Abstract
Protein Phosphatase 1 (PP1) is a major serine/threonine-phosphatase whose activity is dependent on its binding to regulatory subunits known as PP1 interacting proteins (PIPs), responsible for targeting PP1 to a specific cellular location, specifying its substrate or regulating its action. Today, more than 200 PIPs have been described involving PP1 in panoply of cellular mechanisms. Moreover, several PIPs have been identified that are tissue and event specific. In addition, the diversity of PP1/PIP complexes can further be achieved by the existence of several PP1 isoforms that can bind preferentially to a certain PIP. Thus, PP1/PIP complexes are highly specific for a particular function in the cell, and as such, they are excellent pharmacological targets. Hence, an in-depth survey was taken to identify specific PP1α PIPs in human brain by a high-throughput Yeast Two-Hybrid approach. Sixty-six proteins were recognized to bind PP1α, 39 being novel PIPs. A large protein interaction databases search was also performed to integrate with the results of the PP1α Human Brain Yeast Two-Hybrid and a total of 246 interactions were retrieved.
Introduction
The correct targeting and localization of proteins to specific subcellular compartments represent an important biological mechanism for regulating cellular function. These processes are of pivotal importance to correct cell development and differentiation, apoptosis, intercellular communication, proteostasis, and metabolism. Therefore, elucidating the constituent molecular parts of these signaling events, where functionally related proteins are arranged in close proximity, represents a fundamental step toward understanding the function of biological systems.
Among posttranslational modifications, reversible protein phosphorylation mediates most of signal transduction pathways in living cells, through the action of protein kinases and phosphatases (Cohen, 2001). These events occur under tight and transient regulation and abnormal phosphorylation mechanisms lead to disorders such as cancer, diabetes, heart failure,and neurological degeneration (Cohen, 2001; da Cruz e Silva et al., 1995a; Fardilha et al., 2010; Gandy and Greengard, 1994; Neumann, 2002; Sridhar et al., 2000).
Of all Ser/Thr protein phosphatases, Protein Phosphatase 1 (PP1) forms a major class and is highly conserved among all eukaryotes (Lin et al., 1999). Three genes are known to encode PP1 catalytic subunits, termed PP1α, PP1β, and PP1γ, with diversity increased by alternative splicing (da Cruz e Silva et al., 1995b). PP1 regulates a variety of cellular events through the dephosphorylation of multiple substrates and its multifunctionality is due to its association with different regulators and/or targeting subunits (Bollen, 2001; Ceulemans and Bollen, 2004; Fardilha et al., 2010, 2011a) known as PP1 Interacting Proteins (PIPs). The PP1 isoforms are highly conserved across their large catalytic domain, but are divergent at the N and C termini. Thus, PIPs bind to the unique C terminus to direct their isoform specific activities. To exert their dephosphorylation reactions that are important in time and space, the diverse functions of PP1 must be independently regulated. For this reason, PIPs are believed to be much more specific for individual functions and are therefore better targets for specific pathways (Ceulemans and Bollen, 2004; Cohen, 2002; Fardilha et al., 2010, 2011a; Virshup and Shenolikar, 2009).
The large majority of PIPs contain a degenerate, so-called RVxF-motif that conforms to the consensus sequence [R/K]-X0-1-[V/I]-[F/W], where X denotes any residue except proline (Bollen, 2001; Wakula et al., 2003). This motif binds with high affinity to a hydrophobic channel that is remote from the catalytic site of PP1 (L288-M290-C291) (Gibbons et al., 2005). The binding of the RVxF-motif by itself has no major effects on the conformation or activity of PP1 (Egloff et al., 1997). However, RVxF-mediated anchoring of PP1 promotes the occupation of secondary, lower affinity binding sites, and this often does affect the activity and/or substrate specificity of PP1 (Bollen, 2001; Wakula et al., 2003). The RVxF-motif is present in about one-third of all eukaryotic proteins but only a small fraction are PIPs. It seems that RVxF-consensus sequences function as PP1 interaction sites only when they are present in a flexible and exposed loop that can be modeled into a β-strand (Wakula et al., 2003).
Other PP1-binding motifs (PP1-BMs) have been described, F-X-X-R-X-R, present in several PP1 interactors (Ayllon et al., 2002), and the MyPhoNE motif, RXXQ[VIL][KR]X[YW], present in MYPT-1 (Terrak et al., 2004). An additional generic PP1-binding motif was identified, the SILK-motif: [GS]-IL-[KR]. It was first described for I2, a specific PP1 inhibitor (Hurley et al., 2007; Lin et al., 2005). This motif is present in nearly 10% of proteins containing the RVxF-motif and is normally N-terminal to it. The SILK and RVxF-motifs are functionally interchangeable and can both be essential for PP1 anchoring. More recently, work from Bollen and coworkers allowed the redefinition of the RVxF motif and its flanking residues based on the sequences of 143 PIPs: [KRL]-[KRSTAMVHNQ]-[VI]-{FIMYDP}-[FW] (Hendrickx et al., 2009).
The existence of common binding sites for PIPs explains why a relatively small protein such as PP1 can interact with numerous different regulatory proteins and why the binding of most regulatory subunits is mutually exclusive. The relative abundance of each PP1 isoform may be an important factor in determining the composition of numerous PP1 holoenzymes and the relative contribution of each PP1 isoform to different biological functions.
The broad in vitro substrate specificity of PP1 leads to the idea that the enzymatic specificity is mainly dictated by the PIPs. Thus, a complete understanding of PP1 function requires the identification of the associated subunits that direct PP1 specific functions, as well as functional analysis of PP1 holoenzymes. A variety of approaches has identified more than 100 mammalian proteins known to interact with PP1 (Fardilha et al., 2010). These PIPs function as inhibitors, substrate specifiers, and substrate targeting proteins, or a combination thereof. Sometimes PP1 interactors are themselves substrates for associated PP1 (Bollen, 2001; Ceulemans and Bollen, 2004; Fardilha et al., 2010, 2011a). Given the number of protein phosphatases and phosphoprotein substrates encoded in the human genome, a large number of PIPs surely remain to be discovered. Moreover, relatively little is known about isoform specific PP1 regulators. Recently, we have characterized the human testis PP1γ interactome and have shown that there are isoform tissue-specific PIPs (Fardilha et al., 2011b). Some PIPs were identified when PP1γ1 was used as bait while others were only obtained when the bait was PP1γ2. Even more interesting was the fact that the majority of PIPs obtained with a single bait were with the unique C-terminal of PP1γ2 (Fardilha et al., 2011b). Thus, clearly, there exists a PP1 isoform specificity in what concerns PIPs binding that is highly relevant for PP1 isoform particular function.
The majority of the putative PP1 interactions proposed derived primarily from biochemical approaches, high-throughput Yeast Two-Hybrid (YTH) screens, mass spectrometry and in silico screenings (Bennett et al., 2006; Fardilha et al., 2011b; Flores-Delgado et al., 2007; Hendrickx et al., 2009; Hrabchak and Varmuza, 2004; Moorhead et al., 2008; Trinkle-Mulcahy et al., 2006). The YTH system provides a sensitive method for detecting relatively weak and transient protein interactions (Fields and Song, 1989). High-throughput YTH screens, which generated most of the binary protein interaction data currently available, are providing samples of complete interactomes. Even though the resulting interaction mapping lacks sufficient coverage and dynamic information for a complete interactome, they greatly increased our knowledge, although understanding the global organization of proteomes is still far from complete.
Of all mammalian tissues, the brain expresses the highest levels of protein kinases and phosphatases, and PP1 is highly expressed both in neurons and glia (da Cruz e Silva et al., 1995b; Ouimet et al., 1995). It is increasingly evident that protein phosphorylation is a fundamental process associated with memory, learning, and brain function, with prominent roles in the processing of neuronal signals and in short-term and long-term modulation of synaptic transmission (Graff et al., 2010; Koshibu et al., 2009). Because PP1α is known to be highly enriched in the brain (da Cruz e Silva et al., 1995b) the main goal of this work was to identify the proteins expressed in human brain that interact with PP1α by the YTH method. Indeed, we identified 66 PIPs of which 39 represent novel interactions. Also, we integrated the YTH results with protein–protein interactions data from several sources (previously PIPs described in the literature and public Web repositories) and developed physical maps to validate in silico the novel interactions obtained in our YTH. The PP1α interactome thus obtained allowed the identification of novel key proteins in signaling pathways that were not previously taught as such, addressing novel functions to PP1α in the brain.
Materials and Methods
Human brain library screening by Yeast Two-Hybrid
The PP1α cDNA was directionally subcloned into EcoRI/BamHI digested pAS2-1 (GAL4 binding domain expression vector) to produce pAS-PP1α. This expression vector was first used to confirm the expression of the resulting fusion proteins (GAL4-PP1α) in yeast strain AH109. For library screening, the yeast strain AH109 transformed with pAS-PP1α, was mated with yeast strain Y187 expressing the human brain cDNA library (from an adult male brain, Clontech, HL4004AH) in the pACT-2 vector (Gal4 activation domain expression vector). Half the mating mixture was plated onto high stringency medium (quadruple dropout medium (QDO): SD/-Ade/-His/-Leu/-Trp) and the other half onto low stringency medium (triple dropout medium (TDO): SD/-His/-Leu/-Trp), and the plates were incubated at 30°C. Colonies obtained in the low stringency plates were replica plated onto high stringency medium. Finally, all high stringency surviving colonies were plated onto selective medium containing X-α-Gal and incubated at 30°C to check for MEL-1 expression (indicated by the appearance of a blue color). All the YTH reagents were purchased from Clontech, Saint-Germain-en-Laye, France. All other nonspecified reagents were purchased from Sigma-Aldrich, Portugal.
