP confirms recent data showing an association between severity of CAP and genomic pneumococcal load in blood [32]. In turn, this validates our pathophysiological and novel diagnostic concept of transition from asymptomatic carriage to disease at a critical NP colonization density.
There are limited data on the association of colonization density with disease in children. Higher prevalences of NP colonization were detected in Chinese children with pneumonia than in those without pneumonia [33]. High-density NP colonization (≥106 CFU/mL of any bacterial organism at bacterial culture) was more common in Vietnamese children with pneumonia (49%) than among children with acute bronchitis (29%) or healthy children (17%), whereas no difference was found for any particular organism or in the prevalence of low-density NP colonization between any of the groups [34]. Recently, the pneumococcal load in NP samples measured with a multiplex PCR was higher in children with radiologically confirmed CAP than in those with other lower respiratory tract infections or healthy children, even though no clinically useful cutoffs could be calculated for the diagnosis of pediatric pneumonia [13], possibly because children have higher densities of pneumococcal carriage than adults [35].
There are some limitations to our data. Not all patients had a complete set of diagnostic assays, but we have no reason to suspect systematic selection bias. Furthermore, HIV-infected controls less frequently smoked and had higher proportions of current cotrimoxazole prophylaxis and HAART and higher CD4 cell counts than HIV-infected patients with CAP. CD4 cells provide antibody-independent protection from murine pneumococcal NP colonization [36], whereas smoking is a risk factor for pneumococcal carriage [37] and disease [38]. However, regardless of the presence or absence of previous antimicrobial therapy, HAART, smoking or CD4 categories, patients with CAP always had significantly higher lytA rtPCR copy numbers than asymptomatic controls.
We focused on HIV-infected persons and restricted our control group to a convenience sample of HIV-infected asymptomatic controls, reflecting the overwhelming burden of HIV infection among African adults with CAP [1, 39]. Our pneumonia population was highly representative of patients with CAP in sub-Saharan Africa. It is speculative whether our results can be generalized to patients with acute CAP and active tuberculosis . Even though patients with known active tuberculosis were not enrolled, Mycobacterium tuberculosis was the second most common organism, also as a coinfection, which is typical for sub-Saharan Africa [1]. This is consistent with a recent report documenting a 43% reduction of hospitalization for tuberculosis in children vaccinated with pneumococcal conjugate vaccine in Soweto [40]. To further strengthen the generalizability to settings where tuberculosis is less prevalent, a separate analysis excluding patients with a concomitant diagnosis of tuberculosis showed a very similar performance of our proposed NP density cutoff.
As expected, only a few adults with CAP were HIV uninfected, among whom this quantitative rtPCR cutoff also increased the proportion of pneumococcal etiology similar to that seen in HIV-infected adults. Because the mean colonization density did not differ between HIV-uninfected and HIV-infected patients with CAP, and because we expect similar or indeed lower colonization densities and frequencies of asymptomatic colonization in asymptomatic HIV-uninfected compared with HIV-infected adults [41], this assay also seems promising in HIV-uninfected adults. We consider this a proof-of-concept study, which requires validation in different settings in patients who are elderly or have other risk factors for CAP.
In conclusion, we propose a new method to diagnose pneumococcal pneumonia in adults. Using an easily obtainable NP specimen and a rapid and accurate culture-independent assay, quantitative rtPCR markedly increases the proportion of CAP attributed to pneumococcus. This may be useful both for direct patient care, enabling more targeted therapy and for epidemiologic assessment of the vaccine-preventable burden of pneumococcal disease in adults. Further confirmation is needed of the association between high NP colonization density and clinical disease before an NP count of ≥8000 copies/mL can indeed be considered diagnostic of pneumococcal CAP in patients with a compatible clinical picture.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://www.oxfordjournals.org/our_journals/cid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Authors’ contributions.
