Abstract
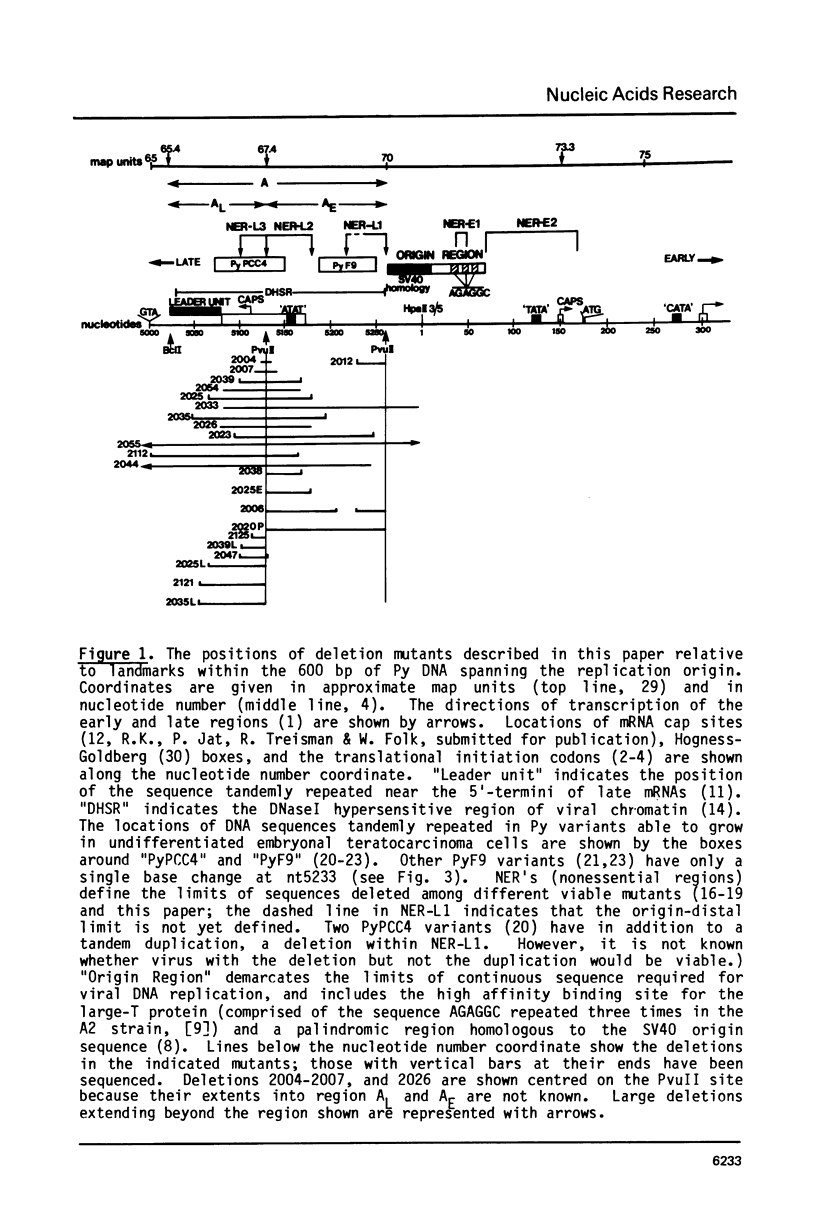
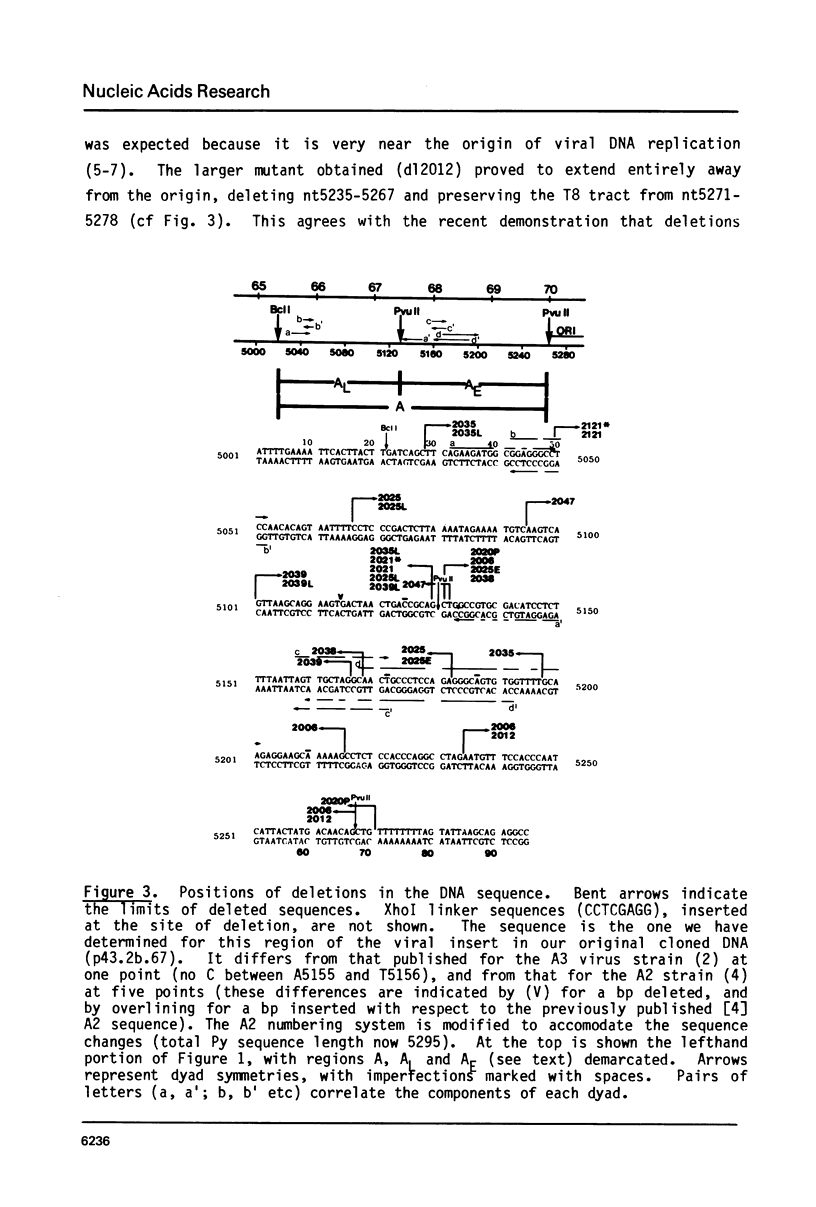
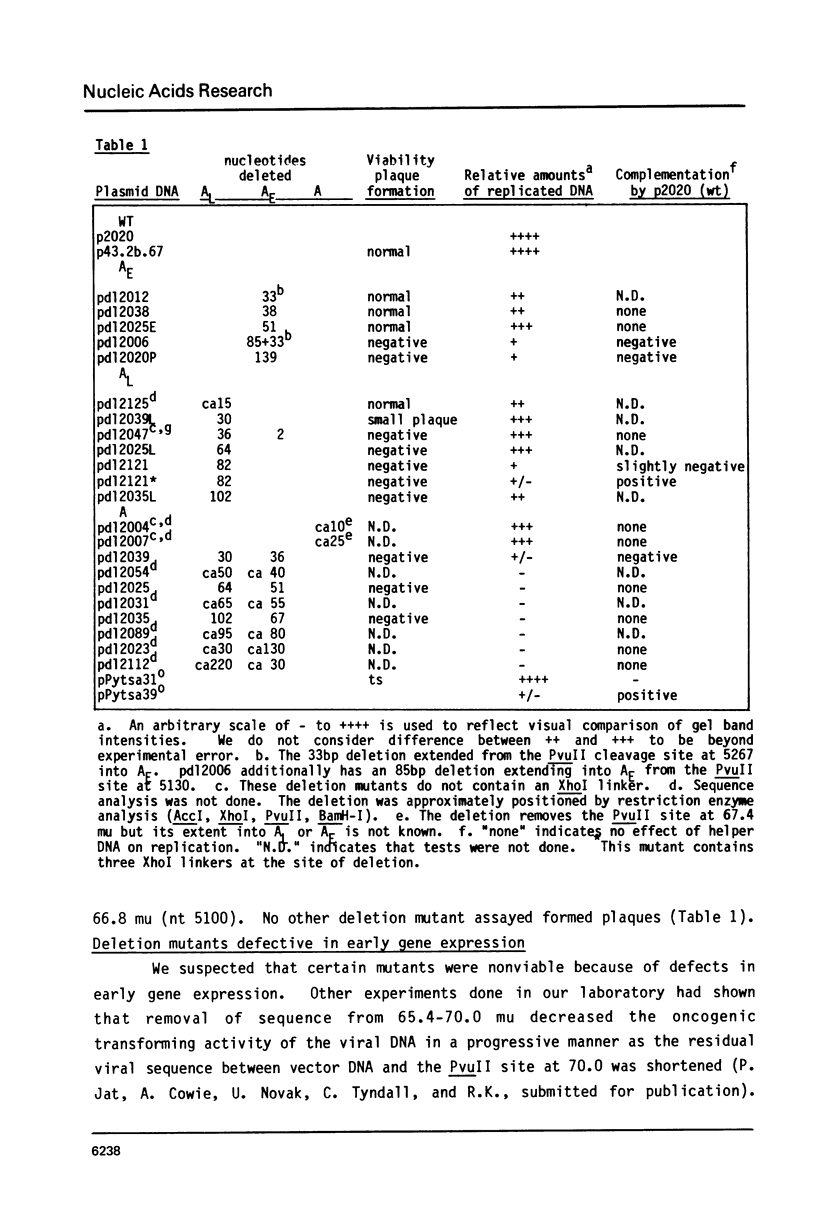
Deletion mutants within the Py DNA region between the replication origin and the beginning of late protein coding sequences have been constructed and analysed for viability, early gene expression and viral DNA replication. Assay of replicative competence was facilitated by the use of Py transformed mouse cells (COP lines) which express functional large T-protein but contain no free viral DNA. Viable mutants defined three new nonessential regions of the genome. Certain deletions spanning the PvuII site at nt 5130 (67.4 mu) were unable to express early genes and had a cis-acting defect in DNA replication. Other mutants had intermediate phenotypes. Relevance of these results to eucaryotic "enhancer" elements is discussed.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Folk W. R. Deletion mutants of polyoma virus defining a nonessential region between the origin of replication and the initiation codon for early proteins. J Virol. 1979 Nov;32(2):530–535. doi: 10.1128/jvi.32.2.530-535.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig M. M., Thomas T., Folk W. R. Regulatory mutants of polyoma virus defective in DNA replication and the synthesis of early proteins. Cell. 1980 Jun;20(2):401–409. doi: 10.1016/0092-8674(80)90626-1. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brockman W. W., Nathans D. The isolation of simian virus 40 variants with specifically altered genomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):942–946. doi: 10.1073/pnas.71.