Recovery of plasmids from yeast and sequence analysis
Yeast plasmid DNA was recovered and used to transform Escherichia coli XL1-Blue. Plasmid DNA was obtained from each resulting bacterial colony and digested with the restriction enzyme HindIII (NEB, Ipswich, MA, USA) to identify the corresponding library plasmids. DNA sequence analysis was performed using an Automated DNA Sequencer (Applied Biosystems, Carlsbad, CA, USA) using the GAL4-AD primer—TACCACTACAATGGATG (Clontech). The DNA sequences obtained were compared to the GenBank database, using the BLAST algorithm, to identify the corresponding encoded proteins.
Databases search of PIPs
The human specific PP1α interactors available in eight online databases: BioGRID, BIND, STRING, HPRD, IntAct, MINT, Reactome-FLS, and InnateDB, were retrieved. Afterward, an exhaustive analysis to the PIPs was made and the proteins were grouped. The PP1α interaction map was made using Cytoscape (Shannon et al., 2003).
Results
Identification of 66 proteins by Yeast Two-Hybrid Screening of a human brain cDNA library
In order to identify PP1α Interacting Proteins expressed in the human brain, an YTH screen of a human brain cDNA library was carried out using full-length human PP1α. The screen yielding 298 positive clones from a total of 2×107 clones screened, corresponding to 66 different protein–protein interactions. After partial or complete sequence analysis (depending on the length of the positive clone's cDNAs), in silico searches of the GenBank database allowed their identification and classification into four separate groups. Two groups are listed on Tables 1 and 2. The third group corresponds to clones putatively encoding novel PIPs with homology to genomic sequences and lists positives where the Genbank sequence similarity, although apparent, did not correspond to an annotated gene (Appendix 1). These cDNA clones may correspond to transcripts derived from novel, previously unidentified genes. The fourth group corresponds to possible false positive hits. Table 1 (known PP1 interacting proteins) lists positives encoding previously identified PIPs, such as Nek2 (Helps et al., 2000b) and PPP1R9B (Allen et al., 1997a). Table 2 (novel PIPs) lists positives encoding known proteins that were not previously associated with PP1 and uncharacterized proteins (present in the database) that are novel PP1 interactors.
Table 1.
Known PP1 Interacting Proteins
| |
|
PP1-BMs |
|
|
|
|
|---|---|---|---|---|---|---|
| Clone ID | No. Clones | RVxF | SILK | Chr | Uniprot accession number | Reference of interaction discovery |
| AATK | 1 | KAVSF | 17 | Q6ZMQ8 | (Gagnon et al., 2007) | |
| AXIN1 | 4 | RVAF/RVEF | SILK | 3 | Q96S65 | (Luo et al., 2007) |
| C1QA | 16 | RSLGF/KGLF | 1 | P02745 | (Fardilha et al., 2011b) | |
| CNST | 25 | RRVRF | SILK | 1 | Q6PJW8 | (Fardilha et al., 2011b) |
| C9orf75 | 45 | KISF / RAIRW | 9 | Q4KMQ1 | (Esteves, 2008; Trinkle-Mulcahy et al., 2006) | |
| KIAA1949 | 4 | KISF | 6 | Q6NYC8 | (Kao et al., 2007; Trinkle-Mulcahy et al., 2006) | |
| LAP1B | 14 | REVRF/ KVNF/ KVKF | SILK | 1 | Q5JTV8 | (Santos, 2009) |
| NEK2 | 2 | KVHF | 1 | P51955 | (Helps et al., 2000a) | |
| PHACTR3 | 2 | RNIF | 20 | Q96KR7 | (Sagara et al., 2003) | |
| PPP1R2 | 1 | KLHY | GILK | 3 | P41236 | (Huang and Glinsmann, 1976) |
| PPP1R3C | 5 | KRVVF / KNVSF /RITF / KIEF | 10 | Q9UQK1 | (Doherty et al., 1996) | |
| PPP1R3D | 2 | RVQF / LRVRF | 6 | O95685 | (Armstrong et al., 1997) | |
| PPP1R3E | 1 | RVRF | 14 | Q9H7J1 | (Ceulemans et al., 2002; Munro et al., 2005) | |
| PPP1R9B | 14 | RKIHF | 17 | Q96SB3 | (Allen et al., 1997a) | |
| PPP1R13A | 17 | RVKF | 1 | Q13625 | (Helps et al., 1995) | |
| PPP1R13B | 5 | LRVRF | 14 | Q96KQ4 | (Helps et al., 1995) | |
| PPP1R3G | 1 | KRVQF | 6 | B7ZBB8 | (Ceulemans et al., 2002) | |
| PPP1R13L | 2 | 19 | Q8WUF5 | (Colland et al., 2004) | ||
| PPP1R15B | 3 | KKVTF | 1 | Q5SWA1 | (Jousse et al., 2003) | |
| PPP1R16A | 1 | KQVLF | 8 | Q96I34 | (Skinner and Saltiel, 2001) | |
| RANBP9 | 20 | RMIHF | 6 | Q96S59 | (Fardilha et al., 2011b) | |
| RIF1 | 9 | KKIAF/ RRVSF | SILK | 2 | Q5UIP0 | (Moorhead et al., 2008; Trinkle-Mulcahy et al., 2006) |
| SH3RF2 | 4 | KTVRF | 5 | Q8TEC5 | (Chen et al., 2009) | |
| STAU1 | 16 | RKVTF | 20 | O95793 | (Monshausen et al., 2002) | |
| WBP11 | 1 | RKVGF / LSVRF | SILK | 12 | Q9Y2W2 | (Llorian et al., 2004) |
| YLPM1 | 8 | RVGF / KRVRW / RAIGF | 14 | P49750 | (Tran et al., 2004; Ulke-Lemee et al., 2007) | |
| ZFYVE9 | 3 | RRVWF / KVIRW | 1 | O95405 | (Bennett and Alphey, 2002; Colland et al., 2004) | |
Number of clones indicate the count of isolated cDNA clones for the respective protein.
PP1-BMs, PP1 binding motifs; Chr, chromosome.
Table 2.
Novel PP1 Interacting Proteins
| |
|
PP1BMs |
|
|
|
|---|---|---|---|---|---|
| Clone ID | No. Clones | RVxF | SILK | Chr | Uniprot accession number |
| ANKRD15 | 2 | 9 | Q14678 | ||
| BTBD10 | 1 | RHVDF | 11 | Q9BSF8 | |
| CLCN2 | 1 | 3 | P51788 | ||
| CEP170 | 1 | RILF | 1 | Q5SW79 | |
| CLTC | 1 | RAIQF | GILR | 17 | Q00610 |
| CKB | 1 | 14 | P12277 | ||
| CNP1 | 1 | KIFF | 17 | P09543 | |
| CNTN1 | 2 | LTITW | 12 | Q12860 | |
| CRK | 1 | 17 | P46108 | ||
| CXXC1 | 1 | 18 | Q9P0U4 | ||
| DCTN1 | 2 | KIKF / KVTF | SILK | 2 | Q14203 |
| DEAF1 | 1 | 11 | O75398 | ||
| FRMPD4 | 1 | KVRF / KVSF | X | Q14CM0 | |
| GLIPR1L2 | 1 | 12 | Q4G1C9 | ||
| GLTSCR2 | 1 | 19 | Q9NZM5 | ||
| IBTK | 1 | KKVSF | 6 | Q9P2D0 | |
| IIP45 | 1 | RVTF | 1 | Q5JXC2 | |
| JPH3 | 1 | 16 | Q8WXH2 | ||
| KCTD20 | 3 | RHVDF | 6 | Q7Z5Y7 | |
| KIAA0460 | 1 | RVGW | SILK | 1 | Q5VT52 |
| KIAA1377 | 8 | KLRW | SILK | 11 | Q9P2H0 |
| LPIN2 | 1 | 18 | Q92539 | ||
| MAFG | 1 | 17 | O15525 | ||
| MAL2 | 1 | 8 | Q969L2 | ||
| MAP4K4 | 2 | 2 | O95819 | ||
| NDP | 2 | X | Q00604 | ||
| PHC1 | 2 | 12 | P78364 | ||
| PIAS1 | 1 | 15 | O75925 | ||
| PIAS3 | 1 | 1 | Q9Y6X2 | ||
| PREX1 | 1 | KKVCF / KVIF | 20 | Q8TCU6 | |
| PRR16 | 2 | RVRF | 5 | Q569H4 | |
| SLC45A1 | 1 | RNVTF | GILK | 1 | Q9Y2W3 |
| SNCAIP | 6 | LRVTF | 5 | Q9Y6H5 | |
| SorLA-1 | 1 | 11 | Q92673 | ||
| SPRED1 | 1 | RHVSF | 15 | Q7Z699 | |
| UBE2Z | 1 | 17 | Q9H832 | ||
| ULK1 | 1 | 12 | O75385 | ||
| ZBTB11 | 1 | GILK | 3 | O95625 | |
| ZNF827 | 1 | LNVQF | 4 | Q17R98 | |
Number of clones indicate the count of isolated cDNA clones for the respective protein.
