Abstract
Network models combined with gene expression studies have become useful tools for studying complex diseases like Alzheimer's disease. We constructed a “Core” Alzheimer's disease protein interaction network by human curation of the primary literature. The Core network consisted of 775 nodes and 2,204 interactions. To our knowledge, this is the most comprehensive and accurate protein interaction network yet constructed for Alzheimer's disease. An “Expanded” network was computationally constructed by adding additional proteins that interacted with Core network proteins, and consisted of 4,945 nodes and 26,064 interactions. We then mapped existing gene expression studies to the Core network. This combined data model identified the MAPK/ERK pathway and clathrin-mediated receptor endocytosis as key pathways in Alzheimer's disease. Important proteins in the MAPK/ERK pathway that interacted in the Core network formed a downregulated cluster of nodes, whereas clathrin and several clathrin accessory proteins that interacted in the Core network formed an upregulated cluster of nodes. The MAPK/ERK pathway is a key component in synaptic plasticity and learning, processes disrupted in Alzheimer's. Clathrin and clathrin adaptor proteins are involved in the endocytosis of the APP protein that can lead to increased intracellular levels of amyloid beta peptide, contributing to the progression of Alzheimer's.
Introduction
Alzheimer's disease (AD) is the most prevalent form of dementia in aging humans. It is already a significant health problem in the United States, and is predicted by some to become the dominant health problem in this country within 15 years, surpassing cancer and cardiovascular disease in terms of the overall financial burden to the U.S. healthcare system (Burke, 2007; Culmsee, 2006; Walsh and Selkoe, 2004).
Although much progress has been made in identifying the causative factors of genetic diseases where variation in a single gene is the predominant factor (i.e., monogenic diseases), far less progress has been made in determining the genetic causes of polygenic diseases such as AD and other neurodegenerative diseases, cancer, and cardiovascular disease. The difficulties in studying polygenic diseases like AD are due to the presence of multiple genetic variants and their interactions with nongenetic factors (e.g., diet) (Kann, 2007; Lesnick et al., 2007; Schadt and Lum, 2006). The complexities of polygenic diseases, coupled with the large amount of molecular interaction data generated by high-throughput techniques, has led to the increasing use of network models to study polygenic diseases (Barabasi and Oltvai, 2004; Schadt and Lum, 2006; Sieberts and Schadt, 2007). Protein–protein interaction (PPI) network models are useful in identifying key proteins and cellular pathways in a particular disease and provide a framework for investigating the complexities of polygenic diseases like AD. Network models have also found widespread use in integrating data from other sources, such as gene expression data (Barabasi and Oltvai, 2004; Camargo and Azuaje, 2007; Kann, 2007; Lu et al., 2007). Mapping gene expression data to PPI networks provides a more meaningful biological context for differentially expressed genes. This combined approach can identify key cellular pathways or complexes where up- or downregulated gene products cluster, thus identifying potential disease-associated genes that may not be significantly up- or downregulated by themselves. PPI network models combined with gene expression data have recently been used to identify key proteins, pathways, and novel candidate genes in the study of polygenic diseases such as cancer (Wachi et al., 2005), atherosclerosis (King et al., 2005), and Parkinson's disease (Lesnick et al., 2007).
We have applied this combination of PPI networks and gene expression data to the study of AD. We first constructed two PPI networks for AD. The first network (“Core” network) was constructed by a human review of primary literature and Web resources related to AD. All Core network proteins and interactions were known to be associated with AD or AD-related cellular processes. We focused predominantly on studies that used multiple methods to experimentally verify specific PPIs and avoided high-throughput data because of the high rates of false positive and negative results known to be associated with this type of data (Batada et al., 2006; Kann, 2007; Zhu et al., 2007). Although other computational or probabilistic network models of AD have been developed (Krauthammer et al., 2004; Liu et al., 2006; Soler-Lopez et al., 2011), genes and proteins present in these models are not necessarily involved in AD, nor do these networks attempt to include the majority of genes and proteins involved in AD and AD-related cellular processes. To our knowledge, the Core network is the most comprehensive and accurate PPI network model of AD constructed to date. The second network (“Expanded” network) computationally enlarged the Core network by adding additional proteins that interact with Core proteins, using data from the Human Protein Reference Database (HPRD) (Prasad et al., 2009). HPRD contains human-curated interaction data from published literature and is one of the larger and more comprehensive protein interaction databases (Gandhi et al., 2006). Although the Expanded network proteins were not known to be associated with AD at the time of network construction, they could have an effect on AD-related processes due to their interactions with Core network proteins.
We performed an initial analysis of the network structure and protein content of both networks in order to identify key proteins, protein interactions, and molecular pathways involved in AD. We then mapped existing gene expression studies to the Core network in order to identify up- and downregulated genes and where their protein products clustered in the Core network, focusing in particular on differentially regulated genes whose protein products interacted with each other to form connected clusters in the Core network. We identified two key cellular pathways in particular that had clusters of differentially regulated genes whose protein products interacted in the Core network: the MAPK/ERK pathway, which is key in long-term potentiation and synaptic plasticity, and clathrin-mediated receptor endocytosis, which is involved in the internalization of the amyloid precursor protein (APP) that can lead to increased intracellular production of the amyloid beta (Aβ) peptide (Schneider et al., 2008).
Materials and Methods
Core network construction
The Core network was built from a review of the primary literature using Web of Science (thomsonreuters.com) and PubMed (www.ncbi.nlm.nih.gov/pubmed/) and a continuous review of current journal titles related to AD and neuroscience. Other resources used included the Online Mendelian Inheritance in Man (OMIM) Web pages at NCBI (www.ncbi.nlm.nih.gov/omim), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000), the Alzheimer Research forum Website (www.alzgene.org) (Bertram et al., 2007), and the Autworks Website (autworks.hms.harvard.edu/). Approximately 5,000 individual articles that were identified by these sources as containing AD-related PPIs were reviewed in this process.
In some instances, experimental data did not identify every specific protein that is involved in a particular pathway, so intermediate proteins in some pathways may not have been identified. This was an acceptable tradeoff so as to avoid including false interactions. In order to fill in the gaps in well-known pathways, we used additional interactions from either the NCBI Entrez Gene Web pages (www.ncbi.nlm.nih.gov/gene/) or the KEGG database when those pathways were implicated in AD by the primary literature. We tried to minimize these changes, because normal signaling pathways can be disrupted in disease states (Ideker and Sharan, 2008; Kann, 2007). Although a few intermediaries may have been missed, the network still indicated the interaction effects as documented in the original article and the overall pathway and key signaling proteins involved. (The Expanded network, as described below, incorporated some of these missing interactions if they were included in HPRD.)
We also replaced some protein complexes with representative complex members. For example, the Core network includes several protein kinase complexes, such as protein kinases A and C, and receptor complexes, such as NMDA glutamate receptors, that are made up of multiple subunits. In most cases, all or most of the relevant subunits were included in the network, with interactions listed between subunits as appropriate. However, for interactions between the protein kinase or receptor complex and other proteins, if a specific subunit was not listed in the journal article, we chose a subunit as a marker for the complex as a whole and used that subunit in that particular protein–protein interaction. Choice of subunits as markers was based as much as possible on those subunits/proteins known to be common in the central nervous system (CNS) or involved in AD.
