Abstract
The discovery of microRNAs (miRNAs) represents one of the most significant advances in biological and medical sciences in the last decade. Hundreds of miRNAs have been identified in plants, viruses, animals and human beings, and these tiny, non-coding RNA transcripts have been found to play crucial roles in important biological processes involved in human health and disease. Recently, many studies have demonstrated that miR-196 plays critical roles in normal development and in the pathogenesis of human disease processes such as cancer. Several investigations have implemented cell culture and animal models to explore the potential molecular mechanisms of miR-196. This review provides updated information about the structure of the miR-196 gene and the roles of miR-196 in development, cancer and disease formation. Importantly, we discuss the possible molecular mechanisms whereby miR-196 regulates cellular functions including targeting molecules and gene regulation pathways; potential clinical applications are addressed, as well as future directions for investigation. miR-196a may prove to be a novel therapeutic target for several cancers.
Keywords: microRNA, miR-196, development, cancer, HOXB8, HMGA, annexin A1
Introduction
The first two microRNAs (miRNAs) discovered, lin-4 and let-7 from Caenorhabditis elegans, were described in 1993 and 2000, respectively. These two miRNAs, which are both 21 nucleotides long, were found to be endogenous regulators of genes involved in developmental timing [1, 2]. Since then, hundreds of miRNAs have been identified in plants, animals, human beings and viruses by molecular approaches and bioinformatics predictions; meanwhile, miRNAs have emerged as crucial players in regulating gene expression in a variety of organisms [3–5]. miRNAs are a broad class of small non-coding RNAs usually 21–25 nucleotides in length that regulate gene expression at the post-transcriptional level [6].
Most miRNAs are transcribed by RNA polymerase II from individual miRNA genes, from the introns of protein coding genes, or from poly-cistronic transcripts that often encode multiple related miRNAs [7]. These long primary transcripts, usually thousands of nucleotides, generate a stem-loop containing primary miRNA (pri-miRNA). In animals, pri-miRNAs are transcribed from the chromosome and then are cleaved into miRNA precursors (pre-miRNAs) by a multiprotein complex made up of Drosha Ribonuclease III and DGCR8, a double-stranded RNA-binding protein [8–10]. Pre-miRNAs of about 60–70 nucleotides fold into an imperfect stem-loop structure and are then transported to the cytoplasm via an exportin-5 and Ran-GTP-dependent mechanism [11]. Pre-miRNAs are next processed by another endonuclease RNase III enzyme Dicer into an imperfect dsRNA duplex made of two complementary strands approximately 22 nucleotides in length: miRNA and miRNA*. These represent the mature miRNA strand and its complementary strand [8, 12]. The stem loop in pre-miRNAs contributes to the strand selection; however, the miRNA* can also be processed to its mature form and used in gene regulation [13]. It was originally proposed that the double stranded small mature RNAs could be separated by an ATP-dependent helicase and the single-stranded form of the miRNA then incorporated into the RISC (RNA-induced silencing complex) [14, 15]. However, the current concept is that Argonaute family proteins at the core of the RISC receive double stranded small RNAs, and the selection of the RNA strand to be incorporated is controlled by the thermodynamic profile of the small RNA duplex termini [16–18]. Once incorporated, the miRNA-programmed forms of RISC (miRISC) participate in post-transcriptional regulation [19, 20]. As part of the RISC, the mature miRNA guides the complex to its mRNA targets, with which it interacts using complementary base-pairing. In animals, miRNAs typically target sequences in the 3′ untranslated regions (3′UTRs) of mRNA that are partially complementary to the miRNA, leading to repression of protein synthesis [21]. If the complementary base-pairing interaction between the miRNA and target mRNA 39UTRs happens to be perfect or near-perfect, the target mRNA can be cleaved and degraded.
