Abstract
Background.
Several genome scans have explored the linkage of chronic kidney disease phenotypes to chromosomic regions with disparate results. Genome scan meta-analysis (GSMA) is a quantitative method to synthesize linkage results from independent studies and assess their concordance.
Methods.
We searched PubMed to identify genome linkage analyses of renal function traits in humans, such as estimated glomerular filtration rate (GFR), albuminuria, serum creatinine concentration and creatinine clearance. We contacted authors for numerical data and extracted information from individual studies. We applied the GSMA nonparametric approach to combine results across 14 linkage studies for GFR, 11 linkage studies for albumin creatinine ratio, 11 linkage studies for serum creatinine and 4 linkage studies for creatinine clearance.
Results.
No chromosomal region reached genome-wide statistical significance in the main analysis which included all scans under each phenotype; however, regions on Chromosomes 7, 10 and 16 reached suggestive significance for linkage to two or more phenotypes. Subgroup analyses by disease status or ethnicity did not yield additional information.
Conclusions.
While heterogeneity across populations, methodologies and study designs likely explain this lack of agreement, it is possible that linkage scan methodologies lack the resolution for investigating complex traits. Combining family-based linkage studies with genome-wide association studies may be a powerful approach to detect private mutations contributing to complex renal phenotypes.
Keywords: albuminuria, chronic kidney disease, glomerular filtration rate, linkage scans, meta-analysis
Introduction
Genetic factors contribute to the development and progression of chronic kidney disease (CKD). Familial aggregation of kidney disease has been shown in multiple epidemiologic studies [1–3], and traits such as estimated glomerular filtration rate (eGFR) and albuminuria are modestly heritable [4–6]. While the advent of large genome-wide association studies has increased the scope for the identification of kidney disease susceptibility genes [7], with the exception of mapping by admixture linkage disequilibrium (LD) [8–10], both association and linkage approaches have had limited success to date.
A genome-wide linkage scan is an unbiased approach to interrogate the genome with multiple markers that identify evenly spaced locations along each of the human chromosomes in families with multiple subjects manifesting the trait of interest. Linkage analysis tests for co-segregation of the marker with a trait or phenotype of interest within a family implying physical linkage between the marker and a phenotype locus. Since the first genome-wide linkage scan for nephropathy genes in 1998 [11], there have been many studies addressing a range of kidney phenotypes in a variety of study populations, ranging from the general population to populations with underlying comorbidities such as diabetes or hypertension which could predispose to CKD. Multiple linkage peaks have been observed with little overlap both for a single phenotype and for related CKD phenotypes. Many peaks have not reached genome-wide significance, possibly due to linkage and population heterogeneity, small sample sizes and limited power or because they were simply false positives. Some of the major challenges to the linkage approach as applied to complex genetic diseases include the need for large numbers of relative pairs to have sufficient power to detect loci that confer modest effects and a relatively homogenous population with regard to ethnicity, comorbidity and distribution of the trait [12, 13].
One approach to compare genetic evidence across linkage studies is a meta-analysis, preferably with the primary data from each study. We therefore undertook a collaborative genome scan meta-analysis (GSMA) [14–16] of existing genome-wide linkage studies of quantitative CKD traits including albuminuria (albumin creatinine ratio, ACR), eGFR, serum creatinine concentration and creatinine clearance. These measures exhibit a continuous phenotypic spectrum and may offer more power than categorization based on arbitrarily defined thresholds. The GSMA is an exploratory data analysis method, which aims to highlight regions containing susceptibility loci, where further studies can be focused.
Materials and methods
Literature search and eligible whole genome scans
We searched PubMed for whole genome scans on four CKD traits, namely (estimated) glomerular filtration rate, ACR (urinary albumin), serum creatinine and creatinine clearance using combinations of the corresponding medical subject heading terms and terms such as ‘genome scan’ or ‘genome search’ and kidney disease, including the above-mentioned CKD traits (full search strategy provided as Supplement). Searches were limited to the English language and humans and to the period 01 January 1998 to 29 December 2010. We complemented searches by perusing reference lists of eligible papers and narrative reviews. Reference lists of relevant original papers and narrative reviews were studied and experts in the field were contacted. Investigators were invited to contribute necessary data for analyses.
We included whole genome scans on the aforementioned four outcomes, provided that they used >300 markers, irrespective of the statistical analyses employed. We excluded studies that mapped only specific chromosomes or chromosomal regions (all 22 autosomes had to be analyzed), genome-wide association studies, studies with <10 families and studies in people with Mendelian forms of inherited kidney disease.
Definition of outcomes
We included studies that estimated eGFR using the four-variable Modification of Diet in Renal Diseases equation [17] and calculated creatinine clearance with the Cockroft–Gault equation [18]. We accepted all methods of serum creatinine measurement irrespective of whether assay calibration was described or not. Studies that analyzed albuminuria as the phenotype expressed it as the albumin excretion rate, normalized to urinary creatinine (ACR).
Data abstraction
Bibliographic information (first author, journal and year of publication), area of recruitment, whether participants were diseased or healthy, number of probands, number of families or sibpairs (as applicable), reported outcomes, number of microsatellite markers, the name of the commercial marker set and the statistical methods and software used in each publication were recorded. The unit of the analysis was the population stratum, defined by ‘racial’ descent (European excluding Hispanic, African-American, American Indian, Hispanic). We retained subgroups from different recruitment locations as separate strata if they were analyzed separately in the primary studies. Potential overlaps between studies were clarified in communication with study authors.
All identified research teams were invited to participate in this collaborative meta-analysis. Participating teams contributed sufficient statistics for each of their population strata. Whenever such data were not provided, we extracted them from publications, by digitizing graphs of linkage scores per pter (p terminal) distance in each chromosome using specialized software and corroborating with results reported in the text and tables. The reliability and validity of this approach has been previously demonstrated [19]. Whenever multiple analyses were available, we chose multipoint LOD scores over other analyses. We also selected results at the longest follow-up. We chose the more adjusted models over less adjusted ones, in an effort to select the most comparable analyses across studies. Most studies did not report unadjusted results.
Genome scan meta-analysis
We synthesized information across genome scans using methods described previously [15, 16, 20, 21]. Briefly, GSMA analyze chromosomic regions, rather than specific markers. It aligns all 22 autosomes in the genome and breaks them into 120 regions (bins) of ∼30 cM each (a cM is a unit of distance based on recombination rates). These regions (bins) are the units of analysis. Larger chromosomes have more bins than smaller ones (e.g. Chromosome 1 has 10 bins, as it is ∼300 cM long, while Chromosomes 21 and 22 have only two bins each). We name bins according to their position in the corresponding chromosome, e.g. Bin 7.2 would be the second bin on Chromosome 7. In each study, we represent each bin by the most significant P-value (or equivalent statistic, such as the highest LOD peak) that corresponds to it.
