Abstract
Background
Presence of microorganisms in the circulating blood whether continuously or intermittently is a threat to every organ in the body. Approximately 200,000 cases of bacteraemia occur annually with mortality rates ranging from 20–50%. Early diagnosis and appropriate treatment of these infections can make the difference between life and death. The aim of the present study was to determine the bacterial flora of the blood stream infections and their antibiotic susceptibility pattern.
Methods
A cross sectional study was conducted on 260 adult febrile patients in Jimma University Specialized Hospital from 27 October 2009 to 26 March 2010. The positive blood cultures were examined and the organisms were identified as per standard procedures. Antimicrobial testing was performed for all isolates by disk diffusion techniques, according to Clinical Laboratory Standards Institute guide lines. The data was analyzed using SPSS for windows version 16 and Microsoft Office Excel.
Results
From the total of two hundred sixty blood specimens only 23(8.8%) were positive to seven different types of bacteria. The isolated bacteria were: Coagulase negative staphylococci 6(26.1%), S. aureus 5 (21.7%), S. pyogens 3 (13.0%), E. coli 4(17.4%), K. pneumoniae 3(13.0%), Salmonella spp. 1(4.3%), and Citrobacter spp. 1(4.3%). The isolates showed high rates of resistance to most antibiotics tested. The range of resistance for gram positive bacteria were 0% to 85.7%, and for gram negative from 0% to 100%. None of the isolates were resistance to ciprofloxacin and ceftriaxone.
Conclusion
Our study result showed the presence of invasive bacterial pathogens with high rate of resistance to most commonly used antibiotics used to treat bacterial infections. Therefore, timely investigation of bacterial flora of the blood stream infections and monitoring of their antibiotic resistance pattern plays an important role in reduction of the incidence of blood stream infections.
Keywords: invasive bacteria, antimicrobial resistance, Jimma, Ethiopia
Introduction
Bloodstream infection (BSI) remains one of the most important causes of morbidity and mortality through out the world. The disease may be short and self limiting or may result in death or serious morbidity including admission to intensive care or prolonged hospital stay (1). A wide range of bacteria has been described in febrile patients including gram negative bacteria such as E. coli, P. aeruginosa, Klebsiella species Neisseria meningitidis, H. influenzae, and gram positive such as Coagulase negative staphylococci (CNS), S. aureus, S. pneumoniae, S. pyogens, S. agalactiae, and E. faecium, (2,3,4,5,6).
Fever is a common presenting symptom to the emergency department and is attributable to a wide range of clinical diseases or infection (2, 7). The timely and appropriate use of antibiotics is currently the only way to treat bacteremia. However, many bacterial pathogens have become resistant to antibiotic regimens and become a serious public health concern with economic and social implications throughout the world. The infections caused by multi drug resistance organisms are more likely to prolong the hospital stay, increase the risk of death and require treatment with more expensive antibiotics (8).
In Ethiopia, except few reports on blood culture isolates from Tikur Anbessa Hospital (3, 9) data about the role and frequency of invasive bacteria on febrile adult cases and their susceptibility patterns is scarce. Thus providing updated information for local hospitals or regions for empirical choice of antibiotics in the treatment of bacteremia is very useful. Therefore, the present study was aimed at determining the frequency or occurrence of blood bacterial isolates in adult febrile cases and evaluate their drug susceptibility patterns at Jimma university specialized hospital.
Materials and Methods
A prospective cross-sectional study was conducted in Jimma university specialized hospital (JUSH) Jimma, South west Ethiopia, to isolate invasive bacteria from adult febrile cases and assess their antibiotic susceptibility pattern from 27 October 2009 to 26 March 2010.
A total of 260 age ≥18 years old patients with fever (axillary temperature of ≥38°C) from the Out Patient Department were taken as study subject. The sample size was determined based on the prevalence rate of the study done by Asrat and Amanuel (3) and calculated with the formula recommended by Naing et al., (10). Patients who were not voluntary to participate and took antibiotic within the last two weeks of visiting the hospital were excluded from this study.
After providing written informed consent, each patient was examined by physician and all relevant data (demographic, clinical, and laboratory data) were also recorded on questionnaire prepared for this purpose.