More than 700 human miRNAs have been discovered, and it is estimated that these miRNAs regulate approximately 30% of all protein-coding genes [21]. miRNAs have been found to participate in almost every cellular process investigated, including such diverse biological functions and processes as development, differentiation, metabolism, growth, proliferation and apoptosis; they are currently the centre of attention in molecular and cell biology research [1, 6]. Dysregulation of miRNAs is thought to contribute to different human pathologies including cancer, heart and neurodegenerative diseases [7, 22–25]. Recently, miRNA profiling studies have indicated that miRNA-196 (miR-196) is overexpressed in several tumour tissue samples. In addition, increasing numbers of reports indicate that miR-196 plays important roles in development and immunity through targeting of specific genes. In this review, we discuss the known functions and molecular mechanisms of miR-196 in normal development and the pathogenesis of cancer. Further investigation of these newly discovered functional roles of miR-196 may lead to potential clinical applications of miR-196 in the management of several human diseases.
Gene structure and regulation
The gene families for miR-10, miR-196 and miR-615 are located in the regions of homeobox (HOX) clusters within the genome of vertebrates [26]. HOX genes encode homeodomain-containing transcription factors that are essential for embryonic development [27]. In many species, they are organized in tightly linked clusters along the chromosome. Activation and silencing of HOX genes requires preservation of their native order within each cluster, and the evolutionary patterns of both miR-10 and miR-196 closely resemble that of the HOX genes in vertebrates [28]. Three miR-196 genes have been found. The miR-196a-1 gene is located on chromosome 17 (17q21.32) at a site between HOXB9 and HOXB10 genes, and the miR-196a-2 gene is located at a region between HOXC10 and HOXC9 on chromosome 12 (12q13.13) (Fig. 1) [28]. The gene for miR-196b is located in a highly evolutionarily conserved region between HOXA9 and HOXA10 genes, on chromosome 7 (7p15.2) in human beings and chromosome 6 (6qB3) in mice (Table 1 and Fig. 1) [29]. miR-196a-1 and miR-196a-2 genes transcribe the same functional mature miRNA sequence (3′-GGGUUGUUGUACUUUGAUGGAU-5′), whereas miR-196b gene produces a small RNA (3′-GGGUUGUUGUCCUUUGAUGGAU-5′), which differs from the sequence of miR-196a by one nucleotide [28]. Many miRNAs are fairly well-conserved among vertebrates; in fact, they are as conserved as the most conservative phylogenetic footprints [30]. Although miRNAs are very small in size, they carry a strong phylogenetic signal [28, 31]. miR-196 homologues are detectable in a variety of vertebrates.
Fig 1.
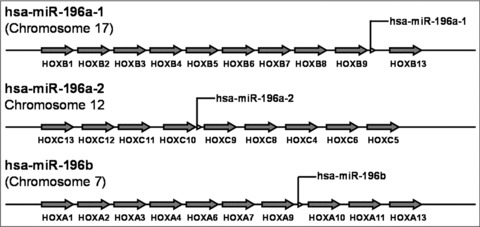
Locations of human miR-196 genes. Information on human miR-196 genes was obtained from the Ensembl Genome Browser (hppt://www.ensembl.org). The miR-196a-1 gene is located in the region between HOXB9 and HOXB13 on chromosome 17, miR-196a-2 in the region between HOXC10 and HOXC9 on chromosome 12, and miR-196b in the region between HOXA9 and HOXA10 coding on chromosome 7.
Table 1.
miR-196 genes and mature sequences*
| miRNA | Gene Location | Mature Sequence |
|---|---|---|
| hsa-miR-196a-1 | Chromosome 17 (17q21.32)(44,064,851–44,064,920) | 5′-UAGGUAGUUUCAUGUUGUUGGG-3′ |
| hsa-miR-196a-2 | Chromosome 12 (12q13.13)(52,671,789–52,671,898) | 5′-UAGGUAGUUUCAUGUUGUUGGG-3′ |
| hsa-miR-196b | Chromosome 7 (7p15.2)(27,175,624–27,175,708) | 5′-UAGGUAGUUUCCUGUUGUUGGG-3′ |
The mature sequences of the hsa-miR-196a-1, hsa-miR-196a-2 and hsa-miR-196b were acquired from the miRBase sequence database (http://microrna.sanger.ac.uk).