W. C. A., S. A. M., P. V. A., and K. P. K. had the idea and initiated the study. W. C. A.,S. A. M., P. V. A., C. C., M. S., and K. P. K. wrote the protocol. W. C. A., N. V. N., T. M., M. W., and A. K. managed the study and collected the data. P. V. A., N. V. N., T. M., M. K., P. Z., A. D., M. S., and K. U. J. were responsible for and performed the assays. W. C. A. performed the analyses. W. C. A., S. A. M., P. V. A., M. S., K. U. J., and K. P. K. interpreted the data. W. C. A. S. A. M., and K. P. K. drafted the manuscript. All authors amended and commented on the final manuscript.
Acknowledgments.
We are indebted to Naseem Ebrahim for technical assistance, Mrs Thulisile Makhanya and Mrs Nomathemba Malele for assistance with specimen collection and Mrs Dipuo Letsapa for data entry.
Financial support.
This work was supported by Centers for AIDS Research (CFAR) Grant National Institutes of Health (NIH) P30 A1050409 to K. P. K. We thank Pfizer for performing the lytA rtPCR, and Binax for supplying ICT, Binax Now Streptococcus pneumoniae free of charge. The funding source had no influence on data analysis, the draft of the manuscript, or the decision to submit the manuscript for publication.
Potential conflicts of interest.
W. C. A. received an honoraria from GlaxoSmithKline (GSK) and support from BRAHMS to attend meetings and fulfilled speaking engagements, and also received institutional grant support from Pfizer. S. A. M. received research funding and honoraria from Pfizer vaccines and GSK and institutional grant support from Wyeth. K. P. K. received consulting and research funding from Pfizer Vaccines and consulting funding from GSK. P. Z., A. D., M. S., and K. U. J. are employees of Pfizer Vaccines.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Scott JA, Hall AJ, Muyodi C, et al. Aetiology, outcome, and risk factors for mortality among adults with acute pneumonia in Kenya. Lancet. 2000;355:1225–30. doi: 10.1016/s0140-6736(00)02089-4. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett JG. Decline in microbial studies for patients with pulmonary infections. Clin Infect Dis. 2004;39:170–2. doi: 10.1086/421498. [DOI] [PubMed] [Google Scholar]
- 3.Klugman KP, Madhi SA, Albrich WC. Novel approaches to the identification of Streptococcus pneumoniae as the cause of community-acquired pneumonia. Clin Infect Dis. 2008;47(Suppl 3):S202–6. doi: 10.1086/591405. [DOI] [PubMed] [Google Scholar]
- 4.Jokinen C, Heiskanen L, Juvonen H, et al. Microbial etiology of community-acquired pneumonia in the adult population of 4 municipalities in eastern Finland. Clin Infect Dis. 2001;32:1141–54. doi: 10.1086/319746. [DOI] [PubMed] [Google Scholar]
- 5.Rimland D, Navin TR, Lennox JL, et al. Prospective study of etiologic agents of community-acquired pneumonia in patients with HIV infection. AIDS. 2002;16:85–95. doi: 10.1097/00002030-200201040-00011. [DOI] [PubMed] [Google Scholar]
- 6.Cutts FT, Zaman SM, Enwere G, et al. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet. 2005;365:1139–46. doi: 10.1016/S0140-6736(05)71876-6. [DOI] [PubMed] [Google Scholar]
- 7.Saito A, Kohno S, Matsushima T, et al. Prospective multicenter study of the causative organisms of community-acquired pneumonia in adults in Japan. J Infect Chemother. 2006;12:63–9. doi: 10.1007/s10156-005-0425-8. [DOI] [PubMed] [Google Scholar]
- 8.Charles PG, Whitby M, Fuller AJ, et al. The etiology of community-acquired pneumonia in Australia: why penicillin plus doxycycline or a macrolide is the most appropriate therapy. Clin Infect Dis. 2008;46:1513–21. doi: 10.1086/586749. [DOI] [PubMed] [Google Scholar]
