Abstract
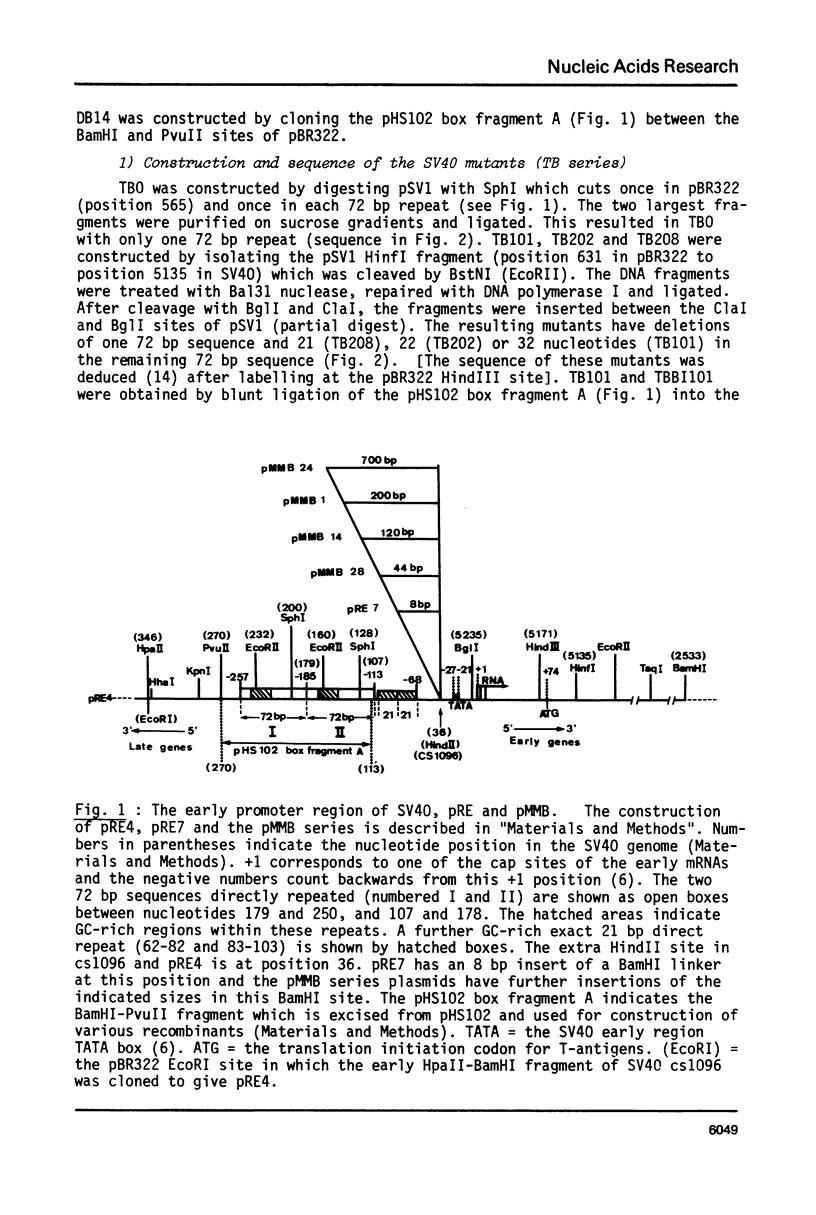
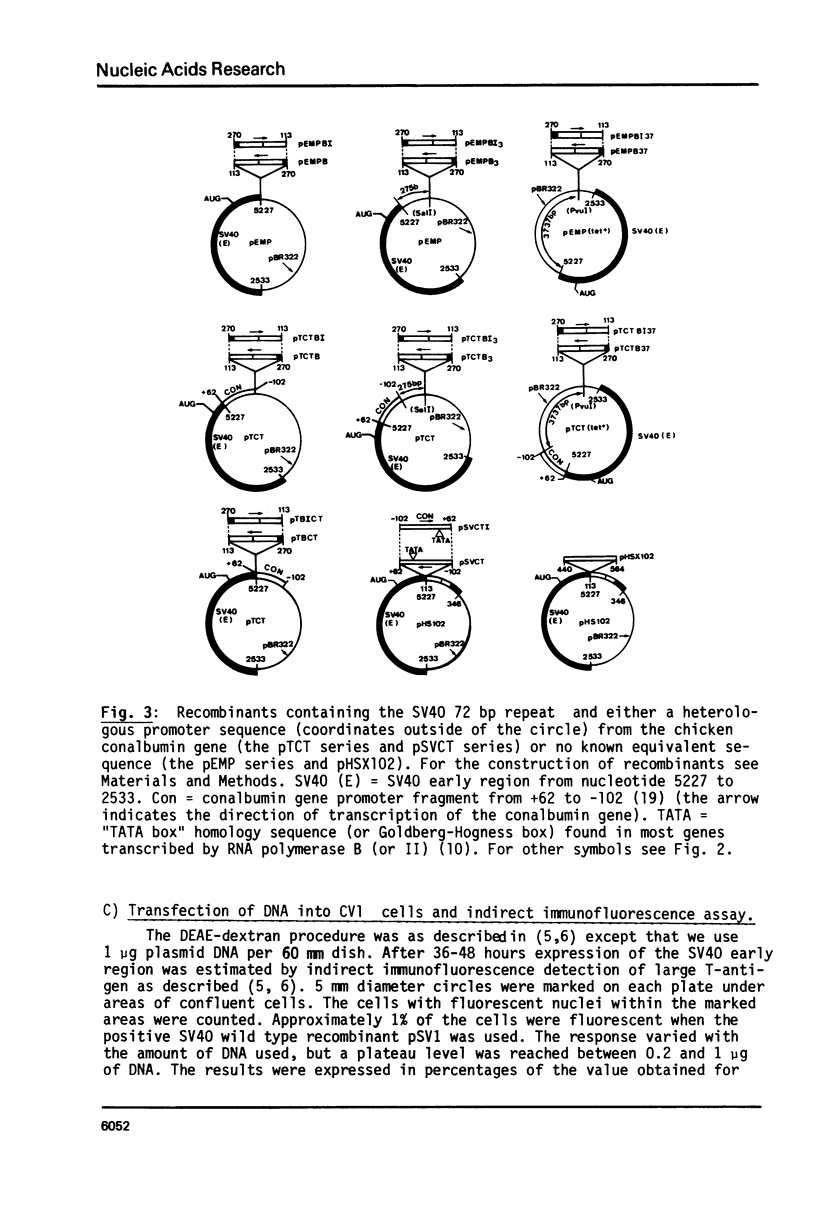
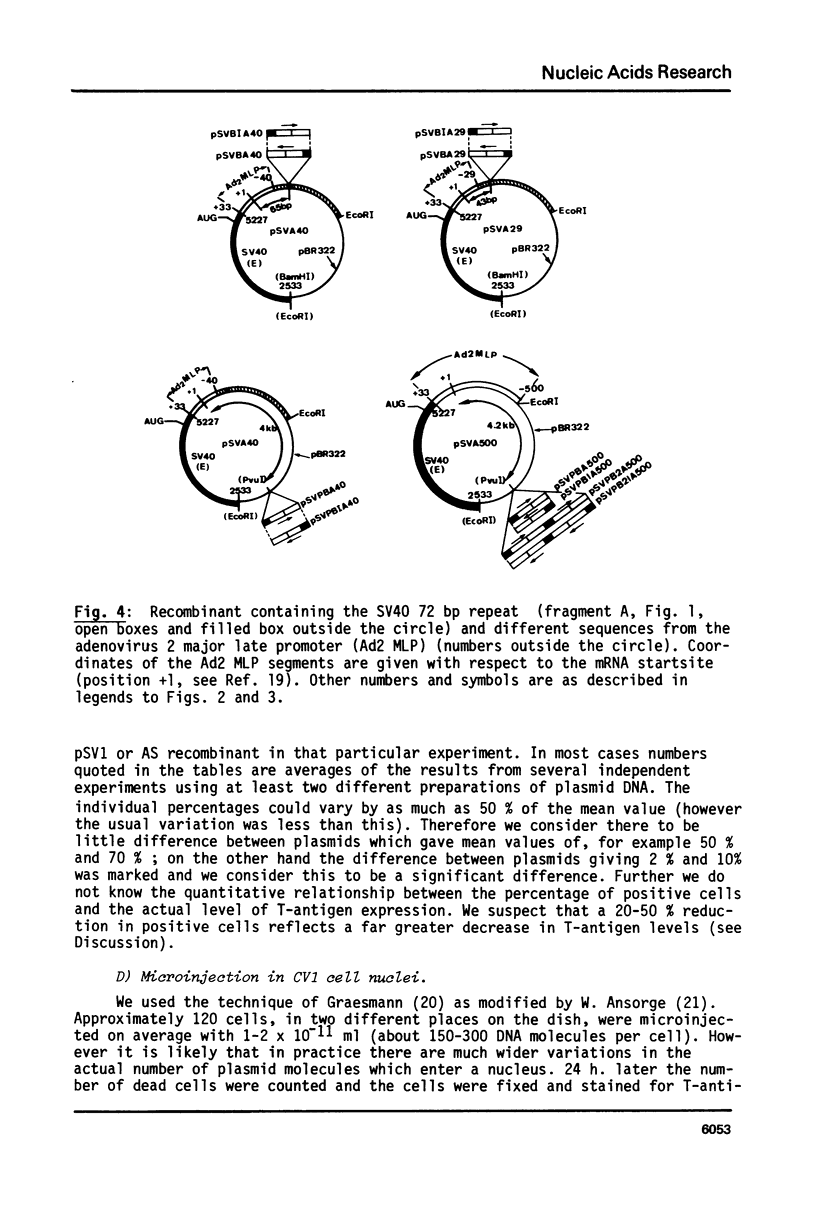
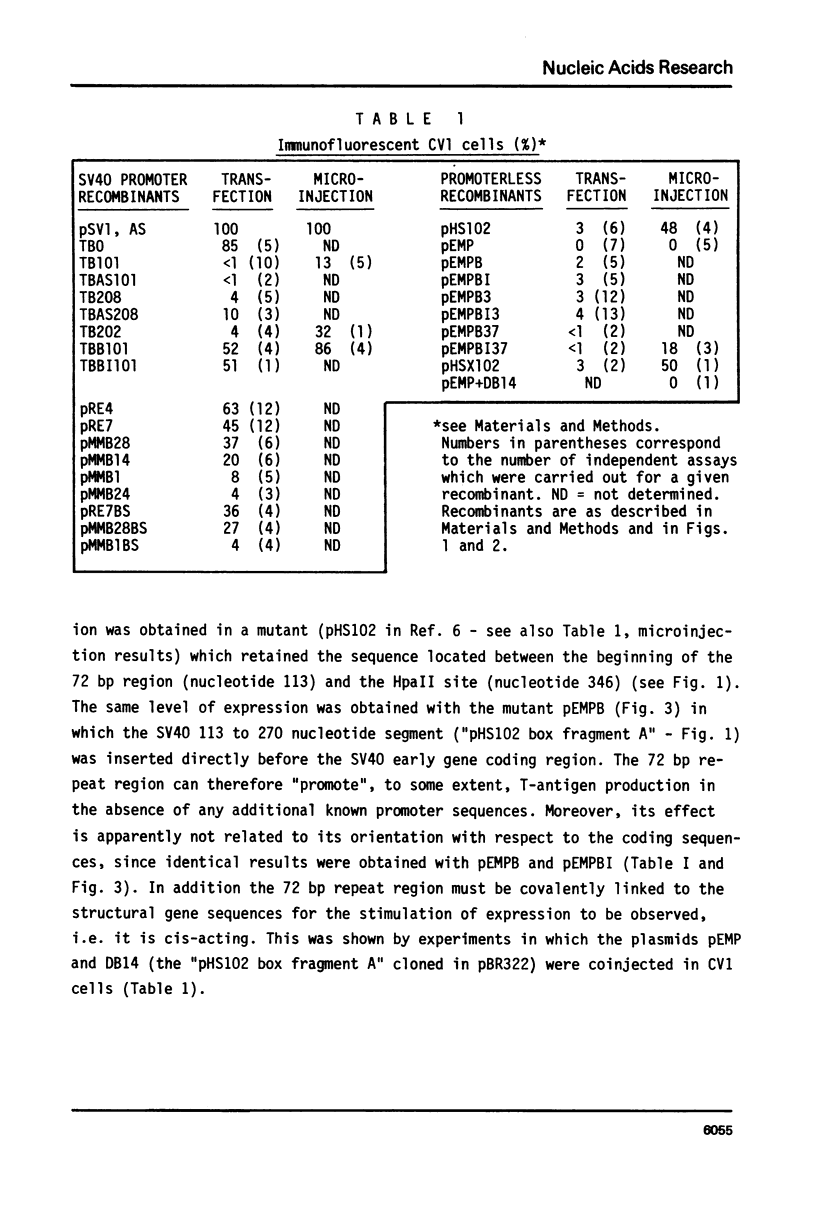
By introduction of recombinant plasmids into monkey CV1 cells, we have unambiguously demonstrated that sequences entirely within the 72 bp repeat, which is located upstream of the SV40 early region, are crucial for T-antigen expression in vivo. We have also shown that a DNA fragment containing the 72 bp repeat, inserted directly before chicken conalbumin or adenovirus-2 major late promoter sequences in chimeric plasmids where these promoters replace that of the SV40 early genes, caused a dramatic increase in the expression of T-antigen in vivo. This effect was independent of the orientation of the 72 bp repeat, but was sensitive to its location within the plasmid, when the 72 bp repeat was separated from the promoter sequences, T-antigen expression was reduced. Insertion of the 72 bp repeat into equivalent plasmids containing no known eukaryotic promoter sequences (plasmids which were not detectably expressed in vivo) gave rise to a measurable, but smaller level of expression. The stimulation of expression by the 72 bp repeat is cis-acting : it required covalent linkage to the recombinant. We discuss the possibility that the 72 bp repeat region in SV40 may act as a bi-directional entry site for RNA polymerase B such that promoter sequences linked to the repeat are more efficiently utilised.