PP1-BMs, PP1 binding motifs; Chr, chromosome.
The YTH screen yielded 298 positive clones (Tables 1 and 2 and Appendix 1) that correspond to 66 different proteins (Tables 1 and 2), not considering PIPs with homology to genomic sequences, some with more than one hit.
Careful analysis of the YTH screen revealed that the most abundant interaction was detected with C9orf75 (Table 1). Thus, 45 positives out of the 298 detected encoded C9orf75, which corresponds to 15% of the positive clones obtained. This protein, also known as Taperin, was associated with autosomal-recessive nonsyndromic hearing loss by target genome capture combined with next-generation capture (Rehman et al., 2010) and by homozygosity mapping (Auluck et al., 2010). Immunolocalization studies of mouse cochlea by Rehman et al. (2010) demonstrated the presence of C9orf75/Taperin at the taper regions of hair cell stereocilia. Nevertheless, the function of C9orf75/Taperin still needs to be elucidated. Interestingly, together, seven well- known PIPs from the 27 correspond to 122 positive clones, 41% of the total interactions. Among them is C1QA (16 positive clones/5%), a protein related to the innate immune response and associated to oxidative stress responses in the brain (Luo et al., 2003; Ten et al., 2010) and PPP1R9B (14 positive clones/5%), also known as Spinophilin, highly abundant in neuronal spines where it is involved in synaptic transmission (Allen et al., 1997b; Feng et al., 2000).
Table 2 presents the list of known proteins or uncharacterized proteins (present only in the database) that have not been previously associated with PP1 and thus represent potential novel PP1 interacting proteins. This group of 39 different PIPs was encoded by 60 cDNAs. Twenty-nine proteins (74%) corresponded to single hits. Two independent positive clones codify to each of seven proteins. KIAA1377, SNCAIP (synuclein, α interacting protein), and KCTD20 (potassium channel tetramerisation domain containing 20) were encoded by eight, six, and three independent positive cDNA clones, respectively. KIAA1377, although of unknown function, is codified by several human brain ESTs (UniGene Hs.156352). SNCAIP, Synphilin-1, predominantly expressed in neurons is located in the cytoplasm and presynaptic nerve terminals and associated with synaptic vesicles, was initially identified as an α-synuclein-interacting protein (Engelender et al., 1999; Ribeiro et al., 2002). However, in several neurodegenerative disorders called α-synucleinopathies, such as Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy, synphilin-1, is mainly localized in neuronal and glial cytoplasmic inclusions (Wakabayashi et al., 2002, 2000). KCTD20 may have a voltage-gated potassium channel activity being involved in potassium ion transport (Gene Ontology GO:0005249) and has also been shown to be expressed in human brain (UniGene Hs.188757).
For all the already known PIPs and for the newly identified (Tables 1 and 2, respectively) a search for the PP1BMs RVxF and SILK was performed. All the proteins already known to be PIPs have the RVxF-motif, except PPP1R13L, and six contain the SILK-motif (Table 1). For the group of proteins that are novel PIPs (Table 2) there are 18 proteins that possess the RVxF motif but 54% does not, and also six have the SILK-motif. Only one PIP, ZBTB11, has a SILK-motif without an RVxF-BM.
Positives whose nucleotide sequence did not align with mRNA or cDNA sequences in the Genbank database are listed in Appendix 1. Thus, because the positives identified were derived from the brain cDNA library, the seven genomic sequences listed probably contain hitherto unidentified genes encoding putative novel PIPs, although the possibility of genomic contamination cannot be excluded.
Only three different possible false positive interactions were detected in our screen corresponding to proteins present in the mitochondria. One clone codifies for the 16s ribosomal RNA, one independent clone codifies for IDH2 (isocitrate dehydrogenase 2), and the other for CYCS (Cytocrome c). These clones might be an artefact from the cDNA library. The last two clones, although being mitochondrial proteins, are codified by nuclear chromosomes, so they may interact with PP1α nonetheless.
Retrieval of PP1α interacting proteins from free access online databases for protein interactions
Eight free access online databases for protein interactions were searched in order to retrieve PP1α interacting proteins (Appendix 2). The specificity of each database is different and with variations in the type and depth of their annotations. STRING is a database of known and predicted protein interactions. Reactome-Fls functions as a data mining resource. MINT focuses on experimentally verified protein–protein interactions mined from the scientific literature by curators. IntAct provides an open source database system and analysis tools for protein interaction data, interactions are derived from literature curation or direct user submissions. BioGRID is an online interaction repository with data compiled through curation efforts. InnateDB database has experimentally verified interactions involved in the innate immune response by integrating known interactions public databases together with curated data. HPRD is a platform to visually depict and integrate information, which is extracted from the literature. Finally, BIND documents molecular interactions by including high-throughput data submissions and hand-curated information gathered from the scientific literature.
After having collected all PP1α interactors from the above databases, the nomenclature for each protein was normalized for the Uniprot accession number in order for the interactions in each database to be compared. In total, we obtained 246 PP1α interactions from the databases and from our YTH screen (Appendix 2). The corresponding number of interactions found in each database are the following: Biogrid, 28; STRING, 113; HPRD, 70; Reactome, 52; InnateDB, 10; IntAct, 18; MINT, 33; and BIND, 30.
Discussion
Protein phosphorylation is critical to the health and vitality of eukaryotic cells and probably their major metabolic control mechanism. Consequently, of note, many disease processes are associated with abnormal phosphorylation of key proteins, thus suggesting a possible common molecular basis for some apparently unrelated and diverse diseases processes. The role played by protein phosphatases in health and disease, and particularly the involvement of PP1, makes it and the proteins that regulate its function (PIPs) excellent targets for pharmacological intervention. Indeed, there are different compounds that act by disrupting PP1/PIPs complex. For example salubrinal, a small molecule that protects cells from endoplasmic reticulum stress, inhibits the formation of PP1-PPP1R15A (Boyce et al., 2005). Treatment of cells with trichostatin A, a deacetylase inhibitor, disrupted HDAC6-PP1 complexes (Brush et al., 2004). In another study, a GADD34-derived peptide competitively disrupted the PP1/GADD34 complex, when added to cells (Kepp et al., 2009). PP1/PIPs complexes seem to be the future targets for several diseases, because PP1 has been associated to several disorders. Nevertheless, the specificity of PP1 targeting should be achieved by two means: PP1 isoform specificity and differential PIP association. These meaning that each PP1 isoform has its tissue and event-specific expression pattern and the same happens to the PIPs, leading to the formation of a specific PP1/PIP complex in a certain place and time event/mechanism. This highly specific complex can then be target by an inhibiting or stimulating molecule. In summary, determining PP1 isoform and tissue specific PIPs is crucial to identify particular complexes. We have previously identified PP1γ1 and PP1γ2 human testis interactome, identifying common and specific interactors in a total of 72 novel interactions (Fardilha et al., 2011b).
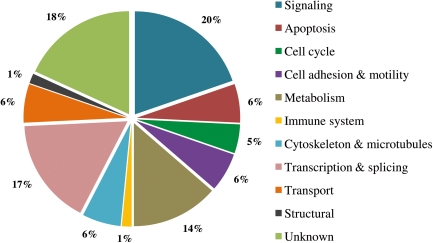
In order to identify potential PP1α interacting and regulating proteins, and to characterize the PP1α human brain interactome a large scale screen for PP1α binding proteins was performed, using the YTH system (Fardilha et al., 2004). A total of 66 proteins were identified from the 298 positive clones obtained that are expressed in human brain and bind PP1α. Furthermore, seven extra proteins codified by cDNA, but present in the database as genomic clones, were also obtained. Moreover, three proteins were considered to be false positive hits because they were mitochondrial proteins. One is, in fact, codified by mitochondrial DNA but the other two are codified by nuclear DNA, and afterward, translocated to the mitochondria. Thus, they could interact with PP1 during this process. These 10 proteins (Appendix 1) were left out of the functional analysis (Fig. 1). For the remainder of the clones (286 clones/96%), corresponding to 66 proteins, the most abundant group are proteins involved in signaling (20%), followed by proteins involved in splicing and transcription (17%) and metabolism (14%). The remaining proteins identified include components of the cytoskeleton, proteins involved in apoptosis, as well as cell cycle and transport (Fig. 1) and 18% still have unknown cellular function. This analyzes reflects the functional diversity of PP1, and all the proteins here mentioned and described illustrate the great multiplicity of cellular pathways and events in which PP1 is involved, and controls targets, and is regulated by its PIPs.