- 9.Musher DM, Montoya R, Wanahita A. Diagnostic value of microscopic examination of Gram-stained sputum and sputum cultures in patients with bacteremic pneumococcal pneumonia. Clin Infect Dis. 2004;39:165–9. doi: 10.1086/421497. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho Mda G, Tondella ML, McCaustland K, et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–6. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters RP, de Boer RF, Schuurman T, et al. Streptococcus pneumoniae DNA load in blood as a marker of infection in patients with community-acquired pneumonia. J Clin Microbiol. 2009;47:3308–12. doi: 10.1128/JCM.01071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdeldaim GM, Stralin K, Olcen P, Blomberg J, Herrmann B. Toward a quantitative DNA-based definition of pneumococcal pneumonia: a comparison of Streptococcus pneumoniae target genes, with special reference to the Spn9802 fragment. Diagn Microbiol Infect Dis. 2008;60:143–50. doi: 10.1016/j.diagmicrobio.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Vu HTT, Yoshida LM, Suzuki M, et al. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J. 2011;30:11–18. doi: 10.1097/INF.0b013e3181f111a2. [DOI] [PubMed] [Google Scholar]
- 14.Yang S, Lin S, Khalil A, et al. Quantitative PCR assay using sputum samples for rapid diagnosis of pneumococcal pneumonia in adult emergency department patients. J Clin Microbiol. 2005;43:3221–6. doi: 10.1128/JCM.43.7.3221-3226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radstrom P, Backman A, Qian N, Kragsbjerg P, Pahlson C, Olcen P. Detection of bacterial DNA in cerebrospinal fluid by an assay for simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and streptococci using a seminested PCR strategy. J Clin Microbiol. 1994;32:2738–44. doi: 10.1128/jcm.32.11.2738-2744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Brien KL, Nohynek H. Report from a WHO Working Group: standard method for detecting upper respiratory carriage of Streptococcus pneumoniae. Pediatr Infect Dis J. 2003;22:e1–11. doi: 10.1097/01.inf.0000049347.42983.77. [DOI] [PubMed] [Google Scholar]
- 17.Stralin K. Usefulness of aetiological tests for guiding antibiotic therapy in community-acquired pneumonia. Int J Antimicrob Agents. 2008;31:3–11. doi: 10.1016/j.ijantimicag.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 18.Lim WS, van der Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: an international derivation and validation study. Thorax. 2003;58:377–82. doi: 10.1136/thorax.58.5.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jokinen J, Scott JA. Estimating the proportion of pneumonia attributable to pneumococcus in Kenyan adults: latent class analysis. Epidemiology. 2010;21:719–25. doi: 10.1097/EDE.0b013e3181e4c4d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonzo TA, Pepe MS. Using a combination of reference tests to assess the accuracy of a new diagnostic test. Stat Med. 1999;18:2987–3003. doi: 10.1002/(sici)1097-0258(19991130)18:22<2987::aid-sim205>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 21.Gray BM, Converse GM, 3rd, Dillon HC., Jr Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis. 1980;142:923–33. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 22.Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–54. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 23.Millar EV, Watt JP, Bronsdon MA, et al. Indirect effect of 7-valent pneumococcal conjugate vaccine on pneumococcal colonization among unvaccinated household members. Clin Infect Dis. 2008;47:989–96. doi: 10.1086/591966. [DOI] [PubMed] [Google Scholar]
- 24.Hammitt LL, Bruden DL, Butler JC, et al. Indirect effect of conjugate vaccine on adult carriage of Streptococcus pneumoniae: an explanation of trends in invasive pneumococcal disease. J Infect Dis. 2006;193:1487–94. doi: 10.1086/503805. [DOI] [PubMed] [Google Scholar]
- 25.Abdullahi O, Nyiro J, Lewa P, Slack M, Scott JA. The descriptive epidemiology of Streptococcus pneumoniae and Haemophilus influenzae nasopharyngeal carriage in children and adults in Kilifi district, Kenya. Pediatr Infect Dis J. 2008;27:59–64. doi: 10.1097/INF.0b013e31814da70c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill PC, Akisanya A, Sankareh K, et al. Nasopharyngeal carriage of Streptococcus pneumoniae in Gambian villagers. Clin Infect Dis. 2006;43:673–9. doi: 10.1086/506941. [DOI] [PubMed] [Google Scholar]