About 5 ml of venous blood was collected aseptically using blood sample collection sets and antiseptics (70% alcohol and 2% tincture iodine) and transferred into a bottle containing sterile Brain heart infusion (BHI) broth (Oxoid Ltd, UK), with 0.025% Sodium Polyanethol Sulfonate (SPS) [Himedia, India]. A minimum blood-to-broth ratio of 1 in 10 was maintained. Blood culture broths were incubated at 370C and checked for sign of bacterial growth daily up to 7 days, and bottles which showed signs of growth were further processed for gram stain and sub-cultured was made onto blood agar (Oxoid Ltd, UK), MacConkey agar (Oxoid Ltd, UK), Manitol salt agar (Oxoid Ltd, UK) and incubated at 37°C for 24 hours (11).
Blood culture broth with no bacterial growth after 7 days were sub-cultured before being reported as a negative result. The isolates were identified with colony morphology, gram staining reaction, biochemical and serological tests (11). Antimicrobial susceptibility testing was performed for the isolated organisms according to Clinical Laboratory Standards Institute (CLSI) guide lines (12, 13). The antibiotic discs and their concentrations were: Penicillin G (P, 10IU), Amoxicillin (AML, 20µg), Ampicillin (AMP, 10µg), Cephalothin (KF, 30µg) Tetracycline (TE, 30µg), Chloramphenicol (C, 30µg), Ciprofloxacin (CIP, 5µg), Trimethoprim+Sulphamethazole (SXT, 25µg) Gentamicin(CN, 10µg), Ceftriaxone (CRO, 30µg), Erythromycin (E, 10µg), Methicillin (MET, 5µg) and Nalidixic acid (NA, 30µg). All the antimicrobials used for the study were obtained from Oxoid, UK. The criteria used to select the antimicrobial agents tested were based on the availability and frequency of prescription for the management of bacterial infections in Ethiopia. The reference strains used as control were E. coli (ATCC 25922), and S. aureus (ATCC 25923). In this study multi-drug resistance was defined as simultaneous resistance to two or more antimicrobial agents.
Both the clinical and laboratory data were entered into a computer and analyzed using SPSS for widows version 16.0 and Microsoft Office Excel. Two-sided P values were calculated using the chi-square test or Fisher's exact test for some variables.
Ethical clearance from Jimma University Ethical Review Board and patient's consent was obtained with their signature after getting enough information on the purpose of the study.
Results
Out of 260 patients 106 (40.8%) were males and 154 (59.2%) females. Their age ranged from 18–61 with median of 26 years. The majority of the patients 158 (60.7%) were between the age of 18 and 29 years. One hundred eighty seven (51.9%) patients were rural and 73 (48.1%) town residents.
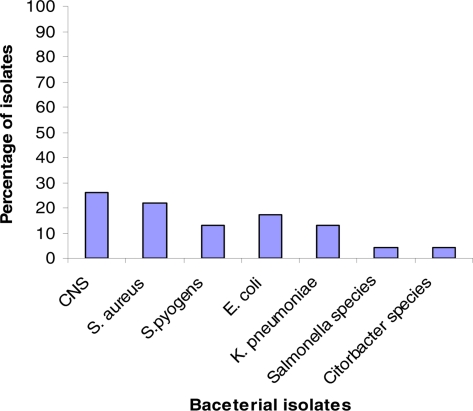
From the 260 blood cultures a total of 23 bacterial strains were isolated from 23 different cases, with a culture positivity rate of 8.8% (Fig. 1). The overall rate of isolation were 60.9% (n= 14) for gram positive bacteria and 39.1% (n= 9) for gram negative bacteria. As it is shown on Fig 1, the predominant isolates were coagulase negative staphylococci (CNS) 6 (26.1%) followed by S. aureus 5 (21.7%) and the least were Salmonella species 1(4.3%), and Citrobacter species (4.3%).
Figure 1.
Frequency of blood bacterial isolates from adult febrile cases in Jimma University
Among the gram positive bacteria high resistance was observed to Penicillin (85.7%) and Ampicillin (71.4%) (Table 1). The gram negative bacteria showed 100% resistance against Ampicilin, 88.9% to Tetracycline, 77.9% to Chloroamphenicol, 77.8% to Co-trimoxazole, and 66.7% against Gentamicin (Table 2). Overall the range of resistance for gram positives was from 0% to 85.7%, and for gram negative from 0% to 100%. However, all bacterial isolates were sensitive to Ceftriaxone and Ciprofloxacin.