FIG. 1.
Distribution of the PP1α regulators according to function. Biological functions attributed to PIPs identified; according to databases by search based on functional motifs present, interacting proteins, cellular localization, and molecular function.
This YTH screen, as already verified in other YTH screens (Fardilha et al., 2011b), shows the inconsistency between the frequency of the cDNAs isolated and the total number of protein interactors identified. For example, the already known PIPs (Table 1) are encoded by 76% (226) of the total cDNAs isolated and account for 41% (27) of the PIPs identified. Neverthless, 20% (60) of the positive clones isolated code for 39 proteins (59%), all novel PIPs (Table 2). In the first case the ratio cDNA/protein is 8.4 and in the second case is 1.6. There are, in the case of the novel PIPs, more proteins identified from single hits (29 proteins). As stated by Fardilha et al. (2011b) this may be the explanation why these novel PIPs were not yet identified, or alternatively, they may have been discarded given that they are less abundant clones in the YTH screens, or they could have just been missed because usually the screens are not that exhaustive. However, as we took an in-depth exhaustive strategy, and decided to sequence all the YTH positive clones, we did not miss the rare interactions or low abundant proteins, difficult to pick up by proteomic methodologies. The identification of both abundant and rare known PIPs in our screens confirms the specificity and reliability of the YTH approach.
All, except one of the previously known PIPs possess an RVxF PP1BM. In contrast, only 46% of the novel PIPs (18 proteins) appear to have the consensus PP1BM. In fact, the number of proteins carrying PP1-BMs may be much higher if one considers the fact that some of the positive clones were not fully sequenced, and thus the analysis had to rely on the amino acid sequences available in the Genbank database, which may not reflect the frequent occurrence of alternatively spliced variants in the brain. The identification of previously identified PIPs and the presence of the PP1BM in the novel PIPs strengthens and validates the results of the YTH screen. In fact, the latter was present in 44 (67%) out of the 66 proteins identified.
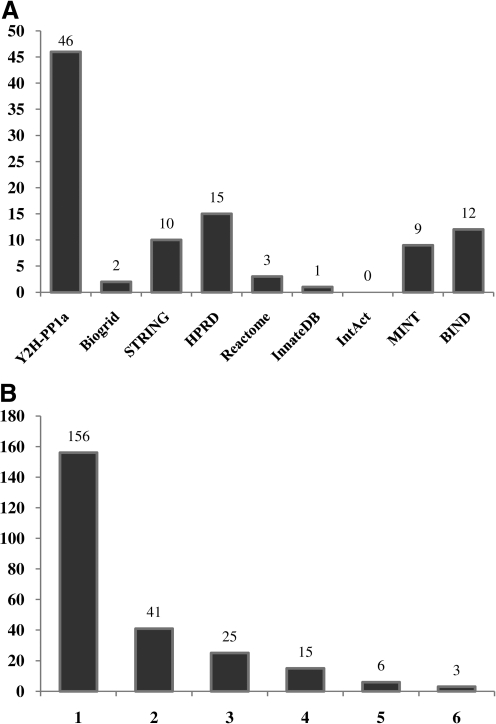
Eight protein–protein interaction databases were searched for PP1α interactions; only human proteins were considered in this study (Appendix 2). After a careful analysis of the interactions and organization of the retrieved results they were compared to the interactions obtained in the human brain PP1α YTH screen. From the PIPs identified in the YTH some were present in the databases searched (Fig. 2A) but other were not, the 39 novel PIPs (Table 2). Yet, the first bar in Figure 2A, corresponding to novel interactions, includes 46 proteins instead of 39. This is because some already characterized interactions are not yet present in the free online databases (e.g., PPP1R3E and PPP1R3G). The highest number of times an interaction appears in a database the stronger is the credibility of the interaction, considering the different criteria used for inclusion of the interactions in the different databases. Of course, this is also dependent on the detection methods of the interaction, some being more reliable than others (Appendix 2). Among the 246 interactions 156 interactions appeared in a single database (Fig. 2B). Forty-one interactions were common to two databases and 25 to three. Fifteen interactions were present in four databases, six interactions in five databases, and finally three interactions were present in six databases. These three PIPs are well-known PIPs: PPP1R15A (or GADD34), PPP1R8 (or NIPP1), and ZFYVE9 (or SARA), which strengths the above idea. The first is involved, together with PP1, in protein synthesis, regulation of calreticulin exposure, and TGF-β signaling (Brush et al., 2003; Kepp et al., 2009; Shi et al., 2004). The second, also with PP1, regulates RNA splicing (Tanuma et al., 2008; Trinkle-Mulcahy et al., 1999). ZFYVE9 is involved in signal transduction enhancing the recruitment of PP1 to the TGF-β receptor 1 (Bennett and Alphey, 2002; Shi et al., 2004).
FIG. 2.
Analysis of the PP1α interacting proteins in the databases searched. (A) Number of PIPs identified in the Yeast Two-Hybrid Screen (Y2H-PP1a) present in the different databases. (B) Number of PIPs identified in all the databases (including the Yeast Two-Hybrid Screen) that are present in only one database; two or more databases.
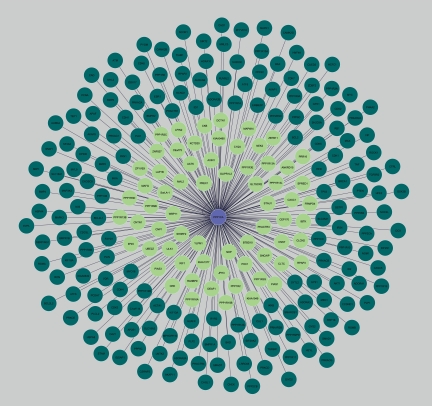
Finally, we used cytoscape (Shannon et al., 2003) to assemble a diagram of the interactome of PP1α (Fig. 3) using the databases interactions (Fig. 3, in dark green) plus our YTH interactions (Fig. 3, in light green). This interaction map clearly shows that our YTH screen contributes in many interactions to the overall picture. Considering the above discussion, we have added 39 new PIPs to the PP1α interactome. Also, we have identified at least three key molecules in the PP1α interactome, which are present in six databases. Indeed, PP1/PPP1R15A is a target for salubrinal in the treatment of Herpes simplex virus infection (Boyce et al., 2005). Thus, we propose that the complexes PP1/PPP1R8 and PP1/ZFYVE9 could also be relevant targets.
FIG. 3.
PP1α interactions mapping. PP1α Interacting proteins retrieved from online databases (dark green) and identified in the Human Brain Yeast Two- Hybrid Screen using PP1α as bait (light green). The total number of interactors is 247 and the proteins obtained in the Yeast Two-Hybrid Screen is 66. Of the 66 proteins 39 are novel PP1α interactors while the remainder are also common to one or more databases (Appendix 2).
Conclusions
In this study we report the identification of 66 proteins expressed in the human brain that bind PP1α. The majority of the detected interactions were novel (39/60%) and the functions of the new PIPs still need to be fully characterized. Only then will the precise roles of PP1α/brain-specific PIPs be fully elucidated. We have tried to do the complete picture of the PP1α interactome taking advantage of databases interaction information. We concluded that some of the PIPs identified in our YTH screen were present in other databases but some were novel. Thus, the present work added 39 novel interactions to PP1α interaction network. Together, our results and drawn conclusions allowed us to identify key PIPs in human brain that bind preferentially PP1α. This study also points to the importance of addressing PP1 isoforms as independent entities and consider PP1α/brain-specific PIP (or other PP1 isoform/PIP tissue specific complex) as an excellent target for specific pharmacologic therapy.
Appendices
Appendix Table A1.
Clones Putatively Encoding Novel PIPs With Homology to Genomic Sequences and Mitochondrial Proteins
| Clone ID | No. clones | Chromosome | NCBI reference sequence |
|---|---|---|---|
| Genomic clones | |||
| C-2190G12 | 1 | 14 | AL_139194 |
| C2genomic | 1 | 2 | NW_001838769 |
| C2genomic | 1 | 2 | NW_001838818 |
| C2genomic | 1 | 2 | NW_001838863 |
| C3genomic | 1 | 3 | NW_001838877 |
| C4genomic | 1 | 4 | NW_001838915 |
| C17genomic | 3 | 17 | NW_001838448 |
| Mitochondrial proteins | |||
| 16S ribosomal RNA | 1 | Mitochondria | AM_263191.1 |
| CYCS | 1 | 7 | NM_018947 |
| IDH2 | 1 | 15 | NM_002168 |
Number of clones indicate the count of isolated cDNA clones for the respective protein.
Appendix Table A2.