GSMA uses the relative ranks of the P-values (or other equivalent statistics) from the linkage analysis over all 120 bins in each study, with the highest rank being the most significant bin (highest peak) and the lowest being the least significant bins. If several bins are tied, they are assigned a common rank. In the GSMA, the bin that has the highest mean rank across all studies is the one where (on average) most high peaks are located. To judge the statistical significance of a bin’s mean rank, we perform a Monte Carlo permutation test [20].
For each bin, we also quantified the heterogeneity in the ranks across available population strata, i.e. a measure of the dispersion of the ranks in the individual studies around their mean. Intuitively, when this dispersion is very low, there is agreement among all studies on the rank of the particular bin. We quantify this dispersion with the Q statistic [15, 16], which is the sum of the squared differences of ranks from their mean. The distribution of the Q statistic is not known; we therefore tested whether the Q statistic implied significantly low heterogeneity based on Monte Carlo permutations.
Subgroup and sensitivity analyses
For each outcome, the main analysis combined all available population strata. We also performed subgroup analyses by ethnicity, baseline health status (general population, diabetes, hypertension or either of the latter two).
The eGFR and creatinine clearance are transformation of serum creatinine; therefore, we also performed a sensitivity analysis where we treated all three outcomes together. To avoid duplication of information, we preferred serum creatinine over eGFR over creatinine clearance, where more than one were reported. Results were very similar with all permutations of the preferred order of outcomes and are thus not reported in further detail. We also performed analyses excluding digitized data and using only data for which primary numerical results were available. Additional sensitivity analyses were carried out using alternate data from studies with more than one analysis (e.g. using results from analyses with fewer adjustments instead of the most adjusted models), but as their results were very similar, they are not reported.
Software and analytic details
All Monte Carlo analyses were run for 100 000 permutations. In main analyses, we weighted bin ranks by the square root of the total number of subjects in the corresponding stratum (study). Because the optimal weighting for GSMA is unclear, we also performed unweighted analyses. Inferences were performed at the α = 0.05 level. Thus, we considered a P-value ≤0.05 as of suggestive significance, and a P-value ≤0.00042 (using Bonferroni corrections for 120 comparisons) as genome-wide significant. Graph digitizing was performed with Engauge Digitizer (version 2.12, Mark Mitchell, 2002). All analyses and graphs were performed in Intercooled Stata 8.2 (Stata Corp. College Station, TX), using the ‘hegesma’ C plugin for Stata (developed by author T.A.T.). The Stata plugin has been verified against the HEGESMA software [16] and also allows for missing bin values.
Results
We included 12 studies corresponding to 22 distinct population (ethnic descent) strata. Table 1 summarizes their characteristics and additional descriptive data is shown in Supplementary Table 2. We obtained data directly from investigators for six studies (13 population strata) [4, 5, 24–26, 28, 29, 33, 34]. For the remaining studies, we extracted data from publications. There were 10 European-descent strata, 5 African-American (AA) strata, 3 Mexican American (MA) strata and 4 American Indian (AI) strata. Six studies had been carried out in general population cohorts and the remaining in populations enriched for diabetes or hypertension.
Table 1.
Characteristics of included studiesa
| Study (ref.) | GFR | Cr | CrCl | ACR | Population | ‘Racial’ descent strata |
Markers (set) | Analysis (software) | |||
| Eur | Af | His | AI | ||||||||
| SAFHSb [22, 23] | Y | Y | Y | Y | General population | – | – | 848c | – | 417 (Marshfield) | VC (SOLAR) |
| FHSd [4, 5] | Y | Y | – | Y | General population | 1055 | – | – | – | 401 (Marshfield 9) | VC (SOLAR) |
| SHFSe [24, 25] | Y | – | – | Y | General population | – | – | – | 1183; 1161; 1153f | ND (ABI PRISM Linkage Mapping Set-MD10 version 2.5) | VC (SOLAR) |
| Utahe [26] | – | Y | Y | – | General population | 850g | – | – | – | 393 (Marshfield 10) | NPL (GENEHUNTER) |
| Eurospanb [27] | – | Y | – | – | General population | ERF: 1388; MICROS: 891; Vis: 580h | ERF: 6008 biallelic; Illumina Infinium Linkage assay, MICROS: 1000 STR’s; deCODE Icelandic Genetic Map, Vis: 747 microsatellite; deCODE Icelandic Genetic Mapi | VC (SOLAR) | |||
| FINDe [28, 29] | Y | – | – | Y | Diabetics | 119 | 218 | 469 | 80 | 404 (Marshfield 8) | HE (SIBPAL) |
| Joslinb [30] | – | – | – | Y | Diabetics | 857 | – | – | – | 383 (Marshfield 12) | VC (SOLAR) |
| SAFDGSb [31] | Y | – | Y | – | Diabetics | – | – | 453 | – | 382 (CIDRj) | VC (SOLAR) |
| AADMb [32] | Y | Y | Y | – | Diabetics | – | 691 | – | – | 372 (CIDR) | VC (SOLAR) |
| DHSe [33] | Y | – | – | Y | Diabetics, hypertensives | 902 | 165 | – | – | 411 (Marshfield 13) | VC (SOLAR) |
| HyperGENe [34] | Y | – | – | Y | Hypertensives | 1040 | 1234 | – | – | 391 (Marshfield) | VC (SOLAR) |
| GENOAb [35] | Y | – | – | Y | Hypertensives | 1022 | 1351 | – | – | 381 (Marshfield 9) | VC (SIMWALK2) |
As clarified in the Materials and methods section, different strata (by racial descent or within the same racial descent) were included as separate entries in the genome search meta-analysis. ‘–’ = not examined/not included, AADM, African-American Diabetes Mellitus Study; Af, African-American descent; AI, American Indian descent; Cr, serum creatinine; CrCl, creatinine clearance; CVD, cardiovascular disease; DHS, Diabetes Heart Study; DM, diabetes mellitus; GFR, estimated glomerular filtration rate; Eur, European descent; FIND, Family Investigation of Nephropathy and Diabetes; FHS, Framingham Heart Study; GENOA, Genetic Epidemiology Network of Arteriopathy; HE, Haseman-Elston regression; His, Hispanic (Mexican American) descent; HTN, hypertensive; HyperGEN, Hypertension Genetic Epidemiology Network Study; ND, no data; NPL, nonparametric linkage; SAFHS, San Antonio Family Heart Study; SAFDGS, San Antonio Family Diabetes/Gall Bladder Study; SHFS, Strong Heart Family Study; VC, variance components linkage analysis; STR, short tandem repeat markers; CIDR, Center for Inherited Disease Research.