Table 1.
Antibiotic resistance of gram positive bacterial isolates from blood culture in Jimma University Specialized Hospital Jimma town, Southwest Ethiopia, October, 2009 to March, 2010.
| Name of isolates | Number of strains (%) resistance to | |||||||||||
| P | AMX | AM | C | CF | CIP | GM | CRO | TE | E | MET | SXT | |
| CNS (n=6) | 4(66.7) | 1(16.7) | 2(33.3) | 2(33.3) | 0(0) | 0(0) | 1(16.7) | 0(0) | 1(16.7) | 0(0) | 2(33.3) | 1(16.7) |
| S. aureus(n=5) | 5(100.0) | 2(40.0) | 5(100.0) | 5(100.0) | 1(20.0) | 0(0) | 3(60.0) | 0(0) | 5(100.0) | 2(40.0) | 5(100.0) | 3(60.0) |
| S. pyogens (n=3) | 3(100.0) | 0(0) | 3(100.0) | 0(0) | 0(0) | 0(0) | 0(0) | 0(0) | 1(33.3) | 0(0) | 0(0) | 0(0) |
| Total (n=14) | 12(85.7) | 3(21.4) | 10(71.4) | 7(50) | 1(7.1) | 0(0) | 4(28.6) | 0(0) | 7(50) | 2(14.3) | 7(50) | 11(78.6) |
Key:- P=Penicillin G, AMX=Amoxicillin, AM =Ampicillin, E= Erythromycin, CF =Cephalothin, TE=Tetracycline, C=Chloramphenicol, CIP=Ciprofloxacin, SXT= Trimethoprim+ Sulphamethazole, GM= Gentamicin, CRO= Ceftriaxone, MET= Methicillin, CNS= Coagulase negative staphylococcus
Table 2.
Antibiotic resistance of gram negative bacterial isolates from blood culture Jimma University Specialized Hospital, Southwest Ethiopia from October 2009 to March 2010
| Name of isolates | Number of strains (%) resistance to | |||||||||
| AMX | AM | C | CF | CIP | GM | CRO | TE | NA | SXT | |
| E. coli (n=4) | 0(0) | 4(100) | 4(100) | 0(0) | 0(0) | 3(75) | 0(0) | 3(75) | 0(0) | 4(100) |
| K. pneumoniae (n=3) | 1(33) | 3(100) | 1(33) | 1(33) | 0(0) | (33) | (0) | 3(100) | 3(100) | 3(100) |
| Salmonella spp. (n=1) | 1(100) | 1(100) | 1(100) | 1(100) | 0(0) | 1(100) | 0(0) | 1(100) | 0(0) | NT |
| Citrobacter spp. (n=1) | 1(100) | 1(100) | 1(100) | 1(100) | 0(0) | 1(100) | 0(0) | 1(100) | 0(0) | 1(100) |
| Total (n=9) | 3(33) | 9(100) | 7(77.8) | 3(33) | 0(0) | 6(66.7) | 0(0) | 8(88.9) | 3(33) | 8(88.9) |
Key: - AMX =Amoxicillin NA=Nalidixic acid, AM =Ampicillin, CF =Cephalothin, TE= Tetracycline, C= Chloramphenicol, CIP= Ciprofloxacin, SXT= Trimethoprim+Sulphamethazole, GM= Gentamicin CRO= Ceftriaxone , MET= Methicillin
Most gram positive and gram negative isolates were resistant to more than three antibiotics. Over all five distinct antibiogram (resistance pattern) were observed in CNS-four in S. aureus, two in S. pyogenes and the antibiogram of gram negative bacteria were two in E. coli and two in K. pneumonae (Table 3). The resistance pattern of CNS varied from 3–6 drugs. Five of the total CNS isolates were multi drug resistant (resistant to 3 or more drugs). All the S. aureus isolates were multi drug resistant varying from 5–10 drugs whereas two and one S. pyogenes isolates were resistant to two and three drugs, respectively. All the gram negative bacteria isolates were multidrug resistant. The single isolate of Salmonella spp. and Citrobacter spp. were resistant to six and seven drugs, respectively.
Table 3.