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980 Nov;22(2 Pt 2):479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Robbins A. K., Nicklin P. M., Osborn K. Polyoma DNA replication: location of the origin in different virus strains. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):219–225. doi: 10.1101/sqb.1974.039.01.028. [DOI] [PubMed] [Google Scholar]
- Deininger P., Esty A., LaPorte P., Friedmann T. Nucleotide sequence and genetic organization of the polyoma late region: features common to the polyoma early region and SV40. Cell. 1979 Nov;18(3):771–779. doi: 10.1016/0092-8674(79)90130-2. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRIED M. ISOLATION OF TEMPERATURE-SENSITIVE MUTANTS OF POLYOMA VIRUS. Virology. 1965 Apr;25:669–671. doi: 10.1016/0042-6822(65)90098-x. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Folk W. R., Wang H. C. Closed circular DNAs with tandem repeats of a sequence from polyoma virus. Virology. 1974 Sep;61(1):140–155. doi: 10.1016/0042-6822(74)90249-9. [DOI] [PubMed] [Google Scholar]
- Freeman A. E., Gilden R. V., Vernon M. L., Wolford R. G., Hugunin P. E., Huebner R. J. 5-Bromo-2'-deoxyuridine potentiation of transformation of rat-embryo cells induced in vitro by 3-methylcholanthrene: induction of rat leukemia virus gs antigen in transformed cells. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2415–2419. doi: 10.1073/pnas.70.8.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Fujimura F. K., Deininger P. L., Friedmann T., Linney E. Mutation near the polyoma DNA replication origin permits productive infection of F9 embryonal carcinoma cells. Cell. 1981 Mar;23(3):809–814. doi: 10.1016/0092-8674(81)90445-1. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M. Amplification of a specific region of the polyoma virus genome. Nature. 1975 Jul 17;256(5514):175–179. doi: 10.1038/256175a0. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Dulbecco R. Characterization of polyoma virus T antigen. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1259–1263. doi: 10.1073/pnas.74.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Katinka M., Vasseur M., Montreau N., Yaniv M., Blangy D. Polyoma DNA sequences involved in control of viral gene expression in murine embryonal carcinoma cells. Nature. 1981 Apr 23;290(5808):720–722. doi: 10.1038/290720a0. [DOI] [PubMed] [Google Scholar]
- Katinka M., Yaniv M., Vasseur M., Blangy D. Expression of polyoma early functions in mouse embryonal carcinoma cells depends on sequence rearrangements in the beginning of the late region. Cell. 1980 Jun;20(2):393–399. doi: 10.1016/0092-8674(80)90625-x. [DOI] [PubMed] [Google Scholar]
- Langbeheim H., Shih T. Y., Scolnick E. M. Identification of a normal vertebrate cell protein related to the p21 src of Harvey murine sarcoma virus. Virology. 1980 Oct 30;106(2):292–300. doi: 10.1016/0042-6822(80)90252-4. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Pipas J. M., Pearson-White S., Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980 Sep 19;209(4463):1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Smith M. J., Canaani E., Robbins K. C., Tronick S. R., Zain S., Aaronson S. A. Nucleotide sequence analysis of the transforming region and large terminal redundancies of Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5234–5238. doi: 10.1073/pnas.77.9.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sekikawa K., Levine A. J. Isolation and characterization of polyoma host range mutants that replicate in nullipotential embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1100–1104. doi: 10.1073/pnas.78.2.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Soeda E., Kimura G., Miura K. Similarity of nucleotide sequences around the origin of DNA replication in mouse polyoma virus and simian virus 40. Proc Natl Acad Sci U S A. 1978 Jan;75(1):162–166. doi: 10.1073/pnas.75.1.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Treisman R. Characterisation of polyoma late mRNA leader sequences by molecular cloning and DNA sequence analysis. Nucleic Acids Res. 1980 Nov 11;8(21):4867–4888. doi: 10.1093/nar/8.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Novak U., Favaloro J., Kamen R. Transformation of rat cells by an altered polyoma virus genome expressing only the middle-T protein. Nature. 1981 Aug 13;292(5824):595–600. doi: 10.1038/292595a0. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Hutchinson M. A., Eckhart W. Isolation and characterization of polyoma virus genomes with deletions between the origin of viral DNA replication and the site of initiation of translation in the early region. J Virol. 1979 Nov;32(2):517–522. doi: 10.1128/jvi.32.2.517-522.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]