Meta-analysis data extracted from digitized graphs.
Data directly obtained from investigators.
Three populations from Oklahoma, Arizona and Dakota, respectively (USA) [25]. For GFR, the corresponding numbers are 1210, 1235 and 1220 [24].
Population enriched for CVD. There are three sequential longitudinal waves with 1516, 1193 and 850 analyzed subjects, respectively. Main analyses used the last wave, sensitivity analyses used the first or second wave instead.
Three isolated populations in Europe: MICROS study (Italy), ERF study (the Netherlands) and Vis study (Croatia).
For map alignment, as the three studies had different genotypes, the deCODE genetic map was used as a reference to align individual maps so that a common linkage analysis could be performed on the same map, based on 1000 markers.
The CIDR genetic map is similar to the Marshfield Genetic map.
Estimated glomerular filtration rate
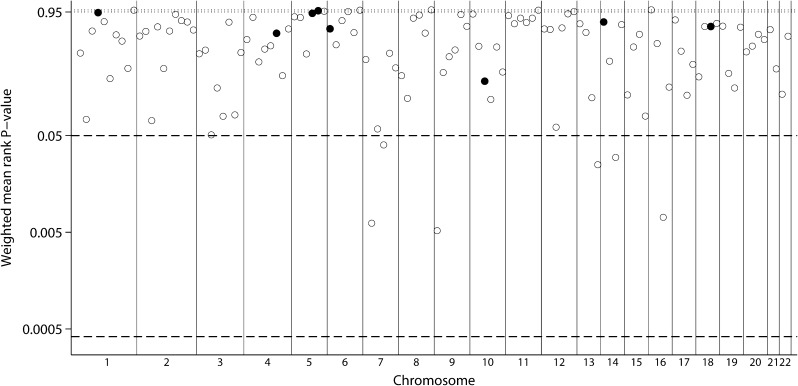
For the outcome of eGFR, we included data from 14 strata in our analysis. In weighted analyses that account for the size of each study, no bin reached genome-wide significance (P-value threshold ≤0.00042) (Figure 1, Table 2). Six bins (7.2, 7.4, 9.1, 13.4, 14.3 and 16.3, with corresponding chromosomal locations indicated in Table 2) reached suggestive significance (P-value threshold ≤0.05). There was no evidence for low between-scan heterogeneity (i.e. ranks were in agreement across population strata beyond what is expected by chance). Bins 9.1 and 7.2 had the smallest P-values for the mean rank (P = 0.0052 and P = 0.0062, respectively). Unweighted analyses were similar. Seven bins (the six from the weighted analyses and Bin 15.4) reached suggestive significance. Bins 16.3 and 9.1 had the lowest P-values (P = 0.0063 and P = 0.0102, respectively). Again, none of the bins with suggestive significance showed evidence for low between-scan heterogeneity.
Fig. 1.
GSMA for GFR. Weighted average ranks from 14 population strata. Vertical lines separate autosomes. The horizontal dashed reference line indicates weighted average ranks at the 95% significance level. Open circles represent bins where no evidence for low between-scan heterogeneity was found with the heterogeneity metric (unadjusted for the bin’s mean rank, at the 0.05 level). Filled circles indicate evidence for low heterogeneity (P ≤ 0.05).
Table 2.
Bins attaining suggestive significance (P ≤ 0.05) in weighted analysesa
| Phenotype; bin | Chromosomal location | Average rank |
P-value for low heterogeneity | |
| Observed | P-value | |||
| eGFR | ||||
| 9.1 | 9p24.3–9p22.3 | 84 | 0.0052 | 0.4145 |
| 7.2 | 7p15.3–7p13 | 83 | 0.0062 | 0.6830 |
| 16.3 | 16q12.2–16q23.1 | 82 | 0.0072 | 0.5705 |
| 13.4 | 13q33.1–13q34 | 78 | 0.0250 | 0.6052 |
| 14.3 | 14q23.3–14q32.12 | 77 | 0.0298 | 0.5067 |
| 7.4 | 7q11.23–7q22.3 | 76 | 0.0401 | 0.2298 |
| Serum creatinine | ||||
| 9.6 | 9q34.11–9q34.3 | 88 | 0.0017 | 0.4266 |
| 7.3 | 7p13–7q11.23 | 86 | 0.0029 | 0.8663 |
| 16.4 | 16q23.1–16q24.1 | 81 | 0.0119 | 0.8982 |
| 9.4 | 9q21.32–9q31.2 | 80 | 0.0151 | 0.7710 |
| 10.3 | 10p11.23–10q22.1 | 79 | 0.0185 | 0.5645 |
| 9.5 | 9q31.2–9q34.11 | 78 | 0.0278 | 0.4132 |
| 9.2 | 9p22.3–9p21.1 | 78 | 0.0306 | 0.9721 |
| 9.3 | 9p21.1–9q21.32 | 78 | 0.0308 | 0.9793 |
| 11.4 | 11q13.3–11q22.1 | 77 | 0.0376 | 0.8685 |
| 11.3 | 11p12–11q13.3 | 76 | 0.0452 | 0.7713 |
| Creatinine clearance | ||||
| 7.3 | 7p13–7q11.23 | 93 | 0.0195 | 0.6463 |
| 6.6 | 6q25.3–6q27 | 91 | 0.0310 | 0.5524 |
| 16.4 | 16q23.1–16q24.1 | 91 | 0.0315 | 0.6064 |
| 10.4 | 10q22.1–10q23.32 | 90 | 0.0337 | 0.9859 |
| 2.6 | 2q21.1–2q24.1 | 88 | 0.0409 | 0.7208 |
| ACR | ||||
| 10.4 | 10q22.1–10q23.32 | 82 | 0.0145 | 0.9425 |
| 7.5 | 7q22.3–7q34 | 82 | 0.0154 | 0.3294 |
| 3.6 | 3q21.2–3q25.32 | 81 | 0.0190 | 0.3643 |
| 7.4 | 7q11.23–7q22.3 | 81 | 0.0205 | 0.5862 |
| 1.2 | 1p36.21–1p35.2 | 79 | 0.0305 | 0.1693 |
GFR, glomerular filtration rate. The threshold for genome-wide significance would be P-value ≤0.05/120 = 0.00042. No bin attained this level in the main analysis.