Resistance antibiogram pattern of the blood bacterial isolates Jimma University Specialized Hospital, Southwest Ethiopia from October 2009 – March 2010.
| Blood isolates | Resistance antibiogram | No |
| Gram positive | ||
| CNS | P,AM,C P,AM,MET P,C,TE P,AM,GM P,AM,C,CF,MET,SXT |
1 1 1 1 1 |
| S. aureus | P,AM,C,TE,MET P,AM,C,GM,TE,MET,SXT P, AMX,AM,C,GM,TE,E,MET,SXT P,AMX,AM,C,CF,GM,TE,E,ME,SXT |
2 1 1 1 |
| S. pyogens | P,AM P,AM,TE |
2 1 |
| Gram negative | ||
| E. coli | AM,C,GM,SXT AM,C,GM,TE,SXT |
1 3 |
| K. pneumonae | AMX,AM,TE,NA AM, C, CF,GM,TE,SXT,NA |
1 2 |
| Salmonella spp. | AMX,AM,C,CF,TE,SXT | 1 |
| Citrobacter spp. | AMX,AM,C,CF,GM,TE,SXT | 1 |
Key: Penicillin G = P, AMX =Amoxicillin NA=Nalidixic acid, AM =Ampicillin, E= Erythromycin, KF =Cephalothin, TE= Tetracycline, C= Chloramphenicol, SXT= Trimethoprim+Sulphamethazole, CN= Gentamicin, MET= Methicillin, CNS= Coagulase negative staphylococcus
Discussion
Blood stream infections cause significant morbidity and mortality worldwide and are among the most common healthcare-associated infections. The isolated bacteria are numerous and their associated disease need urgent and invasive management with antimicrobial drugs (1).
The isolation rate (8.8%) in this study is lower compared to other studies reported in Addis Ababa hospital (21.4%) (3) and in another African and Asian countries like Malawi 17.6% (14), Nepal 23.1% (15), 34% in Gambia (16) and 40% in India (17), respectively. However, there are reports from other parts of the world that agree with the present findings such as in Nepal 10 % (19) and in India 9.94% (6). In the present study gram positive bacteria were the most leading isolates (60.9%) as compared to gram negative (39.1%) which is comparable with previous study done in Addis Ababa, where 62.6% of the isolates were gram positive and 37.4% gram negative, respectively (7). But different from the study results done in Nepal where 89.19% of the total isolates were gram negative and 10.81% gram positive (18). The possible explanation for the difference could be the difference in blood culture system, the study design, geographical location, nature of patient population, epidemiological difference of the etiological agents, and seasonal variation.
Among blood isolates CNS (26.1%) was the most common organism, followed by S. aureus (21.7%) and E. coli (17.4%) and similar findings have been reported in other studies (4, 5, 19). CNSs have been long regarded as non pathogenic but their important role as pathogens and their increasing incidence in different infection like bacteremia related to indwelling devices, central nervous systems shunt infections, native or prosthetic valve endocardiatis, urinary tract infections had been reported and it is also an important cause of morbidity and mortality in immunocopromised individuals (20). Other investigators have also reported an increase of true bacteremia by CNS in recent years (5). In the present study the second predominant isolates were S. aureus, 21.7%, which contrast to the study conducted in USA where the prevalence was 2.6% (21) but comparable with the study results done in Iowa, USA 27.4% of S. aureus bacteremia (22). S. pyogenes accounted 13.0% in the present study which is higher than reported by Usha Arora and Pushpa Devi, 1.24% (23). In this study, E. coli was accounted for 17.4% which agreed with study done in Australia (19%) (24) and higher than other studies (3, 4, 5, 19). K. pneumoniae accounted for 13.0% of the total blood isolates which is almost similar with study results in India done at different times (6, 19). The isolation rate of Salmonella species was 4.3% and comparable with the study done in Addis Ababa, 3.8% (3) and in Jimma, 5.3% (25).