High-mobility group A (HMGA) proteins are able to directly bind to the miR-101b and miR-196a-2 upstream region and regulate the expression of these miRNAs. Several miRNAs such as miR-196a-2, miR-101b, miR-331 and miR-29a are significantly decreased in homozygous hmga1-knockout murine embryonic fibroblasts in comparison with wild-type cells [32]. Chromatin immunoprecipitation assay showed that HMGA1 can bind regions upstream of these miRNAs. Also, a series of reporter transgenes (‘sensors’) in mouse embryos have been created for visualizing the tissue-specific expression of several miRNAs during embryogenesis, including miR-10a and miR-196a, which are both encoded by genes embedded in HOX clusters. It was found that miR-196a was influenced by regulatory controls imposed on the HOX clusters. However, the HOXB9 gene, located immediately upstream of miR-196a-1, actually has more restricted expression in the head and anterior truck of embryos than miR-196; this suggests that miR-196 family members are not regulated simply by control elements shared with the nearest HOX genes [33]. Furthermore, miR-196a negatively regulates target gene HOXB8, indicating that its restricted expression pattern probably reflects a role in the HOX complex expression and function [33].
Target molecules
Identification of putative mRNA targets is important in order to understand the specific functions of miRNAs. However, this can be very challenging because miRNAs are usually imperfectly complementary to the 3′UTR region of their targets mRNAs. In animals, the most consistent requirement for miRNA and target mRNA interaction is a contiguous and perfect base pairing of the 5′miRNA nucleotides 2 to 8, called the ‘seed’ region. In many cases, the seed region seems to determine which targets the miRNA will recognize. In other cases, additional determinants are required. For example, a specific mRNA 3′UTR sequence can have a reasonable complementarity to the 3′ half of a miRNA which allows the interaction to be stabilized, while mismatches are present in the central region of the miRNA-mRNA [21]. However, it is possible that a single mRNA may be regulated by multiple miRNAs, while a single miRNA may target multiple transcripts within a cell type; this amplifies the scope of putative miRNA regulation of gene expression. Thus, the particular cellular environment of a given miRNA will determine its functions in a specific cell type [34]. A given miRNA such as miR-196 may regulate different genes in different types of cells or under different conditions. Regulation mechanisms controlling specific miRNA and mRNA interaction are currently under active investigation.
Mammals have four HOX clusters (HOX A to D) containing a total of 39 genes organized into 13 paralogous subgroups [35]. With the exception of a single G:U mismatch, pairing between miR-196a and the human HOXB8 39UTR is perfect (Fig. 2). This conserved, near-perfect pairing suggests that HOXB8 mRNA is targeted by miR-196a for cleavage. Indeed, miR-196 is known to direct the cleavage of HOXB8 mRNA in mouse embryos and also regulates the expression of HOXC8, HOXD8 and HOXA7 by transcriptional inhibition; however, the mechanisms are not fully understood [28, 36]. The individual target sites of the HOXB8 mRNA 3′UTR are conserved across several vertebrate species (Fig. 2). The short genomic distances between miR-196 and miR-10 and their targets are remarkable; the target genes are located in close proximity to these miRNAs [26, 37]. miR-10c is encoded 25 kb from its target sites in HOXB3a and 48 kb from those in HOXB1a, and a miR-196 paralogue is located only 18 kb from HOXB8 and HOXC8 and 14 kb from HOXA7 [26]. In this case, the miRNA may act in an auto-regulatory fashion on parts of its original precursor [26]. Kawasaki and colleagues showed that miR-196 inhibits HOXB8 expression within myeloid differentiation of HL60 cells. Transfection with the vector-expressed miR-196 repressed the expression of HOXB8 by cleaving the mRNAs. Meanwhile, myeloid differentiation of HL60 cells was enhanced [38]. Thus, the miRNAs appear to fine-tune the expression of the HOX genes by regulating the HOX genes themselves and also by regulating the downstream targets of HOX genes [39].
Fig 2.