For network visualization and analysis, PPI data were imported into the software program Cytoscape v2.6.3 (Shannon et al., 2003) and constructed as an undirected graph. In all cases, the NCBI official symbol was used to identify network proteins. The Core network was completed in December of 2009. (A link to a Cytoscape compatible file of the Core network is included as Supplemental Figure S1. Cytoscape software can be downloaded from cytoscape.org)
Expanded network construction
The Expanded network was constructed in December 2009 using the software program MiMi v3.0.1 (Gao et al., 2009), a plug-in for Cytoscape, which accesses the major on-line databases containing PPI data. Program settings were specified to use HPRD as the reference database, with all Core network proteins as input, and to select all input proteins plus their first neighbors as the undirected output network. The network expansion was limited to first neighbors of Core network proteins in order to focus on proteins that could have a direct impact on known AD-related proteins and to limit the size of the Expanded network. Besides these additional proteins and their interactions, the Expanded network added additional interactions between Core proteins not identified in the literature as being associated with AD and thus not included in the Core network. (A link to a Cytoscape compatible file of the Expanded network is included as Supplemental Figure S2. Cytoscape software can be downloaded from cytoscape.org)
Analysis of network parameters and structure
Network parameters for both Core and Expanded networks were calculated in order to analyze network structure and to identify key nodes. (See Table 1 for a definition of network and node parameters used for this and subsequent network analyses.) Calculation and analysis of network parameters was done using the plug-in programs CentiScape v1.1 (Scardoni et al., 2009) and NetworkAnalyzer v2.6.1 (Assenov et al., 2008) for Cytoscape, and the igraph library (Csardi and Nepusz, 2006) in the software program R (Team, 2009).
Table 1.
Definitions of Network and Node Parameters Used for Network Analysis (Barabasi and Oltvai, 2004; Ideker and Sharan, 2008; Lin et al., 2009; Zhu et al., 2007)
|
Parameter |
Type |
Description |
|---|---|---|
| Degree | Node | Number of interactions |
| Average degree | Network | Average number of interactions for all nodes in a network |
| Power law coefficient | Network | Exponent in equation describing network degree distribution |
| Closeness centrality | Node | Reciprocal of sum of all shortest paths between a particular node and all other network nodes |
| Average closeness centrality | Network | Closeness centrality averaged for all network nodes |
| Average clustering coefficient | Network | Connectivity of all immediate neighbors of a particular node averaged for all network nodes |
| Average shortest path | Network | Average of all shortest paths (in number of edges) between all pairs of nodes in network |
| Density | Network | Ratio of actual number of edges in a network to possible number of edges |
| Diameter | Network | Longest shortest path across network |
Core network parameters were compared to populations of randomly generated networks and rewired versions of the Core network. Generation of random and rewired networks and analysis was performed using igraph to produce rewired networks and the Cytoscape plug-in Random Networks v1.0 (sites.google.com/site/randomnetworkplugin/Home) to create random networks using a Barabasi-Albert algorithm that produces a scale-free network. One thousand networks of each type were created for comparison to Core network parameters. Rewired networks had the same number of nodes and edges as the Core network, but edges were randomly shuffled between nodes. The random networks also had the same number of nodes (775) as the Core network, and were constructed to also have a similar number of edges in order to make comparisons of network parameters valid. The p-values for comparison of network parameters were generated by taking the proportion of the 1,000 random or rewired networks whose relevant parameter was greater than or less than, as appropriate, the Core network parameter under analysis.
Parameters for the Core and Expanded networks were compared to a set of 12 PPI networks taken from Web resources and primary literature:
Five networks, consisting of all listed PPIs as of 11/25/09 for human, A. thaliana, C. elegans, D. melanogaster, and M. musculus from Biogrid on-line database (Stark et al., 2006)
Five networks, consisting of four human PPI networks and one yeast PPI network from Cytoscape software package v. 2.6.3
Two disease networks, consisting of one human ataxia PPI network (Lim et al., 2006) and one mouse model of asthma PPI network (Lu et al., 2007) from published journal articles.
Summary statistics for this sample of PPI networks (mean, minimum, maximum, and standard deviation) were generated using the software program R.
Gene expression data analysis
Gene expression data from four studies (Blalock et al., 2004; Nunez-Iglesias et al., 2010; Williams et al., 2009) (unpublished study conducted by Chen et al.) were mapped onto the Core network. Data from these studies are available on the NCBI Gene Expression Omnibus (GEO) page (www.ncbi.nlm.nih.gov/geo/). The Blalock et al. (2004) study is GSE1297, the Nunez-Iglesias et al. (2010) study is GSE16759, the Williams et al. (2009) study is GSE12685, and the Chen et al. study is GSE18309. The Blalock study consisted of 31 hippocampal tissue samples from control, incipient, moderate, and severe AD; the Nunez-Iglesias study had 16 samples from parietal lobe cortex; the Williams study had 14 samples from synaptoneurosomes prepared from frontal cortex; and the Chen study had 9 samples taken from peripheral blood mononuclear cells. Half of the control samples and half of the AD samples from the Nunez-Iglesias study were analyzed for miRNA expression; those samples were not included in this analysis. Three of the samples from the Chen study were from patients with a diagnosis of mild cognitive impairment rather than AD; those samples were also not used in this analysis. Raw data was downloaded as .CEL files and preprocessed in R with the affy package (Gautier et al., 2004) using the expresso function with mas as the background correct method, quantiles as the normalization method, mas as the pmcorrect method, and medianpolish as the summary method (Choe et al., 2005; Miller et al., 2008). An R script was used to extract the Core network genes from the overall expression dataset. Preprocessed data was mapped to the Core network in Cytoscape as a node attribute. Data for the Blalock study, which consisted of separate microarray analyses for incipient, moderate, and severe AD were mapped separately for each disease level. We selected subpopulations of genes to be mapped to the network by defining downregulated genes as those genes showing a decrease in the log 2 expression value of 0.5 or more compared to control samples and upregulated genes as those genes having a log 2 increase in expression values of 0.5 or more compared to control samples. If there were multiple transcripts listed for any particular gene in the up- or downregulated lists, we selected the lowest log 2 values of the replicates for the downregulated list, and the highest log 2 values for the upregulated list. We then mapped these two sets of genes to the Core network, using the network structure to identify significant clusters of related proteins in particular pathways with coordinated changes in levels of expression. This approach has been used successfully to identify key disease-associated pathways in which individual gene expression levels vary by as little as 20% (Subramanian et al., 2005), a smaller change than we used as our minimum fold cutoff. Once mapped to the original network, subnetworks were extracted consisting of all Core network nodes from that study or level of AD severity that were up- or downregulated. From these subnetworks, we also identified connected components of up- or downregulated nodes that were first neighbors in the original Core network.
We used the Wilcoxon rank-sum test, performed in R, to compare the relative levels of connectivity between these subnetworks. Comparisons were done using the average degree of each subnetwork of up- or downregulated nodes. Note that the degree used was the degree of the node in the subnetwork, not its degree in the original Core network. We used all up- or downregulated nodes in each subnetwork in calculating average degree, not just those nodes in a connected component, so many nodes had a degree of zero. The average degree of each group of nodes was calculated using the CentiScape plug-in in Cytoscape.