- 27.Avni T, Leibovici L, Paul M. PCR diagnosis of invasive candidiasis: systematic review and meta-analysis. J Clin Microbiol. 2010;49:665–70. doi: 10.1128/JCM.01602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madhi SA, Klugman KP. A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med. 2004;10:811–13. doi: 10.1038/nm1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berendt RF, Long GG, Walker JS. Influenza alone and in sequence with pneumonia due to Streptococcus pneumoniae in the squirrel monkey. J Infect Dis. 1975;132:689–93. doi: 10.1093/infdis/132.6.689. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg D, Givon-Lavi N, Newman N, Bar-Ziv J, Dagan R. Nasopharyngeal carriage of individual Streptococcus pneumoniae serotypes during pediatric pneumonia as a means to estimate serotype disease potential. Pediatr Infect Dis J. 2011;30:227–33. doi: 10.1097/INF.0b013e3181f87802. [DOI] [PubMed] [Google Scholar]
- 31.Madhi SA, Klugman KP. World Health Organisation definition of “radiologically confirmed pneumonia” may under-estimate the true public health value of conjugate pneumococcal vaccines. Vaccine. 2007;25:2413–19. doi: 10.1016/j.vaccine.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Rello J, Lisboa T, Lujan M, et al. Severity of pneumococcal pneumonia associated with genomic bacterial load. Chest. 2009;136:832–40. doi: 10.1378/chest.09-0258. [DOI] [PubMed] [Google Scholar]
- 33.Levine OS, Liu G, Garman RL, Dowell SF, Yu S, Yang YH. Haemophilus influenzae type b and Streptococcus pneumoniae as causes of pneumonia among children in Beijing, China. Emerg Infect Dis. 2000;6:165–70. doi: 10.3201/eid0602.000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anh DD, Huong Ple T, Watanabe K, et al. Increased rates of intense nasopharyngeal bacterial colonization of Vietnamese children with radiological pneumonia. Tohoku J Exp Med. 2007;213:167–72. doi: 10.1620/tjem.213.167. [DOI] [PubMed] [Google Scholar]
- 35.McCool TL, Weiser JN. Limited role of antibody in clearance of Streptococcus pneumoniae in a murine model of colonization. Infect Immun. 2004;72:5807–13. doi: 10.1128/IAI.72.10.5807-5813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trzcinski K, Thompson CM, Srivastava A, Basset A, Malley R, Lipsitch M. Protection against nasopharyngeal colonization by Streptococcus pneumoniae is mediated by antigen-specific CD4+ T cells. Infect Immun. 2008;76:2678–84. doi: 10.1128/IAI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenberg D, Givon-Lavi N, Broides A, Blancovich I, Peled N, Dagan R. The contribution of smoking and exposure to tobacco smoke to Streptococcus pneumoniae and Haemophilus influenzae carriage in children and their mothers. Clin Infect Dis. 2006;42:897–903. doi: 10.1086/500935. [DOI] [PubMed] [Google Scholar]
- 38.Nuorti JP, Butler JC, Farley MM, et al. Cigarette smoking and invasive pneumococcal disease. Active Bacterial Core Surveillance Team. N Engl J Med. 2000;342:681–9. doi: 10.1056/NEJM200003093421002. [DOI] [PubMed] [Google Scholar]
- 39.Nyamande K, Lalloo UG, John M. TB presenting as community-acquired pneumonia in a setting of high TB incidence and high HIV prevalence. Int J Tuberc Lung Dis. 2007;11:1308–13. [PubMed] [Google Scholar]
- 40.Moore DP, Klugman KP, Madhi SA. Role of Streptococcus pneumoniae in hospitalization for acute community-acquired pneumonia associated with culture-confirmed Mycobacterium tuberculosis in children: a pneumococcal conjugate vaccine probe study. Pediatr Infect Dis J. 2010;29:1099–104. doi: 10.1097/inf.0b013e3181eaefff. [DOI] [PubMed] [Google Scholar]
- 41.Paul J. Royal Society of Tropical Medicine and Hygiene Meeting at Manson House, London, 12 December 1996. HIV and pneumococcal infection in Africa: microbiological aspects. Trans R Soc Trop Med Hyg. 1997;91:632–7. doi: 10.1016/s0035-9203(97)90500-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.