Serum creatinine concentration
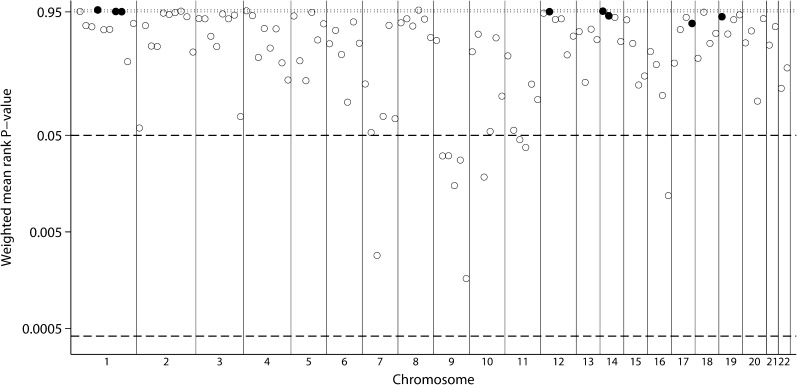
Weighted analyses among 11 genome scan strata resulted in 10 bins that reached suggestive significance (Figure 2, Table 2), although none reached genome-wide significance. The lowest P-values for the mean rank were found for bins 9.6 and 7.3 (P = 0.00165 and 0.00285), respectively. Interestingly, five of six bins in the ninth chromosome (9.2 through 9.6) were among the 10 bins. None of the significant bins had evidence for low between-scan heterogeneity. Inferences in unweighted analyses were the same with the exception of Bin 11.4 which did not reach suggestive significance.
Fig. 2.
GSMA for serum creatinine. Weighted average ranks from 11 population strata. Layout similar to Figure 1.
Creatinine clearance
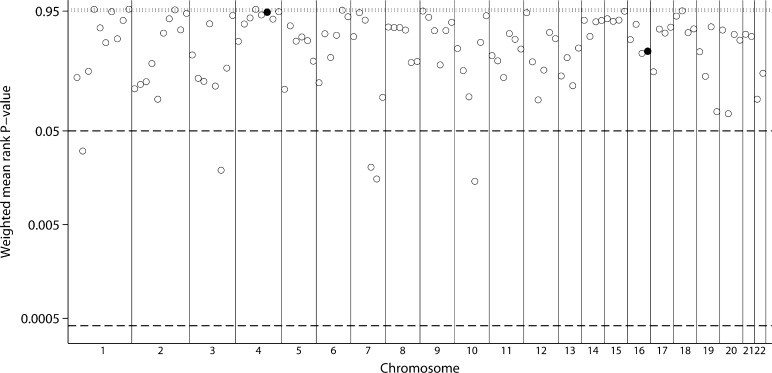
For the outcome of creatinine clearance, four genome scan strata were included in the meta-analyses. In weighted analyses, five bins reached suggestive significance for mean rank (Figure 3; Table 2) with bins 7.3 and 6.6 having the lowest P-values (P = 0.0195 and 0.0310, respectively). No bin reached genome-wide significance. There was evidence for statistically significant high between-scan heterogeneity with all three metrics for Bin 10.4. None of the significant bins had evidence for low between-scan heterogeneity. Inferences in unweighted analyses were the same with the exception of Bin 2.6 which did not reach suggestive significance.
Fig. 3.
GSMA for creatinine clearance. Weighted average ranks from four population strata. Layout similar to Figure 1.
Albumin creatinine ratio
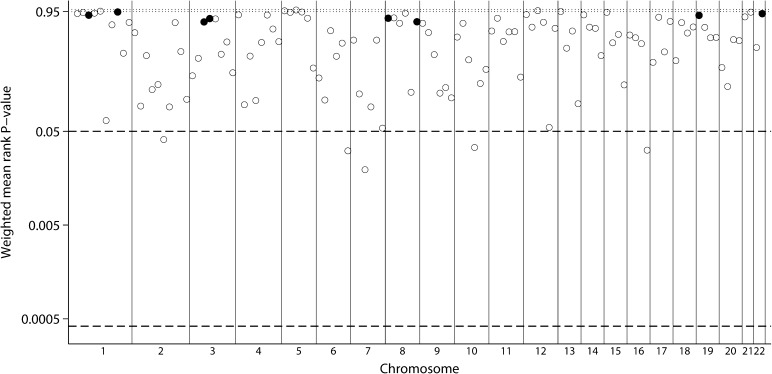
The meta-analysis for ACR was based on 11 genome scan strata. Five bins reached suggestive significance (1.2, 3.6, 7.4, 7.5 and 10.4), but none achieved genome-wide levels and none had significantly low between-scan heterogeneity. The smallest P-values were found for bins 10.4 and 7.5 (P = 0.0145 and 0.0154, respectively; Figure 4, Table 2). Unweighted analyses were very similar, with six bins reaching suggestive significance (the aforementioned 5 and 20.2), again with no evidence for low between-scan heterogeneity.
Fig. 4.
GSMA for ACR. Weighted average ranks from 11 population strata. Layout similar to Figure 1.
Subgroup analyses
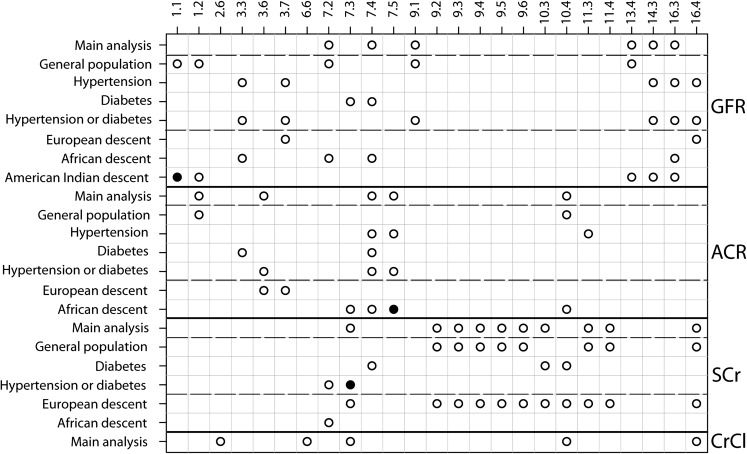
Figure 5 summarizes results of weighted main and subgroup analyses across all four phenotypes (the corresponding figure for unweighted analysis was similar—data not shown). Inferences on significance are not corrected for multiple comparisons. As shown in the figure, several bins were repeatedly identified as of suggestive statistical significance across subgroups and even across phenotypes. For example, Bin 7.4 reached suggestive significance in nine analyses for the outcomes of eGFR, ACR or serum creatinine concentration, Bin 7.3 reached at least suggestive significance in six analyses for all four outcomes, Bin 10.4 reached suggestive significance in six analyses for ACR, serum creatinine concentration and creatinine clearance and Bin 16.4 reached suggestive significance in seven analyses for eGFR, serum creatinine concentration and creatinine clearance. Bins 1.1, 7.3, and 7.5 reached genome-wide significance in the subgroups of AI descent for eGFR, hypertension or diabetes for serum creatinine concentration and African descent for ACR, respectively.