Complete List of PP1α Interacting Proteins Identified in the YTH Screen and Present in the Searched Databases
| Protein ID | Uniprot accession number | Interaction detection | Databases |
|---|---|---|---|
| AATK | Q6ZMQ8 | Y2H | Y2H-PP1a |
| ACTA1 | P68133 | AC | BIOGRID |
| ADORA1 | P30542 | P | STRING |
| ADORA2A | P29274 | P | STRING |
| ADORA2B | P29275 | P | STRING |
| ADORA3 | P33765 | P | STRING |
| AKAP1 | Q92667 | E, P | HPRD, STRING |
| AKAP11 | Q9UK44 | AC+B, P, PP | BIOGRID, STRING, HPRD |
| AKAP9 | Q99996 | E+IC, P | HPRD, STRING |
| AKT1 | P31749 | P | STRING |
| ANKRD15 | Q14678 | Y2H, PP | Y2H-PP1a, BIND, HPRD |
| AP2S1 | P53680 | E | Reactome |
| APAF | O14727 | CO-IP, AA+CO-IP, PP | InnateDB, IntAct, HPRD |
| ATF5 | Q9Y2D1 | P | STRING |
| ATF6 | P18850 | P | STRING |
| ATM | Q13315 | AA | IntAct |
| AURKA | O14965 | E | HPRD |
| AURKAB | Q96GD4 | AA, E | IntAct, Reactome |
| AXIN1 | Q96S65 | Y2H, CO-IP, P | Y2H-PP1a, MINT, STRING |
| BAD | Q92934 | CO-IP | IntAct, HPRD, STRING |
| BAX | Q07812 | CO-IP, PP | InnateDB, HPRD |
| BCL2 | P10415 | AA, E, P, PP | IntAct, Reactome, STRING, HPRD |
| BCL2L1 | Q07817 | AC+B, AA, P | BIOGRID, IntAct, STRING, HPRD, |
| BCL2L2 | Q92843 | AC+B, P, PP | BIOGRID, STRING, HPRD |
| BMP2 | P12643 | E, P | Reactome, STRING |
| BMP4 | P12644 | E, P | Reactome, STRING |
| BMP6 | P22004 | P | STRING |
| BMPR1A | P36894 | E | Reactome, STRING |
| BMPR1B | O00238 | E | Reactome |
| BMPR2 | Q13873 | E, P | Reactome, STRING |
| BRCA1 | P38398 | AC+B, CO-IP, P, PP | BIOGRID, IntAct, STRING, HPRD |
| BTBD10 | Q9BSF8 | Y2H, PD | Y2H-PP1a, InnateDB |
| C1orf114 | Q5TID7 | Y2H, PP | MINT, HPRD |
| C1QA | P02745 | Y2H | Y2H-PP1a |
| C9orf75 | Q4KMQ1 | Y2H, PP | Y2H-PP1a, MINT, HPRD |
| CAD | P27708 | PP | HPRD |
| CAMK2B | Q13554 | P | STRING |
| CAMK2G | Q13555 | P | STRING |
| CASC5 | Q8NG31 | PP, Y2H | HPRD, MINT |
| CCDND1 | P24385 | P | STRING |
| CDC5L | Q99459 | AC+B, CO-IP, E, P, PP | BIOGRID, MINT, Reactome, STRING, HPRD |
| CDH1 | P12830 | CO-IP, AA, PP | InnateDB, IntAct, HPRD |
| CDK1 | P06493 | AA, E, P | IntAct, Reactome, STRING |
| CDK2 | P24941 | AA, E | IntAct, Reactome |
| CDK3 | Q00526 | E | Reactome |
| CDK4 | P11802 | AA, E | IntAct, Reactome |
| CDK5 | Q00535 | E | Reactome |
| CDK6 | Q00534 | E | Reactome |
| CDK7 | P50613 | E | Reactome |
| CEP170 | Q5SW79 | Y2H | Y2H-PP1a |
| CHD1 | O14646 | P | STRING |
| CHEK | O96017 | E | Reactome |
| CHI3L1 | P36222 | P | STRING |
| CKB | P12277 | Y2H | Y2H-PP1a |
| CLCN2 | P51788 | Y2H | Y2H-PP1a |
| CLTC | Q00610 | Y2H | Y2H-PP1a |
| CNP1 | P09543 | Y2H | Y2H-PP1a |
| CNST | Q6PJW8 | Y2H | Y2H-PP1a |
| CNTN1 | Q12860 | Y2H | Y2H-PP1a |
| CRK | P46108 | Y2H | Y2H-PP1a |
| CSRNP2 | Q9H175 | Y2H, E, PP | BIND, MINT, Reactome, HPRD |
| CTRL | P40313 | P | STRING |
| CUED2 | Q9H467 | CO-IP | InnateDB |
| CUL1 | Q13616 | CO-IP, PP | IntAct, HPRD |
| CXXC1 | Q9P0U4 | Y2H | Y2H-PP1a |
| DCTN1 | Q14203 | Y2H | Y2H-PP1a |
| DCX | O43602 | E, P+PP | HPRD, STRING |
| DDX17 | Q92841 | P | STRING |
| DEAF1 | O75398 | Y2H | Y2H-PP1a |
| EED | O75530 | B | BIOGRID, STRING |
| EIF2AK2 | P19525 | AC+B, P, PP | BIOGRID, STRING, HPRD |
| EIF2S1 | P05198 | P+IM | STRING |
| EP300 | Q09472 | E | Reactome |
| ERBB2IP | Q96RT1 | E | Reactome |
| ESR1 | P03372 | AC | IntAct |
| EVPL | Q92817 | P | STRING |
| EXOSC8 | Q96B26 | P | STRING |
| FRMPD4 | Q14CM0 | Y2H | Y2H-PP1a |
| FXYD1 | O00168 | E, P | HPRD, STRING |
| GIYD2 | P50224 | P | STRING |
| GLIPR1L2 | Q4G1C9 | Y2H | Y2H-PP1a |
| GLTSCR2 | Q9NZM5 | Y2H | Y2H-PP1a |
| GPX1 | P07203 | E | Reactome |
| GSK3B | P49841 | AC, E, P | BIOGRID, Reactome,STRING |
| GSTP1 | P09211 | P | STRING |
| GYS1 | P13807 | P+IM | STRING |
| GYS2 | P54840 | P+IM | STRING |
| H2AFX | P16104 | E, P | HPRD, Reactome, STRING |
| HCFC1 | P51610 | AC+B, P | BIOGRID, STRING |
| HEYL | Q9NQ87 | Y2H, PP | MINT,BIND, HPRD |
| HFE2 | Q6ZVN8 | E | Reactome |
| HNF4A | P41235 | CROSSLINK | BIND |
| HOXA10 | P31260 | P | STRING |
| HSPA4 | P34932 | PP | HPRD |
| HSPA8 | P11142 | AC, P | BIOGRID, STRING |
| IBTK | Q9P2D0 | Y2H | Y2H-PP1a |
| ID2 | Q02363 | PP | HPRD |
| IDI1 | Q13907 | P | STRING |
| IIP45 | Q5JXC2 | Y2H | Y2H-PP1a |
| IKKA | O15111 | CO-IP | InnateDB |
| IKKB | O14920 | CO-IP | InnateDB |
| IQGAP1 | P46940 | P | STRING |
| IRS4 | O14654 | P | STRING |
| JPH3 | Q8WXH2 | Y2H | Y2H-PP1a |
| KCNQ1 | P51787 | AC+B, PP | BIOGRID, STRING |
| KCTD20 | Q7Z5Y7 | Y2H | Y2H-PP1a |
| KIAA0460 | Q5VT52 | Y2H | Y2H-PP1a |
| KIAA0649 | Q5T8A7 | Y2H | MINT, BIND |
| KIAA1377 | Q9P2H0 | Y2H | Y2H-PP1a |
| KIAA1949 | Q6NYC8 | Y2H, P, E | Y2H-PP1a, BIND, STRING, HPRD |
| KIF13A | Q9H1H9 | E | HPRD |
| KIF18A | Q8NI77 | Y2H, PP | MINT, HPRD |
| LAP1B | Q5JTV8 | Y2H | Y2H-PP1a, BIND |
| LMTK2 | Q8IWU2 | AC+B+Y2H, P | BIOGRID, STRING |
| LPIN2 | Q92539 | Y2H | Y2H-PP1a |
| LRRC68 | O75864 | Y2H | MINT |
| MAFG | O15525 | Y2H | Y2H-PP1a |
| MAL2 | Q969L2 | Y2H | Y2H-PP1a |
| MAP3K3 | Q99759 | AP, CO-IP | IntAct, MINT |
| MAP4K4 | O95819 | Y2H | Y2H-PP1a |
| MAPK1 | P28482 | E, P | HPRD, Reactome, STRING |
| MAPK3 | P27361 | E, P | HPRD, Reactome, STRING |
| MAX | P61244 | AA | IntAct |
| MEN1 | O00255 | P | STRING |
| MPHOPH10 | O00566 | Y2H | MINT, BIND |
| MRLC3 | P19105 | P | STRING |
| MYC | P01106 | AA | IntAct |
| MYL2 | P10916 | P | STRING |