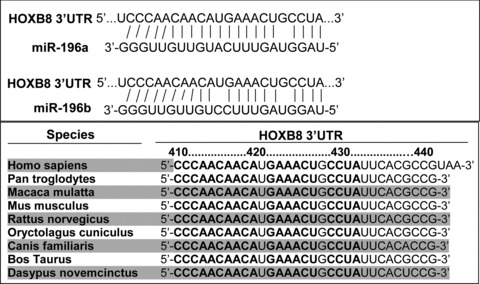
Predicted miR-196 target recognition sites in the HOXB8 3′UTR in human beings and other species. The predicted miR-196 target recognition sites in human beings and other species were obtained from the TargetScanHuman 5.1 program (http://www.targetscan.org).
HMGA2 gene product was identified as a putative target of miR-196a-2, suggesting that HMGA1 can down-regulate the expression of HMGA family members through miR-196a because HMGA1 could up-regulate miR-196a-2 expression [32]. Two potential miR-196a target recognition sites are present at the HMGA2 mRNA 3′UTR, and the nucleotide sequence at these sites are conserved in several species (Fig. 3).
Fig 3.
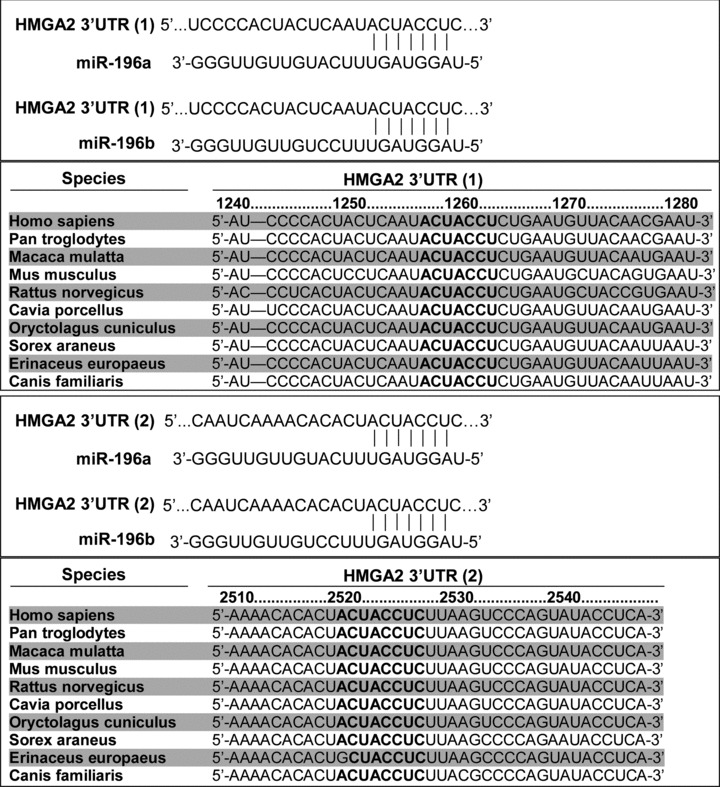
Predicted miR-196 target recognition sites in the HMGA2 3′UTR in human beings and other species.
Luthra et al. reported annexin A1 is another target gene of miR-196a [40]. Annexin A1, also known as lipocortin or p35, is a well-characterized member of the calcium- and phospholipid-binding protein family of annexins. A potential miR-196a target recognition site is present at the annexin A1 mRNA 3′UTR, and the conserved sequence is observed in three species (Fig. 4). However, the homology of miR-196 recognition sites on annexin A1 among species is much lower than that of HOXB8 and HMGA2, described above. It is not clear why this miR-196 target recognition sequence is not conserved in more species. Also, further investigation will be needed to confirm the specific interactions between miR-196 and annexin A1 in different types of cells and under different conditions.
Fig 4.
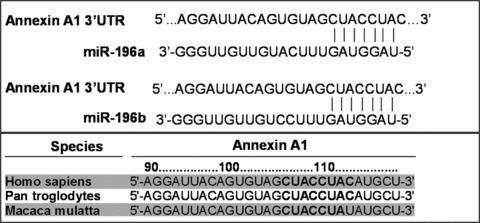
Predicted miR-196 target recognition sites in the annexin A1 3′UTR in human beings and other species.