Results
Network structure and parameters
The completed Core network contained 775 nodes and 2,204 edges. The Expanded network consisted of 4,945 nodes and 26,064 edges. We compared selected Core network parameters (see Table 1 for definitions of network parameters) with two sets of 1,000 networks generated either by rewiring of the Core network or construction of similar-sized random scale-free networks (Table 2). Compared to the rewired graphs, the Core network average clustering coefficient and average shortest path were significantly larger. The average closeness centrality was not statistically significantly different. The density was unchanged, because the rewired graphs had the same number of nodes and edges as the Core graph.
Table 2.
Comparison of Key Core Network Parameters with Summary Statistics from Sets of 1,000 Rewired or Random Networks
| |
|
Rewired |
Random |
||
|---|---|---|---|---|---|
| Parameter | Core | Mean | SD | Mean | SD |
| Number of nodes | 775 | 775 | 0 | 775 | 0 |
| Number of edges | 2,204 | 2204 | 0 | 2322 | 0 |
| Average clustering coefficient | 0.225 | 0.046 (p<0.001) | 0.002 | 0.017 (p<0.001) | 0.002 |
| Average closeness centrality | 0.313 | 0.142 (p=0.058) | 0.067 | na | na |
| Average shortest path | 3.179 | 3.13 (p<0.008) | 0.019 | 2.68 (p<0.001) | 0.087 |
| Average degree | 5.623 | na | na | 5.99 | 0.00 |
| Power law coefficient | 1.302 | na | na | 1.81 (p<0.01) | 0.078 |
| Density | 0.007 | 0.007 | 0.000 | na | na |
See Table 1 for a description of network parameters. The p-values in parentheses indicate whether Core parameters were statistically different from rewired/random graph values. Boxes marked with “na” mean calculation of that particular parameter was not available in the programs being used.
Compared to the random networks, the Core network average clustering coefficient and average shortest path were again significantly larger, whereas the power law coefficient of the degree distribution was significantly smaller.
We also compared key network parameters for both the Core and Expanded networks to a random sample of 12 other existing PPI networks (Table 3). The Core network had a larger average clustering coefficient and average closeness centrality, and a smaller average shortest path length compared to the range of values for the other networks. The power law coefficient of the Core network was toward the low end of the range of the other networks, whereas the density was toward the high end of the range. The Expanded network, with the exception of its average degree, had parameter values that were within the range of the other networks.
Table 3.
Comparison of Key Network Statistics for Core and Expanded Networks with Summary Statistics from a Random Sampling of 12 Other PPI Networks
| |
AD Networks |
Other Networks |
||||
|---|---|---|---|---|---|---|
| Parameter | Core | Expanded | Mean | Min | Max | SD |
| Number of nodes | 775 | 4945 | 3395.7 | 419 | 8674 | 2614.4 |
| Number of edges | 2204 | 26064 | 9579.8 | 1089 | 31656 | 10,600.3 |
| Average clustering coefficient | 0.225 | 0.154 | 0.086 | 0.006 | 0.195 | 0.063 |
| Diameter | 9 | 9 | 14.2 | 4 | 29 | 7.1 |
| Average closeness centrality | 0.313 | 0.048 | 0.076 | 0.025 | 0.182 | 0.053 |
| Average shortest path | 3.179 | 3.618 | 5.126 | 3.390 | 8.449 | 1.616 |
| Average degree | 5.623 | 10.542 | 4.419 | 2.229 | 6.815 | 1.548 |
| Power law coefficient | 1.302 | 1.700 | 1.679 | 1.135 | 1.987 | 0.243 |
| Density | 0.0070 | 0.0020 | 0.0023 | 0.0001 | 0.0110 | 0.0029 |
See Table 1 for a description of parameters. “Min” is the minimum value of the indicated parameter among all the other networks, while “Max” is the maximum value. “SD” is the standard deviation of the parameter.
Key proteins in core and expanded networks
We identified the top 25 proteins by degree in the Core and Expanded networks (Table 4). As the degree indicates the number of direct interactions a protein has, a high degree can indicate proteins with an important role in the network; disruption of these proteins could have a significant impact on the network's functioning (Barabasi and Oltvai, 2004; Batada et al., 2006; Kann, 2007). All of the top 25 proteins by degree in the Expanded network, with the exception of E1A binding protein p300 (EP300), were also present in the Core network, although in some cases with significant increases or decreases in degree.
Table 4.
Top 25 Proteins in Core and Expanded Networks by Degree
|
Core network |
Expanded network |
||||
|---|---|---|---|---|---|
| Rank | Protein | Degree | Rank | Protein | Degree |
| 1 | Aβ | 254 | 1 | YWHAG | 248 |
| 2 | APP | 174 | 2 | TP53 | 244 |
| 3 | PSEN1 | 97 | 3 | CREBBP | 202 |
| 4 | MAPT | 88 | 4 | SRC | 196 |
| 5 | GSK3B | 52 | 5 | GRB2 | 193 |
| 6 | BACE1 | 49 | 6 | EP300 | 190 |
| 7 | PRKCG | 41 | 7 | SMAD3 | 179 |
| 8 | PSEN2 | 38 | 8 | ESR1 | 172 |
| 9 | GRIN1 | 38 | 9 | PRKCA | 170 |
| 10 | AKT1 | 37 | 10 | CSNK2A1 | 166 |
| 11 | CDK5 | 34 | 11 | SMAD2 | 163 |
| 12 | MAPK8 | 34 | 12 | MAPK1 | 160 |
| 13 | NFKB1 | 32 | 13 | EGFR | 156 |
| 14 | PIK3R1 | 31 | 14 | TGFBR1 | 154 |
| 15 | MAP2K1 | 30 | 15 | SMAD4 | 152 |
| 16 | PRKACG | 30 | 16 | TRAF2 | 151 |
| 17 | MAP2K2 | 29 | 17 | FYN | 150 |
| 18 | TP53 | 29 | 18 | AR | 135 |
| 19 | CASP3 | 29 | 19 | RB1 | 134 |
| 20 | RARB | 28 | 20 | CTNNB1 | 129 |
| 21 | MAPK3 | 27 | 21 | PIK3R1 | 128 |
| 22 | APOE | 27 | 22 | CASP3 | 127 |
| 23 | TNF | 27 | 23 | YWHAZ | 125 |
| 24 | MAPK1 | 26 | 24 | PRKACA | 123 |
| 25 | LRP1 | 22 | 25 | CDC2 | 120 |
Bolded proteins in the Expanded network degree column indicate proteins that are not present in the Core Network.