| MYL9 | P24844 | P | STRING |
| MYO16 | Q9Y6X6 | E, P | HPRD,STRING |
| N33 | Q13454 | Y2H, PP | BIND, MINT, HPRD |
| NCOR1 | O75376 | AC | BIOGRID |
| NDP | Q00604 | Y2H | Y2H-PP1a |
| NEK2 | P51955 | E, P, Y2H | Y2H-PP1a, HPRD, Reactome,STRING |
| NOC2L | Q9Y3T9 | B | BIOGRID |
| NOM1 | Q5C9Z4 | Y2H, PP | MINT, HPRD |
| OAZ1 | P54368 | E | Reactome |
| PCDH7 | O60245 | E | HPRD |
| PCNA | P12004 | PP | HPRD |
| PHACTR1 | Q9C0D0 | Y2H+AC, P, PP | BIOGRID, STRING, HPRD |
| PHACTR3 | Q96KR7 | Y2H+AC+FWB, E | Y2H-PP1a, STRING, BIND, HPRD, BIOGRID |
| PHACTR4 | Q8IZ21 | E, P | HPRD, STRING |
| PHC1 | P78364 | Y2H | Y2H-PP1a |
| PHKA2 | P46019 | P | STRING |
| PIAS1 | O75925 | Y2H | Y2H-PP1a |
| PIAS3 | Q9Y6X2 | Y2H | Y2H-PP1a |
| PKN1 | Q16512 | P | STRING |
| PLCL2 | Q9UPR0 | Y2H, PP | BIND, MINT, HPRD |
| PLIN | O60240 | E, P | Reactome, STRING |
| PLK1 | P53350 | E | Reactome |
| PLP1 | P60201 | P | STRING |
| PLP2 | Q04941 | P | STRING |
| PPP1CA | P62136 | PP | HPRD |
| PPP1CB | P62140 | E | Reactome |
| PPP1CC | P36873 | E, P+IM+PP | Reactome,STRING |
| PPP1R10 | Q96QC0 | Y2H, B, E | BIND, BIOGRID, MINT,STRING, HPRD |
| PPP1R11 | O60927 | P, E | STRING, HPRD |
| PPP1R12A | O14974 | P | STRING |
| PPP1R12B | O60237 | P | STRING |
| PPP1R13A | Q13625 | Y2H, PP | Y2H-PP1a, MINT, HPRD, BIND |
| PPP1R13B | Q96KQ4 | Y2H, PP | Y2H-PP1a, MINT, HPRD, BIND |
| PPP1R13L | Q8WUF5 | Y2H, PP | Y2H-PP1a, HPRD, BIND |
| PPP1R15A | O75807 | Y2H, AC, E, P, PP | BIND, BIOGRID, MINT, Reactome, STRING, HPRD |
| PPP1R15B | Q5SWA1 | Y2H, PP | BIND, Y2H-PP1a, MINT, HPRD |
| PPP1R16A | Q96I34 | Y2H | Y2H-PP1a |
| PPP1R1A | Q13522 | P | STRING |
| PPP1R1B | Q9UD71 | E+IC, E, P | HPRD,Reactome, STRING |
| PPP1R2 | P41236 | E, Y2H, E, P | Y2H-PP1a, Reactome, STRING, HPRD |
| PPP1R2P9 | O14990 | E, P | HPRD,STRING |
| PPP1R3A | Q16821 | P | STRING |
| PPP1R3B | Q86XI6 | Y2H, P, PP | BIND, MINT, STRING, HPRD |
| PPP1R3C | Q9UQK1 | Y2H, P | Y2H-PP1a, STRING |
| PPP1R3D | O95685 | Y2H, P | Y2H-PP1a, STRING |
| PPP1R3E | Q9H7J1 | Y2H | Y2H-PP1a |
| PPP1R3G | B7ZBB8 | Y2H | Y2H-PP1a |
| PPP1R8 | Q12972 | Y2H, E, P+IM, PP, B | MINT, Reactome, STRING, HPRD, BIND, BIOGRID |
| PPP1R9A | Q9ULJ8 | E,P+IM | HPRD, STRING |
| PPP1R9B | Q96B17 | Y2H, AC+B, P+IM, PP | Y2H-PP1a, BIOGRID, STRING, HPRD, MINT |
| PPP2CA | P67775 | E,P+IM | HPRD, Reactome,STRING |
| PPP2CB | P62714 | E | Reactome |
| PPP2R4 | Q15257 | P+IM | STRING |
| PPP2R5D | Q14738 | P+IM | STRING |
| PPP4C | P60510 | E | Reactome |
| PPP5C | P53041 | E | Reactome |
| PPP6C | O00743 | E | Reactome |
| PREX1 | Q8TCU6 | Y2H | Y2H-PP1a |
| PRKACA | P17612 | P | STRING |
| PRKACB | P22694 | P | STRING |
| PRKACG | P22612 | P | STRING |
| PRKAG1 | P54619 | IM | STRING |
| PRKAR2A | P13861 | P | STRING |
| PRKAR2B | P31323 | P | STRING |
| PRKCD | Q05655 | E | HPRD |
| PRKCE | Q02156 | P | STRING |
| PRR16 | Q569H4 | Y2H | Y2H-PP1a |
| PTEN | P60484 | CO-IP, AA+CO-IP, E, PP | InnateDB, IntActReactome, HPRD |
| PYGM | P11217 | P | STRING |
| RANBP9 | Q96S59 | Y2H | Y2H-PP1a |
| RB1 | P06400 | AA+CO-IP, E, P, PP | IntAct, Reactome, STRING,HPRD |
| RGMA | Q96B86 | E | Reactome |
| RGMB | Q6NW40 | E | Reactome |
| RIF1 | Q5UIP0 | Y2H | Y2H-PP1a |
| ROCK1 | Q13464 | P | STRING |
| RON | Q04912 | CO-IP | InnateDB |
| RPAP2 | Q8IXW5 | AC | BIOGRID |
| RPAP3 | Q9H6T3 | AC | BIOGRID |
| RRP1B | Q14684 | AC+B | BIOGRID |
| RUVBL2 | Q9Y230 | AC | BIOGRID |
| RYR2 | Q92736 | B, P | BIOGRID,STRING |
| SERPING1 | P05155 | P | STRING |
| SF3A2 | Q15428 | AC | MINT |
| SH2D3A | Q9BRG2 | P | STRING |
| SH2D3C | Q8N5H7 | P | STRING |
| SH3RF2 | Q8TEC5 | Y2H | Y2H-PP1a |
| SIRT2 | Q8IXJ6 | E | Reactome |
| SKP1 | P63208 | AA+CO-IP, PP | IntAct, HPRD |
| SLC18A1 | P54219 | P | STRING |
| SLC18A2 | Q05940 | P | STRING |
| SLC45A1 | Q9Y2W3 | Y2H | Y2H-PP1a |
| SMAD1 | Q15797 | E | Reactome |
| SMAD7 | O15105 | E, P | Reactome, STRING |
| SMARCB1 | Q12824 | AC+B, P | BIOGRID, STRING |
| SNCAIP | Q9Y6H5 | Y2H | Y2H-PP1a |
| SNW1 | Q13573 | CO-IP | MINT |
| SorLA-1 | Q92673 | Y2H | Y2H-PP1a |
| SPRED1 | Q7Z699 | Y2H, PP | Y2H-PP1a, MINT, HPRD, BIND |
| STAM | Q92783 | Y2H, PP | BIND, MINT, HPRD |
| STAU1 | O95793 | Y2H, E, P | Y2H-PP1a, MINT, HPRD, STRING, BIND |
| SYTL2 | Q9HCH5 | Y2H, PP | BIND, MINT, HPRD |
| TEP1 | Q99973 | P | STRING |
| TGFBR1 | P36897 | AC, E, P, CO-IP | BIOGRID, Reactome, STRING, BIND |
| TGFBR2 | P37173 | E, Y2H | Reactome, BIND |
| TIAM1 | Q13009 | P | STRING |
| TNF A | P01375 | AC | InnateDB |
| TP53 | P04637 | Y2H, PP | BIND, MINT, HPRD |
| UBE2Z | Q9H832 | Y2H | Y2H-PP1a |
| ULK1 | O75385 | Y2H | Y2H-PP1a |
| VDR | P11473 | E | HPRD |
| VIP | P01282 | P | STRING |
| WBP11 | Q9Y2W2 | Y2H | Y2H-PP1a |
| YLPM1 | P49750 | PP, Y2H | Y2H-PP1a, HPRD, BIND |
| ZBTB11 | O95625 | Y2H | Y2H-PP1a |
| ZDBF2 | Q9HCK1 | Y2H | BIND |
| ZFYVE16 | Q7Z3T8 | Y2H, E, P, PP | MINT, Reactome, STRING, HPRD, BIND |
| ZFYVE9 | O95405 | Y2H, E, IM, PP | BIND, Y2H-PP1a, MINT, Reactome, STRING, HPRD |
| ZNF827 | Q17R98 | Y2H | Y2H-PP1a |
Y2H-PP1a, human brain PP1α YTH screen; AA, antibody array; AP, affinity purification; B, biochemical; CO-IP, coimmunoprecipitation; E, experimental knowledge based; FWB, Far-Western Blot; IC, inferred by curator; IM, interlogs mapping; P, predicted text mining; PD, pull down; PP, philogenetic profile; Y2H, Yeast Two-Hybrid.