Several other genes have been predicted to be target molecules of miR-196 [41], including S100 calcium-binding protein A9, small proline-rich protein 2C, keratin 5, CLCA family member 2 (a chloride channel regulator), cytochrome P450 (family 4, subfamily B, polypeptide 1), keratin 4, LDOC1, leukotriene A4 hydrolase, pleiotrophin, T-cell differentiation protein, tumour protein D52-like 1, visinin-like 1 and v-ETS erythroblastosis virus E26 oncogene homologue (ERG) [42]. However, most of these target molecules of miR-196 have not been confirmed with experimental approaches. Table 2 shows several target genes of miR-196.
Table 2.
Experimentally verified target genes regulated by miR-196
| Gene | Gen Name | Function | Verification | Reference |
|---|---|---|---|---|
| HOXB8 | Homeobox protein Hox-B8 | Transcription factor | mRNA | 36, 71 |
| HOXC8 | Homeobox protein Hox-C8 | Transcription factor | mRNA, protein | 36, 71, 74 |
| HOXD8 | Homeobox protein Hox-D8 | Transcription factor | mRNA | 36, 71 |
| HOXA7 | Homeobox protein Hox-A7 | Transcription factor | mRNA | 36, 71 |
| HOXB7 | Homeobox protein Hox-B7 | Transcription factor | mRNA | 36, 71 |
| ERG | v-ETS erythroblastosis virus E26 oncogene homologue | Oncogenic ETS transcription factor | mRNA | 42 |
| HMGA2 | HMGA2 | Nuclear architectural factor | mRNA, protein | 32 |
| ANXA1 | Annexin A1 | Apoptosis | mRNA, protein | 40 |
| S100A9 | S100 calcium-binding protein A9 | Calcium binding | mRNA | 41 |
| SPRR2C | Small proline-rich protein 2C | Crosslinker of epidermal differentiation complexes | mRNA | 41 |
| KRTS | Keratin 5 | Structural cytokeratin protein | mRNA | 41 |
Role in development
miR-196 appears to play an important role in development. Its relationship to the HOX gene family, crucial for embryonic development, is well-described. In Drosophila, there is a conserved or possibly convergent interaction between the miR-196 homologue iab-4 and the homeotic Ubx HOX gene [43]. Ronshaugen et al. analysed the iab-4 locus, which produces the miR-iab-4–5p and miR-iab-4–3p and found that miR-iab-4–5p could directly inhibit Ubx activity in vivo. Ectopic expression of miR–iab-4–5p attenuated endogenous Ubx protein accumulation and induced a classical homeotic mutant phenotype: halteres transformed into wings [43]. In chicken, the interaction between miR-196 and HOXB8 has been implicated in the mechanism that abolishes the competence of mesoderm to undergo limb induction by retinoic acid [37]. miR-196 may act at the upstream of HOXB8 and Sonic hedgehog (Shh) in vivo in the context of limb development, specifically in the hindlimb [37].
miR-196a, when overexpressed, is able to induce eye anomalies in Xenopus laevis. Endogenous miR-196a is expressed at the posterior trunk during Xenopus development and its expression level increases in later stages. Overexpression of miR-196a by microinjection of synthetic mammalian miR-196a precursor into Xenopus embryo during early development led to dose-dependent eye anomalies and changes in the expression of several critical genes [44].
HMGA are nuclear architectural factors that play critical roles in a wide range of biological processes. Two variants of HMGA proteins, HMGA1 and HMGA2, have been identified, and both proteins are encoded by two distinct genes and have three DNA binding motifs called AT-hooks [45]. Both HMGA genes are highly expressed during embryogenesis, and there is evidence that the absence of HMGA 2 protein causes growth retardation in mice [46, 47]. It is interesting that HMGA1 is able to regulate miR-196 expression, whereas miR-19a can control HMGA2 mRNA translation, which may affect development.