The rate of resistance for the isolates to antibiotics used is comparatively higher than from the previous reports in Addis Ababa (3, 9) but in agreement with a study done by other investigator elsewhere in the world (19). Overall the range of resistance for gram positives was from 0% to 85.7%, and for gram negative from 0% to 100% which is different from the study result done in Addis Ababa where the rates for gram positive bacteria ranged from 12% to 76%, and for gram negatives ranged from 8% to 46%(3). The increased resistant blood isolates in this study may be an indication of indiscriminate and continuous use of sub-therapeutic doses of commonly available antimicrobials both in the veterinary and public health sectors. Amongst the gram positive organism's high resistance was observed with Ampicillin (71.4%) which is consistent with other study, where the rate of resistance was 74.61 % (19). Further more, high resistance among gram negatives was seen against Ampicillin (82.6%), Tetracycline (65.2%) and Chloroamphenicol (60.9%) which is in agreement with the study findings by Usha Arora and Pushpa Devi in India (19). Almost all isolates were resistant to two or more antimicrobials. All S. aureus were multidrug resistant which is higher than previous study in Jimma (72.9%) (26). All S. aureus isolates were resistant for Penicillin, Ampicillin, chlorampphenicol, Tetracycline and Methicillin. This is in contrast with the study result of Balta and Deribie, where resistance rate against Ampicillin was 87.1%, 70.6 % to chlorampphenicol, and 51.8% against Methicillin (26). The single isolate of Salmonella species and Citrobacter species were found to be resistant to six or more antimicrobials. The high resistance rate of Salmonella species is similar to the study result of Beyene et al where most of the Salmonella isolates were multidrug resistant (25). However, among the antibiotics used for susceptibility testing ceftriaxone and ciprofloxacin were effective against all the isolates.
In Ethiopia the unregulated over-the-counter sale of these antimicrobials, mainly for self-treatment of suspected infection in humans, and to a lesser extent for use in animals without prescription, would inevitably lead to emergence and rapid dissemination of resistance. In addition, availability of cheaper generic drugs of variable quality in the market for treatment of bacterial infections may also contribute to the increased level of resistance. A study on practice of self-medication in Jimma town showed that out of the 152 ill people, 27.6% were self-medicated (27). The relative lesser cost (35.7%) was the major reason for using self medication.
In conclusion, our study result showed the presence of invasive bacterial pathogens with high rate of resistance to most commonly used antibiotics to treat bacterial infections. Therefore, timely investigation of bacterial flora of the blood stream infections and monitoring of their antibiotic resistance pattern plays an important role in reduction of the incidence of blood stream infections.
ACKNOWLEDGMENTS
We are indebted to Jimma University for supporting this study.
References
- 1.Larry GR, Michael LW, Melvin PW. Update on Detection of Bacteremia and Fungemia. Clin Microbiol Rev. 1997;10(3):444–465. doi: 10.1128/cmr.10.3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel RK, Scott AF, James MB, Sanjay S. Brief Report: Incidence, Etiology, Risk Factors, and Outcome of Hospital-acquired Fever. J Gen Intern Med. 2006;21:1184–1187. doi: 10.1111/j.1525-1497.2006.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asrat D, Amanuel Y. Prevalence and antibiotic susceptibility pattern of bacterial isolates from culture in Tikur Anbessa hospital, Addis Ababa, Ethiopia. Ethiop Med J. 2001;39(2):97–104. [PubMed] [Google Scholar]
- 4.James AK, Mark EJ, Deborah CD, Clyde T, Daniel FS, Gregory AV. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Annals of Clinical Microbiology and Antimicrobials. 2004;3(7):1–8. doi: 10.1186/1476-0711-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rina K, Nadeem SR, Kee PN, Parasakthi N. Etiology of blood culture isolates among patients in a multidisciplinary teaching hospital in Kuala Lumpur. J Microbiol Immunol Infect. 2007;40:432–437. [PubMed] [Google Scholar]
- 6.Manjula M, Pyria D, Varsha G. Antimicrobial susceptibility pattern of blood isolates from a teaching Hospital in north India. Jpn J Infec Dis. 2005;58:174–176. [PubMed] [Google Scholar]