Mapping of gene expression data
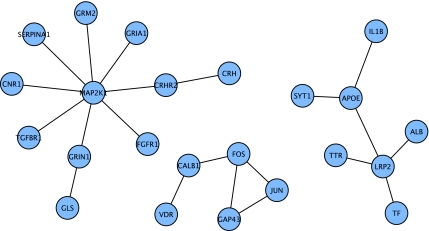
We mapped gene expression data from four studies (Blalock et al., 2004; Nunez-Iglesias et al., 2010; Williams et al., 2009) (unpublished study by Chen et al.) onto the Core network (Table 5). We then looked for clusters of connected proteins (connected components) in the Core network whose underlying genes were either all up- or downregulated in the expression dataset. Mapping of incipient AD downregulated genes from the Blalock study to the Core network revealed three significant connected components in the Core network (Fig. 1), plus two pairs of connected nodes. Upregulated genes from incipient AD had two connected components of five nodes each (Fig. 2), plus additional groups of two and three connected nodes. The largest of the three connected components of downregulated nodes consisted of 11 nodes; the highest degree node in this subnetwork was MAP2K1. The MAP2K1 protein is a key upstream kinase involved in activation of the MAPK/ERK pathway. This pathway is key to several cellular processes, in particular, memory and long-term potentiation (LTP) (Dudai, 2004; Sekine et al., 2006), processes that are disrupted in AD. Other downregulated nodes in this connected component include a variety of receptors: the AMPA glutamate receptor (GRIA1), cannabinoid receptor (CNR1), transforming growth factor-beta (TGF-β) receptor (TGFBR1), metabotropic glutamate receptor (GRM2), and the corticotropin releasing hormone receptor (CRHR2) and its hormone ligand (CRH), all of which mediate activation of the MAPK/ERK pathway.
Table 5.
Comparison of Total Numbers of Up- and Downregulated Genes, Size of Connected Components, and Average Degree in Four Microarray Analyses of AD-Related Genes
| Blalock - incipient | Blalock - moderate | Blalock - severe | Nunez-Iglesias | Williams | Chen | |
|---|---|---|---|---|---|---|
| No. downregulated | 80 | 128 | 167 | 258 | 252 | 180 |
| Average degree | 0.58 | 1.09 | 1.49 | 1.92 | 1.42 | 1.35 |
| Size of largest connected component | 11 | 39 | 81 | 169 | 122 | 88 |
| No. upregulated | 93 | 84 | 198 | 170 | 277 | 277 |
| Average degree | 0.41 | 0.33 | 0.94 | 0.59 | 2.36 | 1.79 |
| Size of largest connected component | 5 | 4 | 24 | 28 | 187 | 159 |
| p-Value | 1.09×10−7 | 7.98×10−5 | 0.095 | 6.42×10−14 | 0.0002 | 0.038 |
“No. downregulated” or “No. upregulated” refer to the number of Core nodes that were identified as being up- or downregulated in the designated study. “Average degree” refers to the average degree of the subnetwork of all up- or downregulated nodes for that study. “Size of largest connected component” refers to the largest number of nodes that were up- or downregulated in a particular study that directly interact with each other in the Core network. The p-values are for Wilcoxon rank-sum test to determine if there is a significant difference in the levels of connectivity (average degree) of up- or downregulated nodes under comparison.
FIG. 1.
Connected components of downregulated nodes from Blalock et al. (2004) for incipient AD that were first neighbors in the Core network.
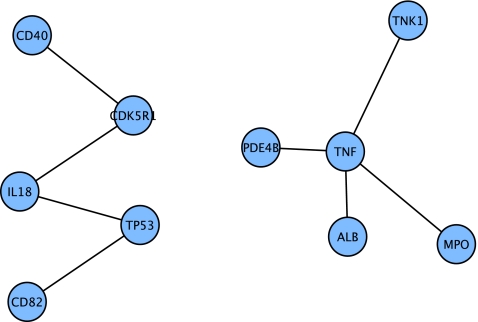
FIG. 2.
Connected components of upregulated nodes from Blalock et al. (2004) for incipient AD that were first neighbors in the Core network.
The next-largest downregulated connected component had seven nodes; the two highest degree nodes were apolipoprotein E (APOE) and low-density lipoprotein receptor-related protein 2 (LRP2). These proteins are key in cholesterol regulation and transport; the APOE gene has been well documented as a risk factor for AD, and binding of the APOE protein to lipoprotein receptors mediates activation of the MAPK/ERK pathway (Hoe et al., 2005).
The other downregulated connected component contained the vitamin D receptor (VDR), calbindin 1 (CALB1), the transcription factors FOS and JUN, and growth-associated protein 43 (GAP43). As with the MAPK/ERK pathway described above, elements of this pathway, in particular, VDR and CALB1, have been shown to be critical for learning and memory (Palop et al., 2003).
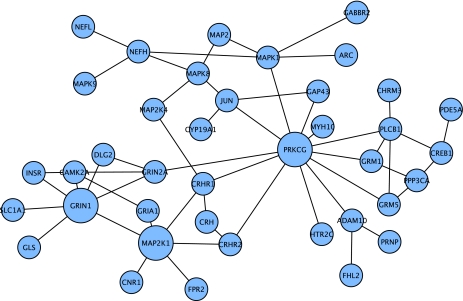
The connected component centered on APOE and LRP2, or the APOE or LRP2 nodes individually, were not found in subsequent mappings of downregulated genes from moderate and severe AD in the Blalock study. In comparison, the small connected component centered on MAP2K1 found in incipient AD expression data continued to expand into moderate (Fig. 3) and severe (Fig. 4) AD, with JUN and GAP43 from the other small connected component also becoming part of this larger downregulated connected component.
FIG. 3.
Connected component of downregulated nodes from Blalock et al (2004) for moderate AD that were first neighbors in the Core network. Larger nodes indicate the highest degree nodes in this connected component.
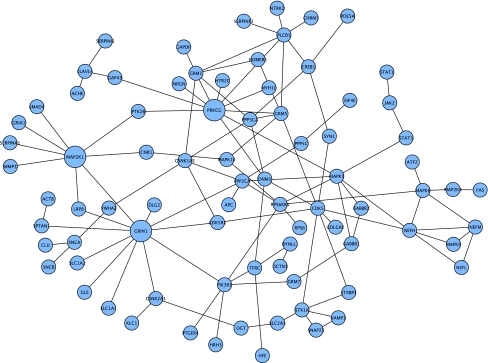
FIG. 4.
Connected component of downregulated nodes from Blalock et al (2004) for severe AD that were first neighbors in the Core network. Larger nodes indicate the highest degree nodes in this connected component.
In moderate AD gene expression in the Blalock study, this connected component of downregulated genes expanded to 39 nodes (Fig. 3). The top nodes by degree in this subnetwork include the key protein kinase C (PRKCG), an NMDA glutamate receptor subunit (GRIN1), and MAP2K1. Additional nodes include another NMDA glutamate receptor subunit (GRIN2A), the transcription factor CREB1, MAPK1 (another key component of the MAPK/ERK pathway), the glutamate transporter SLC1A1, glutaminase (GLS), and key synaptic plasticity (ARC) and synaptic receptor scaffolding (DLG2) nodes. The analysis of key nodes in the Core network by degree, as discussed above, had identified PRKCG as a key protein in the overall Core network. It is also the highest degree node in this connected component of downregulated nodes. Downregulation of PRKCG in turn decreases activity of the MAPK/ERK pathway. PRKCG protein activity increases activity of the alpha-secretase ADAM10, and thus reduces Aβ production, so downregulation of PRKCG would be expected to lead to increased Aβ production (Argellati et al., 2009; Yang et al., 2007).