Role in cancer
miRNAs can modulate critical cellular functions such as proliferation, apoptosis and differentiation. Many miRNA profiling studies have been performed in both normal and diseased tissues. Bloomston et al. defined the expression pattern of miRNA precursors in pancreatic cancer and compared it with those of normal pancreas and chronic pancreatitis using microarray analysis; by looking also at the pattern of miRNA expression with respect to long-term (∼24 months) survival, it was found that miRNA-196a levels are inversely correlated with survival in pancreatic adenocarcinoma patients [48]. It has been reported that an analysis of the expression of miR-196a along with miR-217 can be used to classify benign and malignant pancreatic tissues in pancreatic fine-needle aspirates [49]. Also, the plasma levels of miR-196a along with other miRNAs including miR-21, miR-210 and miR-155 have been found to be significantly higher in patients with pancreatic adenocarcinoma than healthy controls [50]. Thus, miR-196a may represent an important factor in the pathogenesis of pancreatic cancer and may prove useful in the diagnosis, prognosis and/or treatment of this devastating disease.
Up-regulation of miR-196a has also been found in breast cancer. Likewise, we recently reported that the expression of miR-196a was significantly up-regulated by real time PCR analysis in the majority of pancreatic cancer tissues (70%) and cell lines (100%) compared normal pancreatic tissues and cells [51]. Hui et al. performed miRNA expression analysis by using Taqman low density arrays with quantitative real-time PCR confirmation in 40 archival formalin-fixed paraffin-embedded breast lumpectomy specimens, and found that a panel of miRNAs was consistently dysregulated in breast cancer compared with normal breast samples; this included up-regulation of previously reported breast cancer-related miRNAs such as miR-21, miR-155, miR-191 and miR-196a as well as down-regulation of miR-125b and miR-221 [52]. The quality of these data validate conducting global miRNAs profiling using FFPE samples, thereby offering enormous opportunities to evaluate such materials linked to clinical databases in order to rapidly acquire insight into the pathogenic roles of miRNAs.
In leukaemia, levels of miR-10a, miR-10b and miR-196a-1 showed a clear correlation with HOX gene expression in 30 cases of primary adult acute myeloid leukaemia (AML) with normal karyotype, according to quantitative real-time PCR assay. In addition to the HOX genes, nearly 30% of the genes with a high correlation with miR-10a, miR-10b and miR-196a-1 have been shown to have oncogenic potential [53–55]. By quantifying expression of 19 selected miRNA genes, Schotte et al. found that the expression of miR-196b was 500-fold higher in myeloid/lymphoid or mixed lineage leukaemia (MLL)-rearranged cases and 800-fold higher in 5/15 T-lineage acute lymphoblastic leukaemia (T-ALL) cases compared with expression levels in precursor B cell acute lymphoblastic leukaemia (B-ALL) cases [56]. These data indicate that the expression profiles of these miRNA are ALL subtype-specific rather than linked to the differentiation status associated with these subtypes.
Popovic et al. showed that the MLL gene normally regulates expression of miR-196b in a pattern similar to that of the surrounding 5′ HOX genes, HOXA9 and HOXA10, during stem cell differentiation [29]. Again, overexpression of miR-196b was found specifically in patients with MLL-associated leukaemia based on analysis of 55 primary leukaemia samples; these demonstrated increased proliferative capacity and survival, as well as a partial block in differentiation in bone marrow progenitor cells. Meanwhile, leukemogenic MLL fusion proteins cause over expression of miR-196b, which may be necessary for MLL fusion-mediated immortalization. A recent report demonstrated that miR-196b expression is significantly more up-regulated in AML than ALL and that, again, miR-196b up-regulation is negatively associated with overall survival of AML patients [57]. In addition to the miR-196b deregulation observed in leukaemia, miR-196b has been reported to induce changes in myeloid development, and the co-expression of miR-196b and miR-21 reportedly block granulocyte colony-stimulating factor-induced granulopoiesis completely [58].