Fig. 5.
Summary of inferences in main and subgroup analyses across all phenotypes. Shown are all main and subgroup analyses (vertical axis) and the bins that reached at least suggestive significance in any main analysis, genome-wide significance in any analysis or suggestive significance in at least three of the subgroup analyses across all phenotypes (horizontal axis). Bins that reached suggestive significance only in one or two subgroup analyses are not shown. Cells with open circles: suggestive significance (P-value ≤ 0.05); cells with filled circles: genome-wide significance (P-value ≤ 0.00042).
Sensitivity analyses
We also analyzed data on eGFR, creatinine clearance and serum creatinine together. Over the 21 genome scan strata in weighted analyses, 8 bins reached suggestive significance for mean rank (Bins 7.2, 7.3, 9.1, 13.4, 15.4, 16.3, 16.4 and 22.1), with Bins 9.1 and 16.3 having the lowest P-values (P = 0.0020 and 0.0019, respectively). No bin reached genome-wide significance. None of the significant bins had evidence for low between-scan heterogeneity. Inferences in unweighted analyses were very similar (not reported).
Analyses excluding digitized data found no genome-wide significant results. In weighted analyses, the bins with significantly high mean rank for eGFR (n = 10 strata) were 1.2, 3.4, 3.5, 7.2, 7.4, 9.1, 12.3, 13.4, 14.3 and 16.3 with Bin 9.1 having the highest rank. The respective results for serum creatinine (n = 4 strata) were 4.7, 7.2, 7.4, 12.5 and 13.2 with Bin 13.2 having the highest mean rank. For ACR, among eight strata the corresponding bins were 1.2 and 12.3 with 1.2 having the highest mean rank. Only one stratum was available for creatinine clearance (no GSMA possible). Weighted analyses were similar (not reported).
Discussion
Herein we summarized the findings of 22 genome scans from 12 studies for four kidney disease phenotypes. No chromosomal region (bin) reached genome-wide statistical significance in the main analyses which combined across different population strata. Six bins reached suggestive significance for eGFR, 5 for ACR, 10 for serum creatinine concentration and 5 for creatinine clearance in Chromosomes 1, 2, 3, 7, 9, 10, 11, 13, 14 and 16. Four bins, 7.3, 7.4, 10.4 and 16.4, reached suggestive significance for at least two phenotypes. Despite signals of genome-wide significance in subgroup analyses (Bins 1.1, 7.3 and 7.5), our main analyses do not pinpoint specific cytogenetic regions as linked with the examined renal phenotypes. The conducted genome scans did not agree beyond chance in the cytogenetic locations they prioritized.
Comparison of results with findings of primary studies and with GWAS results
Because no single bin attained genome-wide significance in the main analysis, we focused our attention on bins that reached suggestive significance for more than one phenotype (bins in Chromosomes 7, 10 and 16). Table 3 outlines previous genome scans for renal phenotypes with linkage findings which have reached genome-wide significance. Signals appear spread out across the genome although there are repeated signals on Chromosomes 2, 7 and 22 across phenotypes. Notably, apart from regions on Chromosome 7, most high LOD scores in Table 3 are in regions that are not selected in our meta-analysis, in large part attributable to heterogeneity across studies. For example, two studies found genome-wide significance for linkage to eGFR and serum creatinine on Chromosome 2 [29, 33], in the cytogenetic location corresponding to Bin 2.3. This bin was close to reaching suggestive significance in the meta-analysis (Figure 1), but with indications for high between-scan heterogeneity (P-value for heterogeneity ≥0.90).
Table 3.
Observed results from genome scans with LOD scores >3.0a
| Author | Study | Phenotype | Type of population | Ethnicity | Genomic Location (cM) | LOD Score | GSMA Bin | |
| Chr 1 | Schelling [29] | FIND | GFR | Diabetic | Mexican-American | 259 | 3.78 | 1.9 |
| Chr 2 | Freedman [33] | DHS | GFR | Diabetic/hypertensive | African-American and Caucasian | 77 | 4.45 | 2.3 |
| Schelling [29] | FIND | GFR | Diabetic | Mexican-American | 82 | 3.02 | 2.4 | |
| Freedman [33] | DHS | Serum creatinine | Diabetic/hypertensive | African-American and Caucasian | 80 | 3.55 | 2.3 | |
| Hunt [26] | Utah | Serum Creatinine | General population | European-American | 145 | 3.15 | 2.6 | |
| Freedmanb [36] | All-cause ESRD | CKD | African-American | 186 | 3.05 | 2.7 | ||
| Plachac [37] | Joslin | Cystatin | Diabetic | European-American | 202 | 4.1 | 2.8 | |
| Chr 3 | Freedmanb [36] | All-cause ESRD | CKD | African-American | 7 | 3.18 | 3.3 | |
| Chr 5 | Krolewski [30] | Joslin | ACR | Diabetic | European-American | 69 | 3.4 | 5.3 |
| Chr 6 | Freedmanb [36] | All-causeESRD | CKD | African-American | 162.5 | 5.72 | 6.6 | |
| Chr 7 | Turner [35] | GENOA | GFR | Hypertensive | African-American | 43 | 3.65 | 7.2 |
| Schelling [29] | FIND | GFR | Diabetic | Multiethnic | 170 | 3.28 | 7.6 | |
| Schelling [29] | FIND | GFR | Diabetic | Mexican-American | 170 | 4.23 | 7.6 | |
| Plachac [37] | Joslin | Cystatin | Diabetic | European-American | 23 | 4 | 7.1 | |
| Chr 8 | Schelling [29] | FIND | GFR | Diabetic | Mexican-American | 93 | 3.95 | 8.4 |
| Chr 9 | Arar [23] | SAFHS | GFR | Hypertensive | Mexican-American | 75 | 3.87 | 9.3 |
| Chr 10 | Placha . [37] | Joslin | Cystatin | Diabetic | European-American | 114 | 3.6 | 10.4 |
| Chr 12 | Mottl [24] | SHFS | GFR | General population | American Indian (Arizona)d | 39 | 3.5 | 12.2 |
| Chr 13 | Freedmanb [36] | All-cause ESRD | CKD | African-American | 91 | 4.94 | 13.4 | |
| Chr 14 | Leon [34] | HyperGEN | GFR | Hypertensive | African-American and Caucasian | 94 | 3.29 | 14.3 |
| Chr 15 | Leon [34] | HyperGEN | GFR | Hypertensive | African-American and Caucasian | 16 | 3.13 | 15.1 |
| Chr 22 | Krolewski [30] | Joslin | ACR | Diabetic | European-American | 33 | 3.4 | 22.2 |
| Pattaro [27] | Eurospan | Serum creatinine | General population | European-American (MICROS)d | 61 | 3.4 | 22.2 |
DHS, Diabetes Heart Study; GFR; estimated glomerular filtration rate; FIND, Family Investigation of Nephropathy and Diabetes; GENOA, Genetic Epidemiology Network of Arteriopathy; HyperGEN, Hypertension Genetic Epidemiology Network Study; SAFHS, San Antonio Family Heart Study; SHFS, Strong Heart Family Study.