In comparison, the largest connected component of upregulated genes in moderate AD consisted of four nodes: the gamma catalytic subunit of cAMP-dependent protein kinase (PRKACG), endothelial nitric oxide synthase (NOS3), insulin-like growth factor 1 receptor (IGF1R), and tyrosine hydroxylases (TH).
In severe AD from the Blalock study, the downregulated connected component expanded to 81 nodes (Fig. 4). Top nodes by degree in this subnetwork included GRIN1, PRKCG, MAP2K1, as well as two metabotropic glutamate receptors (GRM1 and GRM5). Other key nodes in the downregulated subnetwork included Jun N-terminal kinases (MAPK8 and MAPK9), the kinase CDK5 and its regulator CDK5R1, STAT1, and JAK2, members of the JAK/STAT signaling pathway, and PIK3R1, the regulatory subunit of the phosphoinositide-3-kinase protein complex PI3K, which is a key intermediary in the insulin receptor–AKT1 pathway.
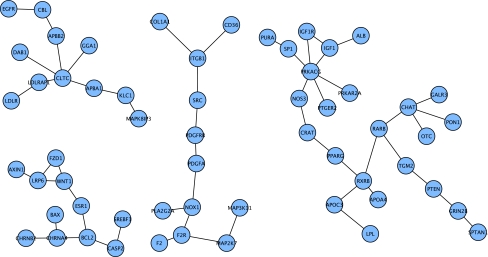
Upregulated nodes from severe AD had four smaller connected components, ranging in size from 11 to 24 nodes (Fig. 5). The top nodes by degree in these subnetworks included PRKACG, clathrin heavy chain (CLTC), choline acetyltransferase (CHAT), and retinoic X receptor B (RXRB). The connected component containing CLTC also contained several clathrin adaptor proteins and receptors, all of which participate in receptor-mediated endocytosis.
FIG. 5.
Connected components of upregulated nodes from Blalock et al (2004) for severe AD that were first neighbors in the Core network.
A similar, although larger (169 nodes) connected component of downregulated nodes was found in the Nunez-Iglesias study (see Supplemental Fig. S3), which also included multiple MAPK/ERK pathway nodes and related receptors. The top nodes by degree in this downregulated subnetwork were APP, GRIN1, and MAPK1. The largest connected component of upregulated nodes consisted of 28 nodes (see Supplemental Fig. S4); the highest degree nodes in this subnetwork were β-catenin (CTNNB1), low-density lipoprotein receptor-related protein 1 (LRP1), and GRIN1, which also appeared in the downregulated subnetwork. Expression arrays often contain probe sets for multiple transcript variants of the same gene, so this most likely indicates a change in expression levels of GRIN1 variants. Given that different variants of receptor subunits affect the overall functional characteristics of a receptor complex, downregulation of one GRIN1 variant and upregulation of another variant could indicate a shift in performance characteristics of the final NMDA receptor complexes.
The Williams study had 252 downregulated nodes, with a connected component of 122 nodes (see Supplemental Fig. S5). The top nodes by degree in this connected component were MAPK8, presenilin 2 (PSEN2), and the transcription factor NFKB1. There were 277 upregulated nodes, with a large connected component of 187 nodes (see Supplemental Fig. S6). The top nodes by degree in this connected component were APP, microtubule-associated protein tau (MAPT), glycogen synthase kinase 3b (GSK3B), and the protein kinase AKT1.
The Chen study had 180 downregulated nodes, with a connected component of 88 nodes (see Supplemental Fig. S7). The top nodes by degree in this connected component were MAPT, PRKACG, MAPK8, MAPK/ERK pathway kinase (MAP2K2), and GRIN1. There were 277 upregulated nodes, with a large connected component of 159 nodes (see Supplemental Fig. S8). The top nodes by degree in the connected component of upregulated nodes were presenilin 1 (PSEN1), MAPT, PRKCG, β-site APP-cleaving enzyme 1 (BACE1), retinoic acid receptor B (RARB), and APOE. As with GRIN1 in the Nunez-Iglesias study, different variants of MAPT appeared in lists of both up- and downregulated nodes in the Chen study.
In terms of the overall number of up- or downregulated genes that corresponded to proteins in the Core network, there was no distinctive pattern among the studies, with some studies reporting more upregulated than down regulated genes, and others reporting the reverse pattern. We also compared the levels of connectivity within all subnetworks of up- or downregulated nodes using a Wilcoxon rank-sum test to compare the average degree of each subnetwork (see Table 5). For the incipient and moderate AD samples from the Blalock study, and the Nunez-Iglesias study, the downregulated node subnetworks had a significantly higher average degree (p << 0.05) than did the corresponding group of upregulated nodes. For the severe AD samples from the Blalock study, although the actual average degree of the downregulated nodes was 59% higher than that of the upregulated subnetwork of nodes (1.49 vs. 0.94 average degree), the results of the Wilcoxon test were not significant (p=0.095). In contrast, the comparison of average degree in the up- and downregulated subnetworks in the Williams and Chen studies showed that the upregulated subnetworks were more highly connected (p=0.0002 and p=0.038, respectively) than were the downregulated subnetworks.
Discussion
The Core network had a larger average clustering coefficient than rewired and random networks, which indicates a high degree of local connectivity. In addition, the Core network had a power law coefficient that was significantly smaller than the random networks; it is also smaller than what is seen for typical biological networks, which generally have a power law coefficient in the range of 2–3 (Barabasi and Oltvai, 2004; Raman, 2010). This small power law coefficient indicates an increased proportion of high-degree proteins (i.e., hubs) in the Core network (Barabasi and Oltvai, 2004). The Core network also had a higher average clustering coefficient, higher average closeness centrality, and a shorter average shortest path length than did a sample of 12 other existing PPI networks. The density of the Core network was toward the upper end of the range of the other 12 networks.
Emerging from these comparisons is a Core network with a high degree of local structure (high average clustering coefficient), but also a high degree of global connectivity (high density, short average path length, and high closeness centrality). This overall connectivity is enhanced by the increased importance of hub proteins, indicated by the small power law coefficient for the degree distribution. This type of network, with high local clustering, a larger proportion of high-degree hubs and a higher level of global connectivity, is often found in networks that respond to extracellular signals (e.g., neurons), as it allows the cell to rapidly react to changing external conditions (Zhu et al., 2007). This rapid reaction is beneficial under normal circumstances, but under pathological conditions such as AD this means that the effects of network disruptions can spread rapidly.