Highly elevated levels of miR-196a have also been detected in oesophageal adenocarcinoma, based on analysis of microdissected paraffin-embedded tissues from 11 patients [41]. This study found that miR-196a may have growth-promoting and anti-apoptotic functions; further, it may be considered a marker of progression from Barrett’s oesophagus to low-grade dysplasia, high-grade dysplasia and oesophageal adenocarcinoma. miR-196a levels were inversely correlated with the predicted target keratin 5, small proline-rich protein 2C and S100 calcium-binding protein A9 mRNA levels in oesophageal adenocarcinoma.
The expression of HMGA proteins is usually low or absent in most of normal adult tissues; however, overexpression of HMGA proteins has been observed in several human malignancies and has been found to correlate with occurrence of metastasis and poor prognosis. Moreover, transgenic mice overexpressing either HMGA1 or HMGA2 develop lymphoma and pituitary adenomas, which strongly suggest oncogenic property of these proteins [59–62]. It could promote tumour progression if HMGA1 dominantly up-regulates miR-196 expression; however, if miR-196 significantly inhibits HMGA2 protein synthesis, it could inhibit tumour progression. Thus, ultimate impact of miR-196 on the tumour pathogenesis will be dependent on the balance of regulation of miR-196 expression and miR-196 targeting molecules (oncogenes or tumour suppression genes) in specific cells. These regulation mechanisms of miR-196 expression and functions are largely unknown and warrant further investigation.
Increased levels of miR-196a have also been found in 12 cancer cell lines of oesophageal, breast and endometrial origin and 10 oesophageal adenocarcinomas; furthermore, levels of miR-196a have been demonstrated to have a significant inverse correlation with annexin A1 mRNA levels in these cancers [40]. Annexin A1 has been postulated to act as tumour suppressor in different cancers such as prostate, breast, oesophageal and B-cell non-Hodgkin’s lymphoma [63–67]. As annexin A1 could be a target molecule of miR-196a, increased miR-196a may mediate down-regulation of annexin A1, contributing to tumorigenesis. However, overexpression of annexin A1 has been reported in pancreatic, hepatic and stomach cancers, suggesting that the functional role of annexin A1 may be tissue- and cell type-specific [68–70]. Of note, the majority of pancreatic cancer tissues and cell lines demonstrate a high expression level of miR-196a [49]. The relationship and functional interactions of miR-196a and annexin A1 warrant further investigation.
miR-196a may also have pro-oncogenic functions in colorectal cancer. The miR-196a expression profile in colorectal cancer, mucosal samples and diverse cancer cell lines has been quantified by RT-PCR, and miR-196b has been found to be up-regulated in colonic cancer. High levels of miR-196a were observed to activate the Akt signalling pathway; promote cancer cell detachment, migration, invasion and chemosensitivity; and increase the development of lung metastases in mice after tail vein injection [71, 72].
Although most studies on miR-196 suggest its oncogenic function in cancer, it is important to point out that miR-196 may play a tumour suppressive role as well. For example, strongly reduced expression of miR-196a was observed in melanoma cells when compared to healthy control melanocytes, and the low miR-196a expression in melanoma cells led to high HOXB7 gene expression, which subsequently raised ETS-1 and BMP4 activity and thereby enhanced migratory potential of these melanoma cells [73]. Moreover, a recent report by Li et al. showed that enforced expression of miR-196a or miR-196b reduced in vitro invasion and in vivo spontaneous metastasis of breast cancer cells, indicating that the members of miR-196 family are potent metastasis suppressors [74]. The mechanisms of action of miR-196 in different cancer types are largely unknown. It may depend on the molecules that miR-196 targets. If miR-196 has a dominant effect on the inhibition of oncogenic molecules, miR-196 will play a tumour suppressor function; although if miR-196 mainly targets tumour suppressors, miR-196 will demonstrate oncogenic effects.