- 7.Yung A. Approach to undifferentiated fever in adults. In: Cameron P, Jelinek G, Kelly A, et al., editors. Textbook of adult emergency medicine. 1st ed. Edinburgh: Churchill Livingston; 2000. pp. 303–307. [Google Scholar]
- 8.JoAnn D. Antibiotic resistance: The ongoing challenge for effective drug therapy. JAAPA. 2009;22(3):18–22. doi: 10.1097/01720610-200903000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Gedebou M, Tassew A, Azene G. Blood culture isolates from and Addis Ababa Hospital: Frequency and antibiotic sensitivities. East Afr Med J. 1984;61:190–196. [PubMed] [Google Scholar]
- 10.Naing L, Winn T, Rusli BN. Practical Issues in Calculating the Sample Size for Prevalence Studies. Archives of Orofacial Sciences. 2006;1:9–14. [Google Scholar]
- 11.World Health Organization, author. Manual for the laboratory identification and antimicrobial susceptibility testing of bacterial pathogens of public health concern in the developing world. WHO/CDS/CSR/RMD/; 2003. [April, 2010]. p. 6. Available on line at http://www.who.int/csr/resources/publications/drugresist/WHO_CDS_CSR_RMD_2003_6/en/ [Google Scholar]
- 12.Bauer AW, Kirby MM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. AM J Clin Pathol. 1966;36:493–496. [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute, author. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. Pennsylvania: CLSI; 2005. Supplemental tables. CLSI Publication M100-S15, M2-A8 and M7-A6. [Google Scholar]
- 14.Lennox KA, L Clifford M, Okey N, et al. A Hospital-Based Prevalence Survey of Bloodstream Infections in Febrile Patients in Malawi: Implications for Diagnosis and Therapy. JID. 2000;181:1414–1420. doi: 10.1086/315367. [DOI] [PubMed] [Google Scholar]
- 15.Amatya NM, Sheresha B, Lekhak B. Etiological agents of bacteremia and antibiotic susceptibility pattern in Kathmandu model hospital. J Nepal Med Assoc. 2007;46(167):112–118. [PubMed] [Google Scholar]
- 16.Philip CH, Charles OO, Usman AI, et al. Bacteraemia in patients admitted to an urban hospital in West Africa. BMC Infectious Diseases. 2007;7(2):1–8. doi: 10.1186/1471-2334-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuma S, Chitnis DS. Antibiogram study over bacterial isolated from cases of bacteremia. Indian J Med Sci. 1996;50:325–329. [PubMed] [Google Scholar]
- 18.David RM, Christopher WW, Mark DZ, et al. The Etiology of Febrile Illness in Adults Presenting to Patan Hospital in Kathmandu, Nepal. Am J Tro Med Hyg. 2004;70(6):670–675. [PubMed] [Google Scholar]
- 19.Usha A, Pushpa D. Bacterial profile of blood stream infections and antibiotics resistance pattern of isolates. JK Science. 2007;9(4):186–190. [Google Scholar]
- 20.Johannes H, Donald AG. Coagulase-negative staphylococcus: Role as pathogens. Ann Rev Med. 1999;50:223–236. doi: 10.1146/annurev.med.50.1.223. [DOI] [PubMed] [Google Scholar]
- 21.David WB, Kenneth S, Elizabeth M, et al. Predicting Bacteremia in Patients with Sepsis Syndrome. J Infect Dis. 1997;176:1538–1551. doi: 10.1086/514153. [DOI] [PubMed] [Google Scholar]
- 22.Daniel JD, Kathleen L, Patti R, et al. Accuracy and Appropriateness of Antimicrobial Susceptibility Test Reporting for Bacteria Isolated from Blood Cultures. J Clin Microbiol. 2004;42(2):2258–2260. doi: 10.1128/JCM.42.5.2258-2260.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souvenir D, Anderson DE, Palpant S, et al. Blood cultures positive for coagulase-negative staphylococci: Antisepsis, Pseudobacteremia, and therapy of patients. J Clin Microbiol. 1998;36:1923–1926. doi: 10.1128/jcm.36.7.1923-1926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mark W D, Gary L, Jhumur R, Dale AF, Nicholas MA, Bart JC. Epidemiology of community-acquired and nosocomial bloodstream infections in tropical Australia: a 12-month prospective study. Tropical Medicine and International Health. 2004;9(7):795–804. doi: 10.1111/j.1365-3156.2004.01269.x. [DOI] [PubMed] [Google Scholar]
- 25.Beyene G, Asrat D, Engers H, Wain J. Phenotypic and Molecular characterization of Salmonella in Ethiopia. Ethiopia: Addis Ababa University; 2009. PhD dissertations. [Google Scholar]
- 26.Balta B, Fetene D. Nasal carriage of methicillen resistant Staphylococcus aureus strains among inpatients of Jimma Hospital, Southwest Ethiopia. Ethiop J Health Sci. 2003;13(2):107–116. [Google Scholar]
- 27.Worku S, G/Mariam A. Practice of Self-medication in Jimma Town. Ethiop J Health Dev. 2003;17(2):111–116. [Google Scholar]