We identified the key proteins based on high degree in the Core and Expanded networks. Many of the top 25 Core network nodes by degree are known to have substantial involvement in AD: Aβ, APP, PSEN1/2, MAPT, GSK3B, BACE1, GRIN1, CDK5, APOE, and low-density lipoprotein receptor-related protein 1(LRP1). Aβ is the top node by degree in the Core network, interacting with over 250 network proteins. Besides these well-known proteins, the list of high degree nodes from the Core network identified other key proteins that are part of important signaling pathways. PIK3R1 is part of the insulin receptor (INSR)/PI3K/AKT1 signaling pathway that is involved in glucose/insulin metabolism (Burke, 2007; Griffin et al., 2005), inhibition of the MAPK8 pathway (Burke, 2007), protection of cells against Aβ toxicity (Griffin et al., 2005), inhibition of apoptosis (Burke, 2007), and regulation, directly and indirectly via GSK3B, of phosphorylation and subsequent activity of MAPT (Dickey et al., 2008; Griffin et al., 2005). The map kinases MAPK1/3, along with associated protein kinases MAP2K1/2, are members of the MAPK/ERK pathway, another key signaling pathway that is involved in long-term potentiation and memory consolidation in the hippocampus (Dudai, 2004), processes that are severely affected in AD, as well as in cell proliferation, development, and transcriptional regulation (Sekine et al., 2006). Downstream targets of the MAPK1/3 signaling pathway include CREB1, NFKB1, MAPT, ARC, the γ-secretase complex member nicastrin (NCSTN) and GSK3B, all of which are part of the Core network.
We also determined that all of the top 25 proteins by degree in the Core network formed a connected component. In general, undirected biological networks tend to have few connections between high degree nodes (i.e., disassortative); this protects the network from disruption (Maslov and Sneppen, 2002). Due to the direct interactions between these top proteins, the Core network is more susceptible to disruption, and thus in AD, mutations to, or changes in expression levels of these key nodes, could rapidly affect the rest of the network.
All of the top 25 proteins by degree in the Expanded network are present in the Core network with the exception of EP300. EP300 is a homolog of the CREB binding protein (CREBBP) and interacts with numerous transcriptional regulatory factors. It also has histone acetyltransferase activity, interacts with the tumor suppressor protein p53 (TP53), and binds to CREB1 (Barco and Kandel, 2006). Francis et al. (2007) found that wild-type PSEN1 enhanced the transcriptional activity of EP300, but that PSEN1 with AD-associated mutations did not, suggesting a potential role for EP300 in AD-related processes.
Several proteins that were in the Core network list of top 25 proteins by degree also appeared in the Expanded network top 25 proteins by degree, including TP53, caspase 3 (CASP3), PIK3R1, and MAPK1. This could indicate an even greater role in AD for these proteins due to their large number of interactions.
One of the reasons we constructed the Expanded network was to identify additional proteins that could be involved in AD. Only a few of the top Core proteins by degree appeared in the list of top 25 proteins by degree in the Expanded network, indicating that most of the proteins that are recognized as being crucial to AD pathogenic processes and were highly connected in the Core network were not necessarily key hubs in the larger Expanded network. Similarly, several Core proteins with low degree showed a significant increase in degree in the Expanded network, indicating that these proteins could play a larger than previously appreciated role in AD. For example, YWHAG, a member of the important 14-3-3 protein family, had the largest increase in degree between the Core and Expanded networks and also had the largest degree of all nodes in the Expanded network. The 14-3-3 protein family is a group of key signaling proteins involved in important CNS functions such as memory and learning, response to stress, apoptosis, and neurotransmitter production (Ferl et al., 2002; Limviphuvadh et al., 2007). Several other proteins of the 14-3-3 family appeared in the Expanded network, including YWHAZ (degree 125, also in the Core network), YWHAB (degree 111), YWHAE (degree 65), YWHAQ (degree 65), and YWHAH (degree 50), suggesting a potentially important role for these proteins in AD-related processes.
To further analyze the Core network, we mapped gene expression data from four studies onto the network. Mapping of the Blalock study data, which included gene expression data from incipient, moderate, and severe AD, identified a connected component of downregulated nodes, centered on the MAPK/ERK pathway and related receptors, that continued to increase in size with disease progression. In severe AD, this connected component consisted of 81 downregulated nodes. The Nunez-Iglesias study had a large connected component of 169 downregulated nodes that also contained key proteins of the MAPK/ERK pathway. Downregulated nodes showed a significantly higher level of connectivity to each other relative to upregulated nodes in the Blalock and Nunez-Iglesias studies. The progression of AD, particularly in the Blalock study, showed a small number of nodes centered on the MAPK/ERK pathway and related receptors that were initially downregulated. This small connected component of downregulated nodes gradually increased in size with disease progression, affecting a widening number of key receptor-mediated signaling pathways, related proteins, and downstream targets, such as CREB1, which are key in regulation of subsequent gene expression in the CNS. Given the importance of the MAPK/ERK pathway and related proteins in memory and LTP in the hippocampus, one can see the relationship with key phenotypic characteristics such as impairment of memory and higher cognitive functions present in AD patients. The analysis of key network parameters as previously described also highlighted the importance of the MAPK/ERK pathway in the Core network and in AD.
Although upregulated nodes in the Blalock study did not show the same level of connectivity as did the downregulated nodes, there were still some significant connected components of upregulated nodes, particularly in severe AD. The largest of these consisted of 24 nodes, with PRKACG as the high degree node. Other nodes in this connected component included the insulin-like growth factor 1 (IGF1), its receptor (IGF1R), albumin (ALB), and endothelial nitric oxide synthase (NOS3). PRKACG, NOS3, and IGF1R also appeared as an upregulated triplet connected component in moderate AD in the Blalock study. IGF1, ALB, and IGF1R are involved in uptake and processing of Aβ peptides (Subramanian et al., 2005), so their upregulation in severe AD is possibly a response to increased Aβ levels. Another part of this connected component centers on CHAT, a key enzyme in acetylcholine synthesis, and three proteins that modulate CHAT activity (CALR3, PON1, and OTC). Given the loss of cholinergic neurons in AD, upregulation of this cluster could be a cellular response to AD pathology.
Another of the connected components of upregulated nodes in severe AD in the Blalock study centered on the clathrin heavy chain protein (CLTC). Other nodes in this connected component included a group of clathrin adaptor proteins (APBB2, GGA1, DAB1, APBA1, and LDLRAP1). These proteins are involved in intracellular trafficking and clathrin-mediated receptor endocytosis. LDLRAP1 mediates endocytosis of the LDL receptor (LDLR), which was also part of this upregulated cluster. APBA1, in conjunction with the MAPK8 interacting protein (MAPK8IP3), and APBB2, in conjunction with the adaptor protein CBL, act as clathrin adaptors involved in the trafficking and endocytosis of APP. MAPK8IP3 and CBL were also in this upregulated cluster. Upregulation of this connected component could increase clathrin-mediated receptor endocytosis. Endocytosis of the APP protein has been shown to increase β- and γ-secretase processing of APP and thus increase intracellular Aβ (Schneider et al., 2008), so the upregulation of clathrin and related adaptor proteins could potentially lead to increased intracellular pathological effects of Aβ. In a recent study, Marquer et al. (2011), showed a connection between high levels of cholesterol, a hallmark of sporadic AD, with increased APP and BACE1 clustering in lipid rafts of neurons and subsequent rapid clathrin-mediated APP endocytosis, which led to increased levels of intracellular Aβ production. Several genes identified by recent association studies as being linked to AD are also involved in receptor-mediated endocytosis and/or clathrin interactions (Naj et al., 2011). These include CD2-associated protein CD2AP (in Expanded network), bridging integrator 1 BIN1 (in Expanded network), and phosphatidylinositol binding clathrin assembly protein PICALM (in Core network). Our data plus the results of these recent association studies point to the potential importance of clathrin-mediated endocytosis in AD.