Acknowledgments
This work was supported by the Centre for Cell Biology of the University of Aveiro, by grants from Fundação para a Ciência e Tecnologia of the Portuguese Ministry of Science and Higher Education to SLCE (SFRH/BD/41751/2007), SCD (SFRH/BD/21559/2005), and EFCS (LSHM-CT-2007-037950).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Allen P.B. Ouimet C.C. Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci USA. 1997a;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen P.B. Ouimet C.C. Greengard P. Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci USA. 1997b;94:9956–9961. doi: 10.1073/pnas.94.18.9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C.G. Browne G.J. Cohen P. Cohen P.T. PPP1R6, a novel member of the family of glycogen-targetting subunits of protein phosphatase 1. FEBS Lett. 1997;418:210–214. doi: 10.1016/s0014-5793(97)01385-9. [DOI] [PubMed] [Google Scholar]
- Auluck P.K. Caraveo G. Lindquist S. α-Synuclein: Membrane interactions and toxicity in Parkinson's disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- Ayllon V. Cayla X. Garcia A. Fleischer A. Rebollo A. The anti-apoptotic molecules Bcl-xL and Bcl-w target protein phosphatase 1alpha to Bad. Eur J Immunol. 2002;32:1847–1855. doi: 10.1002/1521-4141(200207)32:7<1847::AID-IMMU1847>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Bennett D. Alphey L. PP1 binds Sara and negatively regulates Dpp signaling in Drosophila melanogaster. Nat Genet. 2002;31:419–423. doi: 10.1038/ng938. [DOI] [PubMed] [Google Scholar]
- Bennett D. Lyulcheva E. Alphey L. Hawcroft G. Towards a comprehensive analysis of the protein phosphatase 1 interactome in Drosophila. J Mol Biol. 2006;364:196–212. doi: 10.1016/j.jmb.2006.08.094. [DOI] [PubMed] [Google Scholar]
- Bollen M. Combinatorial control of protein phosphatase-1. Trends Biochem Sci. 2001;26:426–431. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- Boyce M. Bryant K.F. Jousse C. Long K. Harding H.P. Scheuner D., et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Brush M.H. Weiser D.C. Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol Cell Biol. 2003;23:1292–1303. doi: 10.1128/MCB.23.4.1292-1303.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush M.H. Guardiola A. Connor J.H. Yao T.P. Shenolikar S. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J Biol Chem. 2004;279:7685–7691. doi: 10.1074/jbc.M310997200. [DOI] [PubMed] [Google Scholar]
- Ceulemans H. Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- Ceulemans H. Stalmans W. Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. Bioessays. 2002;24:371–381. doi: 10.1002/bies.10069. [DOI] [PubMed] [Google Scholar]
- Chen C.Y. Lai N.S. Yang J.J. Huang H.L. Hung W.C. Li C., et al. FLJ23654 encodes a heart protein phosphatase 1-binding protein (Hepp1) Biochem Biophys Res Commun. 2010;391:698–702. doi: 10.1016/j.bbrc.2009.11.123. [DOI] [PubMed] [Google Scholar]
- Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268:5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- Cohen P.T. Protein phosphatase 1—targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Colland F. Jacq X. Trouplin V. Mougin C. Groizeleau C. Hamburger A., et al. Functional proteomics mapping of a human signaling pathway. Genome Res. 2004;14:1324–1332. doi: 10.1101/gr.2334104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cruz e Silva E.F. Da Cruz e Silva O.A. Zaia C.T. Greengard P. Inhibition of protein phosphatase 1 stimulates secretion of Alzheimer amyloid precursor protein. Mol Med. 1995a;1:535–541. [PMC free article] [PubMed] [Google Scholar]
- Da Cruz e Silva E.F. Fox C.A. Ouimet C.C. Gustafson E. Watson S.J. Greengard P. Differential expression of protein phosphatase 1 isoforms in mammalian brain. J Neurosci. 1995b;15:3375–3389. doi: 10.1523/JNEUROSCI.15-05-03375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty M.J. Young P.R. Cohen P.T. Amino acid sequence of a novel protein phosphatase 1 binding protein (R5) which is related to the liver- and muscle-specific glycogen binding subunits of protein phosphatase 1. FEBS Lett. 1996;399:339–343. doi: 10.1016/s0014-5793(96)01357-9. [DOI] [PubMed] [Google Scholar]
- Egloff M.P. Johnson D.F. Moorhead G. Cohen P.T. Cohen P. Barford D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997;16:1876–1887. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S. Kaminsky Z. Guo X. Sharp A.H. Amaravi R.K. Kleiderlein J.J., et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat Genet. 1999;22:110–114. doi: 10.1038/8820. [DOI] [PubMed] [Google Scholar]
- Esteves S.L. Characterization of Human Brain Protein Phosphatase 1 Alpha Interacting Proteins Using the Yeast Two-Hybrid System. Aveiro: Biology Department. University of Aveiro; 2008. [Google Scholar]
- Fardilha M. Esteves S.L. Korrodi-Gregorio L. Da Cruz e Silva O.A. Da Cruz e Silva F.F. The physiological relevance of protein phosphatase 1 and its interacting proteins to health and disease. Curr Med Chem. 2010;17:3996–4017. doi: 10.2174/092986710793205363. [DOI] [PubMed] [Google Scholar]
- Fardilha M. Esteves S.L. Korrodi-Gregorio L. Pelech S. Da Cruz E.S.O.A. Da Cruz E.S.E. Protein phosphatase 1 complexes modulate sperm motility and present novel targets for male infertility. Mol Hum Reprod. 2011a;17:453–456. doi: 10.1093/molehr/gar004. [DOI] [PubMed] [Google Scholar]
- Fardilha M. Esteves S.L. Korrodi-Gregorio L. Vintem A.P. Domingues S.C. Rebelo S., et al. Identification of the human testis protein phosphatase 1 interactome. Biochem Pharmacol. 2011b doi: 10.1016/j.bcp.2011.02.018. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Fardilha M. Wu W. Sa R. Fidalgo S. Sousa C. Mota C., et al. Alternatively spliced protein variants as potential therapeutic targets for male infertility and contraception. Ann N Y Acad Sci. 2004;1030:468–478. doi: 10.1196/annals.1329.059. [DOI] [PubMed] [Google Scholar]
- Feng J. Yan Z. Ferreira A. Tomizawa K. Liauw J.A. Zhuo M., et al. Spinophilin regulates the formation and function of dendritic spines. Proc Natl Acad Sci USA. 2000;97:9287–9292. doi: 10.1073/pnas.97.16.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S. Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Flores-Delgado G. Liu C.W. Sposto R. Berndt N. A limited screen for protein interactions reveals new roles for protein phosphatase 1 in cell cycle control and apoptosis. J Proteome Res. 2007;6:1165–1175. doi: 10.1021/pr060504h. [DOI] [PubMed] [Google Scholar]
- Gagnon K.B. England R. Diehl L. Delpire E. Apoptosis-associated tyrosine kinase scaffolding of protein phosphatase 1 and SPAK reveals a novel pathway for Na-K-2C1 cotransporter regulation. Am J Physiol Cell Physiol. 2007;292:C1809–C1815. doi: 10.1152/ajpcell.00580.2006. [DOI] [PubMed] [Google Scholar]
- Gandy S. Greengard P. Regulated cleavage of the Alzheimer amyloid precursor protein: molecular and cellular basis. Biochimie. 1994;76:300–303. doi: 10.1016/0300-9084(94)90162-7. [DOI] [PubMed] [Google Scholar]
- Gibbons J.A. Weiser D.C. Shenolikar S. Importance of a surface hydrophobic pocket on protein phosphatase-1 catalytic subunit in recognizing cellular regulators. J Biol Chem. 2005;280:15903–15911. doi: 10.1074/jbc.M500871200. [DOI] [PubMed] [Google Scholar]
- Graff J. Koshibu K. Jouvenceau A. Dutar P. Mansuy I.M. Protein phosphatase 1-dependent transcriptional programs for long-term memory and plasticity. Learn Mem. 2010;17:355–363. doi: 10.1101/lm.1766510. [DOI] [PubMed] [Google Scholar]
- Helps N.R. Barker H.M. Elledge S.J. Cohen P.T. Protein phosphatase 1 interacts with p53BP2, a protein which binds to the tumour suppressor p53. FEBS Lett. 1995;377:295–300. doi: 10.1016/0014-5793(95)01347-4. [DOI] [PubMed] [Google Scholar]
- Helps N.R. Luo X. Barker H.M. Cohen P.T. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem J. 2000a;349:509–518. doi: 10.1042/0264-6021:3490509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helps N.R. Luo X. Barker H.M. Cohen P.T. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem J. 2000b;349:509–518. doi: 10.1042/0264-6021:3490509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A. Beullens M. Ceulemans H. Den Abt T. Van Eynde A. Nicolaescu E., et al. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Hrabchak C. Varmuza S. Identification of the spermatogenic zip protein Spz1 as a putative protein phosphatase-1 (PP1) regulatory protein that specifically binds the PP1cgamma2 splice variant in mouse testis. J Biol Chem. 2004;279:37079–37086. doi: 10.1074/jbc.M403710200. [DOI] [PubMed] [Google Scholar]