Full text
PDF

















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., Chambon P. Deletions covering the putative promoter region of early mRNAs of simian virus 40 do not abolish T-antigen expression. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3865–3869. doi: 10.1073/pnas.77.7.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Capecchi M. R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980 Nov;22(2 Pt 2):479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- Cochet M., Gannon F., Hen R., Maroteaux L., Perrin F., Chambon P. Organization and sequence studies of the 17-piece chicken conalbumin gene. Nature. 1979 Dec 6;282(5739):567–574. doi: 10.1038/282567a0. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Lebowitz P., Frisque R. J., Gluzman Y. Identification of a promoter component involved in positioning the 5' termini of simian virus 40 early mRNAs. Proc Natl Acad Sci U S A. 1981 Jan;78(1):100–104. doi: 10.1073/pnas.78.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessmann M., Graessman A. "Early" simian-virus-40-specific RNA contains information for tumor antigen formation and chromatin replication. Proc Natl Acad Sci U S A. 1976 Feb;73(2):366–370. doi: 10.1073/pnas.73.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980 Aug;21(1):115–125. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- JACOB F., ULLMAN A., MONOD J. LE PROMOTEUR, 'EL'EMENT G'EN'ETIQUE N'ECESSAIRE 'A L'EXPRESSION D'UN OP'ERON. C R Hebd Seances Acad Sci. 1964 Mar 16;258:3125–3128. [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. A nucleosome-free region in SV40 minichromosomes. Nature. 1980 May 22;285(5762):263–265. doi: 10.1038/285263a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Miller K. G. Eukaryotic DNA topoisomerases: two forms of type I DNA topoisomerases from HeLa cell nuclei. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3487–3491. doi: 10.1073/pnas.78.6.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelkin B. D., Pardoll D. M., Vogelstein B. Localization of SV40 genes within supercoiled loop domains. Nucleic Acids Res. 1980 Dec 11;8(23):5623–5633. doi: 10.1093/nar/8.23.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P., Corden J., Kédinger C., Chambon P. Promotion of specific in vitro transcription by excised "TATA" box sequences inserted in a foreign nucleotide environment. Nucleic Acids Res. 1981 Aug 25;9(16):3941–3958. doi: 10.1093/nar/9.16.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaife J., Beckwith J. R. Mutational alteration of the maximal level of Lac operon expression. Cold Spring Harb Symp Quant Biol. 1966;31:403–408. doi: 10.1101/sqb.1966.031.01.052. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Sternglanz R., DiNardo S., Voelkel K. A., Nishimura Y., Hirota Y., Becherer K., Zumstein L., Wang J. C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci U S A. 1981 May;78(5):2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Deletion mapping a eukaryotic promoter. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4461–4465. doi: 10.1073/pnas.78.7.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K. N., Shenk T. Definition of the boundaries of the origin of DNA replication in simian virus 40. Nucleic Acids Res. 1978 Oct;5(10):3635–3642. doi: 10.1093/nar/5.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn H., Fiers W. Nucleotide sequence of the Hind-C fragment of simian virus 40 DNA. Comparison of the 5'-untranslated region of wild-type virus and of some deletion Mutants. Eur J Biochem. 1979 Oct;100(1):51–60. doi: 10.1111/j.1432-1033.1979.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn H., Fiers W. Nucleotide sequence of the Hind-C fragment of simian virus 40 DNA. Comparison of the 5'-untranslated region of wild-type virus and of some deletion Mutants. Eur J Biochem. 1979 Oct;100(1):51–60. doi: 10.1111/j.1432-1033.1979.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Sauer G. Localization of a deletion in the DNA of a biologically active simian virus 40 strain. FEBS Lett. 1976 Dec 1;71(2):313–315. doi: 10.1016/0014-5793(76)80959-3. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Chambon P. A T to A base substitution and small deletions in the conalbumin TATA box drastically decrease specific in vitro transcription. Nucleic Acids Res. 1981 Apr 24;9(8):1813–1824. doi: 10.1093/nar/9.8.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Derbyshire R., Guy A., Molko D., Roget A., Téoule R., Chambon P. Specific in vitro transcription of conalbumin gene is drastically decreased by single-point mutation in T-A-T-A box homology sequence. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7024–7028. doi: 10.1073/pnas.77.12.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]