The Williams and Chen gene expression studies showed the opposite pattern from the Blalock and Nunez-Iglesias studies in terms of the average degree of up- and downregulated connected components, with higher connectivity levels in upregulated nodes. Both the Blalock and Nunez-Iglesias studies were taken from hippocampal or parietal lobe tissues of AD patients, whereas the Williams study was performed in vitro on synaptoneurosomes prepared from frontal cortex samples of subjects diagnosed with incipient AD, and the Chen study samples were extracted from peripheral blood mononuclear cells of AD patients. Recent research has shown a weak correlation between brain and blood gene expression levels (Cai et al., 2010).
Although it is difficult to draw global assumptions from a limited sample set, for the four expression studies we mapped to the Core network, some patterns emerged. Overall there appeared to be no clear differences between the total numbers of up- or downregulated genes that mapped to the Core network across any of the studies. However, the expression patterns in AD from those studies that used human CNS samples (Blalock and Nunez-Iglesias) indicated a significant increase in connectivity of downregulated genes relative to upregulated genes, and an increased size of the largest connected components of downregulated nodes over upregulated nodes. The Williams and Chen studies, which were not directly based on CNS samples, showed an increased connectivity of upregulated nodes, along with larger connected components of upregulated nodes relative to downregulated nodes. In either case, we surmised that connected (interacting) proteins in a network, if the underlying genes are concurrently up- or downregulated, would be likely to amplify the effect of shifts in expression levels on cellular processes as opposed to a similar number of up- or downregulated genes whose protein products do not directly interact with each other or appear in the same cellular pathway.
Conclusions
The structure of the Core network model of AD showed a dense, highly connected network with a larger than expected number of high degree hubs. Alrhough this type of network is able to rapidly transduce signals, it is also vulnerable to the disruption of the key hubs, many of which are known to be involved in AD pathology. The top 25 proteins by degree formed a connected component; this direct interaction of key hubs also makes the network susceptible to disruption. Coupled with the widespread pathological interactions of Aβ, the Core network could suffer rapid and widespread disruptions during the progression of AD. This suggests that effective treatment approaches to AD should also be global in their approach, targeting multiple Core network proteins and pathways to prevent or slow this disruption.
We identified key proteins and related cellular pathways in the Core and Expanded networks using an analysis of network parameters for both networks and mapping of expression data to the Core network. Based on the literature used to construct the Core network, we then looked for groups of these key proteins that were members of important pathways known to be involved in AD. The key pathways identified in the Core network included the INSR/PI3K/AKT1 pathway and the MAPK/ERK pathway, in conjunction with glutamate-receptor mediated pathways and related scaffolding proteins. The INSR/PI3K/AKT1 pathway is a significant signaling pathway in the CNS that is involved in glucose/insulin metabolism and energy metabolism, which can be disrupted in AD. Both the MAPK/ERK pathway and glutamate receptor complexes (receptors and related scaffolding proteins) are key in many critical CNS processes, in particular learning, memory, and LTP, which suffer major disruptions in AD. The mapping of gene expression data to the Core network in particular highlighted the importance of the MAPK/ERK pathway and glutamate receptor complexes, several key components of which were downregulated in AD. One potentially important upregulated pathway identified by mapping gene expression data to the Core network centered on clathrin and several clathrin adaptor proteins involved in receptor endocytosis. This suggests a potential increase in clathrin-mediated receptor endocytosis, which could include the endocytosis of APP. This has been shown to lead to increases in intracellular Aβ production. Several recent AD-associated genes identified by association studies are also involved in clathrin-mediated endocytosis or interact with clathrin, highlighting the potential importance of this process in AD.
Although the top proteins in the Expanded network by degree were predominantly Core network proteins, several showed a significant increase in degree from the Core to the Expanded network, such the 14-3-3 family members YWAHG and YWHAZ. In addition, the transcriptional cofactor EP300 appeared in the list of top 25 Expanded network proteins by degree, but is not present in the Core network. This suggests these proteins may play a larger than expected role in AD-associated processes and could be likely targets for further AD research.
Although network models and gene expression data are useful tools in and of themselves, we have demonstrated that the combination of PPI network models and gene expression data can provide additional important information in the study of complex diseases like AD by identifying key genes, proteins, and cellular pathways involved in disease processes. This, in turn, can help to prioritize gene and protein targets for future research.
Supplementary Material
Acknowledgments
The authors thank Loubin Yang for IT help and manuscript review, and Parag Joshi and Gaurav Kaushik for manuscript review. The research was supported in part by NIH Grant P20 RR016454 from the INBRE Program, a Graduate Student Research and Scholarship Committee Grant sponsored by the Idaho State University Office of Research, and a grant from the Molecular Research Core Facility of Idaho State University.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Argellati F. Domenicotti C. Passalacqua M. Janda E. Melloni E. Marinari U.M., et al. Protein kinase C-dependent alpha-secretory processing of the amyloid precursor protein is mediated by phosphorylation of myosin II-B. FASEB J. 2009;23:1246–1251. doi: 10.1096/fj.08-119263. [DOI] [PubMed] [Google Scholar]
- Assenov Y. Ramirez F. Schelhorn S.E. Lengauer T. Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L. Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–115. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Barco A. Kandel E.R. The role of CREB and CBP in brain function. In: Thiel G., editor. Transcription Factors in the Nervous System. Wiley-VCH; Weinheim: 2006. [Google Scholar]
- Batada N.N. Hurst L.D. Tyers M. Evolutionary and physiological importance of hub proteins. Plos Comput Biol. 2006;2:748–756. doi: 10.1371/journal.pcbi.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L. McQueen M.B. Mullin K. Blacker D. Tanzi R.E. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Blalock E.M. Geddes J.W. Chen K.C. Porter N.M. Markesbery W.R. Landfield P.W. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R.E. Inhibition of mitogen-activated protein kinase and stimulation of Akt kinase signaling pathways: two approaches with therapeutic potential in the treatment of neurodegenerative disease. Pharmacol Ther. 2007;114:261–277. doi: 10.1016/j.pharmthera.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C. Langfelder P. Fuller T.F. Oldham M.C. Luo R. van den Berg L.H., et al. Is human blood a good surrogate for brain tissue in transcriptional studies? BMC Genomics. 2010;11:589. doi: 10.1186/1471-2164-11-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo A. Azuaje F. Linking gene expression and functional network data in human heart failure. Plos One. 2007:2. doi: 10.1371/journal.pone.0001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S.E. Boutros M. Michelson A.M. Church G.M. Halfon M.S. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005:6. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G. Nepusz T. The igraph software package for complex network research. InterjComplex Syst. 2006:1695. [Google Scholar]