- Huang F.L. Glinsmann W.H. Separation and characterization of two phosphorylase phosphatase inhibitors from rabbit skeletal muscle. Eur J Biochem. 1976;70:419–426. doi: 10.1111/j.1432-1033.1976.tb11032.x. [DOI] [PubMed] [Google Scholar]
- Hurley T.D. Yang J. Zhang L. Goodwin K.D. Zou Q. Cortese M., et al. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J Biol Chem. 2007;282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- Jousse C. Oyadomari S. Novoa I. Lu P. Zhang Y. Harding H.P., et al. Inhibition of a constitutive translation initiation factor 2alpha phosphatase, CReP, promotes survival of stressed cells. J Cell Biol. 2003;163:767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S.C. Chen C.Y. Wang S.L. Yang J.J. Hung W.C. Chen Y.C., et al. Identification of phostensin, a PP1 F-actin cytoskeleton targeting subunit. Biochem Biophys Res Commun. 2007;356:594–598. doi: 10.1016/j.bbrc.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Kepp O. Galluzzi L. Giordanetto F. Tesniere A. Vitale I. Martins I., et al. Disruption of the PP1/GADD34 complex induces calreticulin exposure. Cell Cycle. 2009;8:3971–3977. doi: 10.4161/cc.8.23.10191. [DOI] [PubMed] [Google Scholar]
- Koshibu K. Graff J. Beullens M. Heitz F.D. Berchtold D. Russig H., et al. Protein phosphatase 1 regulates the histone code for long-term memory. J Neurosci. 2009;29:13079–13089. doi: 10.1523/JNEUROSCI.3610-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q. Buckler E.S.T. Muse S.V. Walker J.C. Molecular evolution of type 1 serine/threonine protein phosphatases. Mol Phylogenet Evol. 1999;12:57–66. doi: 10.1006/mpev.1998.0560. [DOI] [PubMed] [Google Scholar]
- Lin T.H. Tsai P.C. Liu H.T. Chen Y.C. Wang L.H. Hsieh F.K., et al. Characterization of the protein phosphatase 1-binding motifs of inhibitor-2 and DARPP-32 by surface plasmon resonance. J Biochem (Tokyo) 2005;138:697–700. doi: 10.1093/jb/mvi167. [DOI] [PubMed] [Google Scholar]
- Llorian M. Beullens M. Andres I. Ortiz J.M. Bollen M. SIPP1, a novel pre-mRNA splicing factor and interactor of protein phosphatase-1. Biochem J. 2004;378:229–238. doi: 10.1042/BJ20030950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W. Peterson A. Garcia B.A. Coombs G. Kofahl B. Heinrich R., et al. Protein phosphatase 1 regulates assembly and function of the beta-catenin degradation complex. EMBO J. 2007;26:1511–1521. doi: 10.1038/sj.emboj.7601607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. Weber G.A. Zheng J. Gendelman H.E. Ikezu T. C1q-calreticulin induced oxidative neurotoxicity: relevance for the neuropathogenesis of Alzheimer's disease. J Neuroimmunol. 2003;135:62–71. doi: 10.1016/s0165-5728(02)00444-7. [DOI] [PubMed] [Google Scholar]
- Monshausen M. Rehbein M. Richter D. Kindler S. The RNA-binding protein Staufen from rat brain interacts with protein phosphatase-1. J Neurochem. 2002;81:557–564. doi: 10.1046/j.1471-4159.2002.00887.x. [DOI] [PubMed] [Google Scholar]
- Moorhead G.B. Trinkle-Mulcahy L. Nimick M. De Wever V. Campbell D.G. Gourlay R., et al. Displacement affinity chromatography of protein phosphatase one (PP1) complexes. BMC Biochem. 2008;9:28. doi: 10.1186/1471-2091-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Ceulemans H. Bollen M. Diplexcito J. Cohen P.T. A novel glycogen-targeting subunit of protein phosphatase 1 that is regulated by insulin and shows differential tissue distribution in humans and rodents. FEBS J. 2005;272:1478–1489. doi: 10.1111/j.1742-4658.2005.04585.x. [DOI] [PubMed] [Google Scholar]
- Neumann J. Altered phosphatase activity in heart failure, influence on Ca2+ movement. Basic Res Cardiol. 2002;97(Suppl 1):I91–I95. doi: 10.1007/s003950200036. [DOI] [PubMed] [Google Scholar]
- Ouimet C.C. Da Cruz e Silva E.F. Greengard P. The alpha and gamma 1 isoforms of protein phosphatase 1 are highly and specifically concentrated in dendritic spines. Proc Natl Acad Sci USA. 1995;92:3396–3400. doi: 10.1073/pnas.92.8.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A.U. Morell R.J. Belyantseva I.A. Khan S.Y. Boger E.T. Shahzad M., et al. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C.S. Carneiro K. Ross C.A. Menezes J.R. Engelender S. Synphilin-1 is developmentally localized to synaptic terminals, and its association with synaptic vesicles is modulated by alpha-synuclein. J Biol Chem. 2002;277:23927–23933. doi: 10.1074/jbc.M201115200. [DOI] [PubMed] [Google Scholar]
- Sagara J. Higuchi T. Hattori Y. Moriya M. Sarvotham H. Shima H., et al. Scapinin, a putative protein phosphatase-1 regulatory subunit associated with the nuclear nonchromatin structure. J Biol Chem. 2003;278:45611–45619. doi: 10.1074/jbc.M305227200. [DOI] [PubMed] [Google Scholar]
- Santos M. Validation of LAP1B as a Novel Protein Phosphatase 1 Regulator. Aveiro: Biology Department, University of Aveiro; 2009. [Google Scholar]
- Shannon P. Markiel A. Ozier O. Baliga N.S. Wang J.T. Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W. Sun C. He B. Xiong W. Shi X. Yao D., et al. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164:291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J.A. Saltiel A.R. Cloning and identification of MYPT3: a prenylatable myosin targetting subunit of protein phosphatase 1. Biochem J. 2001;356:257–267. doi: 10.1042/0264-6021:3560257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar R. Hanson-Painton O. Cooper D.R. Protein kinases as therapeutic targets. Pharm Res. 2000;17:1345–1353. doi: 10.1023/a:1007507224529. [DOI] [PubMed] [Google Scholar]
- Tanuma N. Kim S.E. Beullens M. Tsubaki Y. Mitsuhashi S. Nomura M., et al. Nuclear inhibitor of protein phosphatase-1 (NIPP1) directs protein phosphatase-1 (PP1) to dephosphorylate the U2 small nuclear ribonucleoprotein particle (snRNP) component, spliceosome-associated protein 155 (Sap155) J Biol Chem. 2008;283:35805–35814. doi: 10.1074/jbc.M805468200. [DOI] [PubMed] [Google Scholar]
- Ten V.S. Yao J. Ratner V. Sosunov S. Fraser D.A. Botto M., et al. Complement component c1q mediates mitochondria-driven oxidative stress in neonatal hypoxic-ischemic brain injury. J Neurosci. 2010;30:2077–2087. doi: 10.1523/JNEUROSCI.5249-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak M. Kerff F. Langsetmo K. Tao T. Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- Tran H.T. Ulke A. Morrice N. Johannes C.J. Moorhead G.B. Proteomic characterization of protein phosphatase complexes of the mammalian nucleus. Mol Cell Proteomics. 2004;3:257–265. doi: 10.1074/mcp.M300115-MCP200. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L. Ajuh P. Prescott A. Claverie-Martin F. Cohen S. Lamond A.I., et al. Nuclear organisation of NIPP1, a regulatory subunit of protein phosphatase 1 that associates with pre-mRNA splicing factors. J Cell Sci. 1999;112(Pt 2):157–168. doi: 10.1242/jcs.112.2.157. [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L. Andersen J. Lam Y.W. Moorhead G. Mann M. Lamond A.I. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulke-Lemee A. Trinkle-Mulcahy L. Chaulk S. Bernstein N.K. Morrice N. Glover M., et al. The nuclear PP1 interacting protein ZAP3 (ZAP) is a putative nucleoside kinase that complexes with SAM68, CIA, NF110/45, and HNRNP-G. Biochim Biophys Acta. 2007;1774:1339–1350. doi: 10.1016/j.bbapap.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Virshup D.M. Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33:537–545. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K. Engelender S. Tanaka Y. Yoshimoto M. Mori F. Tsuji S., et al. Immunocytochemical localization of synphilin-1, an alpha-synuclein-associated protein, in neurodegenerative disorders. Acta Neuropathol. 2002;103:209–214. doi: 10.1007/s004010100451. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K. Engelender S. Yoshimoto M. Tsuji S. Ross C.A. Takahashi H. Synphilin-1 is present in Lewy bodies in Parkinson's disease. Ann Neurol. 2000;47:521–523. [PubMed] [Google Scholar]
- Wakula P. Beullens M. Ceulemans H. Stalmans W. Bollen M. Degeneracy and function of the ubiquitous RVXF motif that mediates binding to protein phosphatase-1. J Biol Chem. 2003;278:18817–18823. doi: 10.1074/jbc.M300175200. [DOI] [PubMed] [Google Scholar]