Studies with dichotomous outcomes, not included in present analysis.
Not analyzed as part of study.
Subgroup in study.
Recent associations have been identified between CKD phenotypes and markers on Chromosome 16 [7] and Chromosome 22 [8–10, 38–40]. In our meta-analysis, we showed suggestive significance for eGFR in several subgroup analyses for bins 16.3 and 16.4, although none of the individual studies in Table 3 had a peak in Chromosome 16. On the other hand, two studies as shown in Table 3, reported linkage peaks in Chromosome 22 [41, 42] and we noted suggestive significance for bin 22.1 when data on eGFR, creatinine clearance and serum creatinine were combined. We did not note any other overlap with loci identified in recent meta-analysis of genome-wide association data for CKD [43, 44]. Kitsios and Zintzaras have investigated the agreement between genome-wide linkage scans and genome-wide association studies and noted ‘genomic convergence’ in 2 of 19 phenotypes. Although convergence could support true genetic effects, lack of congruence may reflect that these studies are designed to answer different questions (association studies are aimed at identifying common variants involved in disease etiology with small effects, whereas linkage approaches attempt to identify rare variants with larger effects) and employ different approaches. Perhaps the two types of research are better viewed as complementary in elucidating the genetics of common complex disease [45, 46].
Issues affecting the results and their interpretation
The genome scan meta-analysis has been applied to other complex phenotypes [21, 41, 42, 47–50] with the identification of several novel loci; loci with genome-wide significance have been identified in autism [21], hypertension [49] and inflammatory bowel disease and celiac disease [38, 50], schizophrenia [39], multiple sclerosis [40] and asthma and atopy [51]. However, diverse linkage signals across scans, possibly suggesting genetic heterogeneity across subsyndromes and subpopulations have been noted as a factor limiting effective synthesis of linkage data.
In our approach, we combined data across strata of different racial descent and health status. GSMA can identify regions that give weak but consistent linkage signals in multiple genome scans [42] but will not identify linked regions that are present in only a subset of scans for example because of population-specific effects [20]. Several studies included in the GSMA increased their statistical power by combining data across racial groups [28, 29, 33, 34]. While we employed a similar approach, we appreciate that combining across racial subgroups also increased heterogeneity. We therefore performed subgroup analyses by race and health status to explore subgroup-specific signals. In contrast to association studies, linkage analyses are family-based and therefore immune to population stratification. However, it is now appreciated that they may be affected by differences in LD patterns and range (admixture LD [52]) as well as allele frequencies across different ethnic groups which could potentially add to the heterogeneity of the linkage signal.
The synthesized outcomes in this study are surrogates of kidney disease. eGFR values depend on the estimating equation and factors such as race [53]; similarly, microalbuminuria can reverse [54] and may not uniformly correlate with severity of CKD; [55] treatment with inhibitors of the renin angiotensin system could minimize variability of both eGFR and ACR; finally, most studies have relied on single measurements which do not capture either the marked day to day variability of ACR or the change in eGFR over time [56]. Any of these factors may have impacted the results of the original study and accordingly the results of the meta-analysis.
Most included studies did not report unadjusted analyses, and therefore it is not possible to explore whether the negative findings of the meta-analysis are related to the choice of the analytic models in the primary studies. For example, if one adjusts for a factor that is in the path between the genetic information and the examined outcome, the relationship of the genetic information and the outcome could be ‘masked’. In studies which reported information from unadjusted analyses, the relative ranking across genetic markers was very similar to those from adjusted analyses.
An additional element of heterogeneity may have been introduced by combining original and digitized data. Digitizing is as best as accurate as the digitized figure. If the figure is of poor quality, digitizing may introduce errors compared to the primary data. In our case, almost all figures were of good quality. Limiting the analyses to the 13 strata from which we had primary data did not change the main findings. Nevertheless, it would be desirable that primary studies publish Supplementary material with tabular data or with complete and good quality graphs.
Finally, we used bin sizes of ∼30 cM. While this choice is somewhat arbitrary, it has some motivations from theory and simulation studies. The question of bin size for GSMA has been empirically explored by Hermanowski et al. [40] and discussed by Wise et al. [14] who recommended a bin width of ∼30 cM, being wide enough to limit the correlation between adjacent bins and not too wide to avoid including distinct peak LOD scores from different studies within the same bin. Overall, the effect of different bin definitions is likely to be minor compared to other sources of error and heterogeneity in the genome-wide linkage studies analyzed [57].
Conclusion
To our knowledge, this is the first study to combine the findings from numerous existing genome linkage scans conducted for continuous traits of renal phenotypes. Overall, even after combining 22 genome scans, little evidence of consistent linkage on specific chromosomal regions was detected. While heterogeneity across populations, methodologies and study designs likely explains this lack of agreement, it is also possible that genome-wide scan methodologies have insufficient resolution for investigating complex renal phenotypes. Used in a complementary role to genome-wide association approaches, they may be valuable for uncovering private gene mutations that contribute to renal phenotypes within families.
Supplementary data
Supplementary data is available online at http://ndt.oxfordjournals.org/.
Acknowledgments
We gratefully acknowledge the help of Adrienne Williams, Julie Zeigler and Sarah Ialacci in collating and supplying data files and Dr Georgios Kitsios for providing information on which chromosomal locations correspond to each bin.