- Culmsee C. Molecular insights into mechanisms of the cell death program: role in the progression of neurodegenerative disorders. Curr Alzheimer Res. 2006;3:269–283. doi: 10.2174/156720506778249461. [DOI] [PubMed] [Google Scholar]
- Dickey C.A. Koren J. Zhang Y.J. Xu Y.F. Jinwal U.K. Birnbaum M.J., et al. Akt and CHIP coregulate tau degradation through coordinated interactions. Proc Natl Acad Sci USA. 2008;105:3622–3627. doi: 10.1073/pnas.0709180105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- Ferl R. Manak M. Reyes M. The 14-3-3s. Genome Biol. 2002;3:3010.3011–3010.3017. doi: 10.1186/gb-2002-3-7-reviews3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis Y.I. Diss J.K.J. Kariti M. Stephanou A. Latchman D.S. p300 activation by Presenilin 1 but not by its M146L mutant. Neurosci Lett. 2007;413:137–140. doi: 10.1016/j.neulet.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Gandhi T.K.B. Zhong J. Mathivanan S. Karthick L. Chandrika K.N. Mohan S.S., et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Gao J. Ade A.S. Tarcea V.G. Weymouth T.E. Mirel B.R. Jagadish H., et al. Integrating and annotating the interactome using the MiMI plugin for cytoscape. Bioinformatics. 2009;25:137–138. doi: 10.1093/bioinformatics/btn501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L. Cope L. Bolstad B.M. Irizarry R.A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Griffin R.J. Moloney A. Kelliher M. Johnston J.A. Ravid R. Dockery P., et al. Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer's disease pathology. J Neurochem. 2005;93:105–117. doi: 10.1111/j.1471-4159.2004.02949.x. [DOI] [PubMed] [Google Scholar]
- Hoe H.S. Harris D.C. Rebeck G.W. Multiple pathways of apolipoprotein E signaling in primary neurons. J Neurochem. 2005;93:145–155. doi: 10.1111/j.1471-4159.2004.03007.x. [DOI] [PubMed] [Google Scholar]
- Ideker T. Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M.G. Protein interactions and disease: computational approaches to uncover the etiology of diseases. Brief Bioinformatics. 2007;8:333–346. doi: 10.1093/bib/bbm031. [DOI] [PubMed] [Google Scholar]
- King J.Y. Ferrara R. Tabibiazar R. Spin J.M. Chen M.M. Kuchinsky A., et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–118. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- Krauthammer M. Kaufmann C.A. Gilliam T.C. Rzhetsky A. Molecular triangulation: bridging linkage and molecular-network information for identifying candidate genes in Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:15148–15153. doi: 10.1073/pnas.0404315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnick T.G. Papapetropoulos S. Mash D.C. Ffrench-Mullen J. Shehadeh L. de Andrade M., et al. A genomic pathway approach to a complex disease: axon guidance and parkinson disease. Plos Genet. 2007;3:984–995. doi: 10.1371/journal.pgen.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. Hao T. Shaw C. Patel A.J. Szabo G. Rual J.F., et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Limviphuvadh V. Tanaka S. Goto S. Ueda K. Kanehisa M. The commonality of protein interaction networks determined in neurodegenerative disorders (NDDs) Bioinformatics. 2007;23:2129–2138. doi: 10.1093/bioinformatics/btm307. [DOI] [PubMed] [Google Scholar]
- Lin W.H. Liu W.C. Hwang M.J. Topological and organizational properties of the products of house-keeping and tissue-specific genes in protein–protein interaction networks. BMC Syst Biol. 2009:3. doi: 10.1186/1752-0509-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. Jiang T.Z. Ma S.D. Zhao H.Z. Li J. Jiang X.P., et al. Exploring candidate genes for human brain diseases from a brain-specific gene network. Biochem Biophys Res Comm. 2006;349:1308–1314. doi: 10.1016/j.bbrc.2006.08.168. [DOI] [PubMed] [Google Scholar]
- Lu X. Jain V.V. Finn P.W. Perkins D.L. Hubs in biological interaction networks exhibit low changes in expression in experimental asthma. Mol Syst Biol. 2007;3:6. doi: 10.1038/msb4100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquer C. Devauges V. Cossec J.-C. Liot G. Lecart S. Saudou F., et al. Local cholesterol increase triggers amyloid precursor protein-Bace 1 clustering in lipid rafts and rapid endocytosis. FASEB J. 2011;25:1295–1305. doi: 10.1096/fj.10-168633. [DOI] [PubMed] [Google Scholar]
- Maslov S. Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Miller J.A. Oldham M.C. Geschwind D.H. A systems level analysis of transcriptional changes in Alzheimer's disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj A.C. Jun G. Beecham G.W. Wang L.-S. Vardarajan B.N. Buros J., et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011 doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez-Iglesias J. Liu C.C. Morgan T.E. Finch C.E. Zhou X.J. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. Plos One. 2010:5. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop J.J. Jones B. Kekonius L. Chin J. Yu G.Q. Raber J., et al. Neuronal depletion of calcium-dependent proteins in the dentate gyrus is tightly linked to Alzheimer's disease-related cognitive deficits. Proc Natl Acad Sci USA. 2003;100:9572–9577. doi: 10.1073/pnas.1133381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T.S.K. Goel R. Kandasamy K. Keerthikumar S. Kumar S. Mathivanan S., et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K. Construction and analysis of protein–protein interaction networks. Autom Exp. 2010;2:2. doi: 10.1186/1759-4499-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardoni G. Petterlini M. Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E. Lum P.Y. Reverse engineering gene networks to identify key drivers of complex disease phenotypes. J Lipid Res. 2006;47:2601–2613. doi: 10.1194/jlr.R600026-JLR200. [DOI] [PubMed] [Google Scholar]
- Schneider A. Rajendran L. Honsho M. Gralle M. Donnert G. Wouters F., et al. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci. 2008;28:2874–2882. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y. Takeda K. Ichijo H. The ASK1-MAP kinase signaling in ER stress and neurodegenerative diseases. Curr Mol Med. 2006;6:87–97. doi: 10.2174/156652406775574541. [DOI] [PubMed] [Google Scholar]
- Shannon P. Markiel A. Ozier O. Baliga N.S. Wang J.T. Ramage D., et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberts S.K. Schadt E.E. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Lopez M. Zanzoni A. Lluis R. Stelzl U. Aloy P. Interactome mapping suggests new mechanistic details underlying Alzheimer's disease. Genome Res. 2011;21:364–376. doi: 10.1101/gr.114280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C. Breitkreutz B.J. Reguly T. Boucher L. Breitkreutz A. Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. Tamayo P. Mootha V.K. Mukherjee S. Ebert B.L. Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.D.C. R: a language and environment for statistical computing (R Foundation for Statistical Computing. Vienna; Austria: 2009. [Google Scholar]
- Wachi S. Yoneda K. Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D.M. Selkoe D.J. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron. 2004;44:181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Williams C. Shai R.M. Wu Y.C. Hsu Y.H. Sitzer T. Spann B., et al. Transcriptome analysis of synaptoneurosomes identifies neuroplasticity genes overexpressed in incipient Alzheimer's disease. Plos One. 2009:4. doi: 10.1371/journal.pone.0004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q. Pan J. Ba M.W. Sun Z.K. Ma G.Z. Lu G.Q., et al. New protein kinase C activator regulates amyloid precursor protein processing in vitro by increasing alpha-secretase activity. Eur J Neurosci. 2007;26:381–391. doi: 10.1111/j.1460-9568.2007.05648.x. [DOI] [PubMed] [Google Scholar]
- Zhu X.W. Gerstein M. Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.