Funding. This work was supported by (i) National Institutes of Health (DK066992) to Dr Rao; (ii) Grant 1 UL1 RR025752-02, Tufts Clinical and Translational Science Institute; (iii) the National Heart, Lung and Blood Institute’s Framingham Heart Study (N01-HC-25195); (iv) Wake Forest analyses were supported by National Institutes of Health grants (R01 DK053591 to D.W.B.), (R01 HL56266 to B.I.F.), (R01 DK070941 to B.I.F.), (DK084149 to B.I.F.) and, in part, by the General Clinical Research Center of the Wake Forest University School of Medicine grant M01 RR07122; (v) HyperGEN study (HL54471, HL54472, HL54473, HL54495, HL54496, HL54497, HL54509, HL54515 and HL55673); (vi) (R01-AG18734); (vii) The FIND study was supported by Research Grants (U01DK57292, U01DK57249, U01DK57295, U01DK57298, U01DK57300, U01DK57303, U01DK57304 and U01DK57329) from the NIDDK and, in part, by the Intramural Research Program of the NIDDK. This work was also supported by the National Center for Research Resources for the General Clinical Research Center grants: Case Western Reserve University (M01-RR-000080), Wake Forest University (M01-RR-07122), Harbor-UCLA Medical Center (M01-RR-00425), College of Medicine-University of California Irvine (M01-RR-00827-29), University of New Mexico HSC (M01-RR-00997) and Frederic C. Bartter (M01-RR-01346).
Genotyping for the original studies was performed by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University, contract no. N01-HG-65403.
Conflict of interest statement. None declared.
References
- 1.Bowden DW. Genetics of kidney disease. Kidney Int. 2003;63(Suppl 83):S8–S12. doi: 10.1046/j.1523-1755.63.s83.3.x. [DOI] [PubMed] [Google Scholar]
- 2.Freedman BI, Spray BJ, Tuttle AB, et al. The familial risk of end-stage renal disease in African Americans. Am J Kidney Dis. 1993;21:387–393. doi: 10.1016/s0272-6386(12)80266-6. [DOI] [PubMed] [Google Scholar]
- 3.Lei HH, Perneger TV, Klag MJ, et al. Familial aggregation of renal disease in a population-based case-control study. J Am Soc Nephrol. 1998;9:1270–1276. doi: 10.1681/ASN.V971270. [DOI] [PubMed] [Google Scholar]
- 4.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol. 2004;15:2457–2461. doi: 10.1097/01.ASN.0000135972.13396.6F. [DOI] [PubMed] [Google Scholar]
- 5.Fox CS, Yang Q, Guo CY, et al. Genome-wide linkage analysis to urinary microalbuminuria in a community-based sample: the Framingham Heart Study. Kidney Int. 2005;67:70–74. doi: 10.1111/j.1523-1755.2005.00056.x. [DOI] [PubMed] [Google Scholar]
- 6.Langefeld C, Beck S, Bowden D, et al. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am J Kidney Dis. 2004;43:796–800. doi: 10.1053/j.ajkd.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 7.Kottgen A, Glazer NL, Dehghan A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman BI, Hicks PJ, Bostrom MA, et al. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int. 2009;75:736–745. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp JB, Smith MW, Nelson GW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imperatore G, Hanson RL, Pettitt DJ, et al. Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Pima Diabetes Genes Group. Diabetes. 1998;47:821–830. doi: 10.2337/diabetes.47.5.821. [DOI] [PubMed] [Google Scholar]
- 12.Altmüller J, Palmer LJ, Fischer G, et al. Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet. 2001;69:936–950. doi: 10.1086/324069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiryluk K. Quantitative genetics of renal function: tackling complexities of the eGFR phenotype in gene mapping studies. Kidney Int. 2008;74:1109–1112. doi: 10.1038/ki.2008.479. [DOI] [PubMed] [Google Scholar]
- 14.Wise LH, Lanchbury JS, Lewis CM. Meta-analysis of genome searches. Ann Hum Genet. 1999;63:263–272. doi: 10.1046/j.1469-1809.1999.6330263.x. [DOI] [PubMed] [Google Scholar]
- 15.Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28:123–137. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- 16.Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 17.Levey AS, Greene T, Kusek JW, et al. A simplified equation to predict glomerular filtration rate from serum creatinine. J Am Soc Nephrol. 2000;11:155A. [Google Scholar]
- 18.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 19.Shadish WR, Brasil IC, Illingworth DA, et al. Using UnGraph to extract data from image files: verification of reliability and validity. Behav Res Methods. 2009;41:177–183. doi: 10.3758/BRM.41.1.177. [DOI] [PubMed] [Google Scholar]
- 20.Levinson DF, Levinson MD, Segurado R, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, part I: methods and power analysis. Am J Hum Genet. 2003;73:17–33. doi: 10.1086/376548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trikalinos TA, Karvouni A, Zintzaras E, et al. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- 22.Arar N, Nath S, Thameem F, et al. Genome-wide scans for microalbuminuria in Mexican Americans: the San Antonio Family Heart Study. Genet Med. 2007;9:80–87. doi: 10.1097/gim.0b013e31803068ec. [DOI] [PubMed] [Google Scholar]
- 23.Arar NH, Voruganti VS, Nath SD, et al. A genome-wide search for linkage to chronic kidney disease in a community-based sample: the SAFHS. Nephrol Dial Transplant. 2008;23:3184–3191. doi: 10.1093/ndt/gfn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mottl AK, Vupputuri S, Cole SA, et al. Linkage analysis of glomerular filtration rate in American Indians. Kidney Int. 2008;74:1185–1191. doi: 10.1038/ki.2008.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mottl AK, Vupputuri S, Cole SA, et al. Linkage analysis of albuminuria. J Am Soc Nephrol. 2009;20:1597–1606. doi: 10.1681/ASN.2008080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt SC, Coon H, Hasstedt SJ, et al. Linkage of serum creatinine and glomerular filtration rate to chromosome 2 in Utah pedigrees. Am J Hypertens. 2004;17:511–515. doi: 10.1016/j.amjhyper.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Pattaro C, Aulchenko YS, Isaacs A, et al. Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int. 2009;76:297–306. doi: 10.1038/ki.2009.135. [DOI] [PubMed] [Google Scholar]
- 28.Iyengar SK, Abboud HE, Goddard KA, et al. Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: the family investigation of nephropathy and diabetes (FIND) Diabetes. 2007;56:1577–1585. doi: 10.2337/db06-1154. [DOI] [PubMed] [Google Scholar]
- 29.Schelling JR, Abboud HE, Nicholas SB, et al. Genome-wide scan for estimated glomerular filtration rate in multi-ethnic diabetic populations: the Family Investigation of Nephropathy and Diabetes (FIND) Diabetes. 2008;57:235–243. doi: 10.2337/db07-0313. [DOI] [PubMed] [Google Scholar]
- 30.Krolewski AS, Poznik GD, Placha G, et al. A genome-wide linkage scan for genes controlling variation in urinary albumin excretion in type II diabetes. Kidney Int. 2006;69:129–136. doi: 10.1038/sj.ki.5000023. [DOI] [PubMed] [Google Scholar]
- 31.Puppala S, Arya R, Thameem F, et al. Genotype by diabetes interaction effects on the detection of linkage of glomerular filtration rate to a region on chromosome 2q in Mexican Americans. Diabetes. 2007;56:2818–2828. doi: 10.2337/db06-0984. [DOI] [PubMed] [Google Scholar]
- 32.Chen G, Adeyemo AA, Zhou J, et al. A genome-wide search for linkage to renal function phenotypes in West Africans with type 2 diabetes. Am J Kidney Dis. 2007;49:394–400. doi: 10.1053/j.ajkd.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Freedman BI, Bowden DW, Rich SS, et al. Genome-wide linkage scans for renal function and albuminuria in Type 2 diabetes mellitus: the Diabetes Heart Study. Diabet Med. 2008;25:268–276. doi: 10.1111/j.1464-5491.2007.02361.x. [DOI] [PubMed] [Google Scholar]
- 34.Leon JM, Freedman BI, Miller MB, et al. Genome scan of glomerular filtration rate and albuminuria: the HyperGEN study. Nephrol Dial Transplant. 2007;22:763–771. doi: 10.1093/ndt/gfl674. [DOI] [PubMed] [Google Scholar]
- 35.Turner ST, Kardia SL, Mosley TH, et al. Influence of genomic loci on measures of chronic kidney disease in hypertensive sibships. J Am Soc Nephrol. 2006;17:2048–2055. doi: 10.1681/ASN.2005121254. [DOI] [PubMed] [Google Scholar]
- 36.Freedman BI, Bowden DW, Rich SS, et al. A genome scan for all-cause end-stage renal disease in African Americans. Nephrol Dial Transplant. 2005;20:712–718. doi: 10.1093/ndt/gfh704. [DOI] [PubMed] [Google Scholar]
- 37.Placha G, Poznik GD, Dunn J, et al. A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes. 2006;55:3358–3365. doi: 10.2337/db06-0781. [DOI] [PubMed] [Google Scholar]
- 38.Forabosco P, Neuhausen SL, Greco L, et al. Meta-analysis of genome-wide linkage studies in celiac disease. Hum Hered. 2009;68:223–230. doi: 10.1159/000228920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng MY, Levinson DF, Faraone SV, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. 2009;14:774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermanowski J, Bouzigon E, Forabosco P, et al. Meta-analysis of genome-wide linkage studies for multiple sclerosis, using an extended GSMA method. Eur J Hum Genet. 2007;15:703–710. doi: 10.1038/sj.ejhg.5201818. [DOI] [PubMed] [Google Scholar]
- 41.Zintzaras E, Kitsios G. Identification of chromosomal regions linked to premature myocardial infarction: a meta-analysis of whole-genome searches. J Hum Genet. 2006;51:1015–1021. doi: 10.1007/s10038-006-0053-x. [DOI] [PubMed] [Google Scholar]
- 42.Zintzaras E, Kitsios G, Harrison GA, et al. Heterogeneity-based genome search meta-analysis for preeclampsia. Hum Genet. 2006;120:360–370. doi: 10.1007/s00439-006-0214-1. [DOI] [PubMed] [Google Scholar]
- 43.Kottgen A. Genome-wide association studies in nephrology research. Am J Kidney Dis. 2010;56:743–758. doi: 10.1053/j.ajkd.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 44.Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roeder K, Bacanu SA, Wasserman L, et al. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shriner D, Baye TM, Padilla MA, et al. Commonality of functional annotation: a method for prioritization of candidate genes from genome-wide linkage studies. Nucleic Acids Res. 2008;36:e26. doi: 10.1093/nar/gkn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denham S, Koppelman GH, Blakey J, et al. Meta-analysis of genome-wide linkage studies of asthma and related traits. Respir Res. 2008;9:38. doi: 10.1186/1465-9921-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Li C, Xu H, et al. Novel non-HLA-susceptible regions determined by meta-analysis of four genomewide scans for ankylosing spondylitis. J Genet. 2008;87:75–81. doi: 10.1007/s12041-008-0010-x. [DOI] [PubMed] [Google Scholar]
- 49.Koivukoski L, Fisher SA, Kanninen T, et al. Meta-analysis of genome-wide scans for hypertension and blood pressure in Caucasians shows evidence of susceptibility regions on chromosomes 2 and 3. Hum Mol Genet. 2004;13:2325–2332. doi: 10.1093/hmg/ddh237. [DOI] [PubMed] [Google Scholar]
- 50.van Heel DA, Fisher SA, Kirby A, et al. Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum Mol Genet. 2004;13:763–770. doi: 10.1093/hmg/ddh090. [DOI] [PubMed] [Google Scholar]
- 51.Bouzigon E, Forabosco P, Koppelman GH, et al. Meta-analysis of 20 genome-wide linkage studies evidenced new regions linked to asthma and atopy. Eur J Hum Genet. 2010;18:700–706. doi: 10.1038/ejhg.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rybicki BA, Iyengar SK, Harris T, et al. The distribution of long range admixture linkage disequilibrium in an African-American population. Hum Hered. 2002;53:187–196. doi: 10.1159/000066193. [DOI] [PubMed] [Google Scholar]
- 53.National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification and stratification. Am J Kidney Dis. 2002;39:S1–S266. [PubMed] [Google Scholar]
- 54.Perkins BA, Ficociello LH, Silva KH, et al. Regression of microalbuminuria in type 1 diabetes. N Engl J Med. 2003;348:2285–2293. doi: 10.1056/NEJMoa021835. [DOI] [PubMed] [Google Scholar]
- 55.Garg JP, Bakris GL. Microalbuminuria: marker of vascular dysfunction, risk factor for cardiovascular disease. Vasc Med. 2002;7:35–43. doi: 10.1191/1358863x02vm412ra. [DOI] [PubMed] [Google Scholar]
- 56.Mogensen CE, Vestbo E, Poulsen PL, et al. Microalbuminuria and potential confounders. A review and some observations on variability of urinary albumin excretion. Diabetes Care. 1995;18:572–581. doi: 10.2337/diacare.18.4.572. [DOI] [PubMed] [Google Scholar]
- 57.Forabosco P, Ng MY, Bouzigon E, et al. Data acquisition for meta-analysis of genome-wide linkage studies using the genome search meta-analysis method. Hum Hered. 2007;64:74–81. doi: 10.1159/000101425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.