miRNAs are likely important players in the pathogenesis of cancer, and dysregulation of miRNA expression in cancer is an important field of investigation. Both genetic and epigenetic alterations may affect miRNA biogenesis and activity [75, 76]. For example, one single-nucleotide polymorphism (SNP) of miR-196a-2 (rs11614913) has been associated with survival in Chinese individuals with lung cancer; those patients with the CC genotype had reduced survival. This particular SNP affects the binding efficiency of miR-196a-2 to its target mRNA [77, 78]. The presence of the CC genotype also correlated with increased susceptibility to lung cancer in the same patient population [78]. Consistently, this variant is also associated with increased gastric cancer risk in Chinese patients [79]. In two separate studies, the TT genotype of this SNP was found to be associated with decreased risk of breast cancer in a Chinese population, whereas the CC genotype again was associated with an increased risk of breast cancer in Chinese patients [80, 81]. Meanwhile, the opposite relationship has been described in glioma, where the CC genotype was associated with a decreased risk [82]. This discrepancy highlights the need for further investigation of this SNP in different tissues, tumours and patient populations. Of note, the major allele in the Caucasian population is the C allele, whereas in the Chinese population, the major allele is T. So, interestingly, the ‘variant’ homozygote CC genotype as described in Chinese population studies is actually the dominant genotype in Caucasians; this exemplifies the limitations of attributing the terms ‘wild-type’ and ‘variant’ to the alleles when describing SNPs.
Other biological functions
MiRNA-196 appears to have several important cellular functions in addition to development and malignancy. Endometriosis is a gynaecological disorder in females in which endometrial-like cells grow in areas outside the uterine cavity. A miRNA expression study identified miR-196b as one of 8 down-regulated miRNA in ectopic endometrial tissues as compared to paired eutopic tissues [83]. miR-196 may also play a role in the pathogenesis of severe congenital neutropenia (SCN), a rare haematological disorder characterized by an abnormally low number of neutrophils. Mutations in several genes, including growth factor independent protein 1 (Gfi1), have been reported to be responsible for SCN. Gfi1 is a transcriptional repressor that is required for normal myelopoiesis, and Gfi1 loss-of-function mutations are found in some patients with SCN. It has been shown that miR-196b is negatively regulated by Gfi1; its expression is up-regulated in Gfi1−/– mice and Gfi1N382S SCN patients. Thus, up-regulation of miR-196 may mediate some of the effects of this disease [83].
miR-196 also has biological functions involving immunology, inflammation and virus defence. Indeed, CD56+ T cells may be able to up-regulate the expression of miR-196a, which shows anti-hepatitis C virus (HCV) properties [84, 85]. Furthermore, antiviral cytokine interferon β can induce numerous cellular miRNAs, including miR-196, which have sequence-predicted targets within HCV genomic RNA [84]. Overexpression of these miRNAs in infected liver cells leads to a marked attenuation of viral replication, demonstrating the importance of miRNAs in anti-viral immunity [86]. Since viruses also play a role in the pathogenesis of several tumour types, miRNAs may mediate viral–host interactions that have important implications not only for infectious diseases but also for cancer pathogenesis.
A recent study revealed that miR-196a plays an important role in mesenchymal stem cell differentiation. Specifically, overexpression of miR-196a inhibited the proliferation of human adipose tissue-derived mesenchymal stem cells while enhancing osteogenic differentiation [87]. Finally, miR-196a may prove useful with respect to storage of blood. miR-196a, along with three other miRNAs, displayed an increased expression in red blood cells up to day 20 after blood collection, after which expression subsequently decreased [88]. Thus, miRNAs may be potentially useful markers to predict storage-related lesions during blood storage.
Summary and perspectives
miR-196 (miR-196a-1, miR-196a-2 and miR-196b) is transcribed from three different genes. Although the mature nucleotide sequence is identical for miR-196a-1 and miR-196a-2, mature miR-196b differs from miR-196a by one nucleotide. The expression levels of miR-196 appear to be strictly controlled under physiologic conditions, whereas dysregulation of miR-196 can be observed in many disease conditions. miR-196 may play critical roles in normal development and cancer pathogenesis by targeting several regulation molecules including HOXB8, HMGA2 and annexin A1. Meanwhile, it may also play roles in viral immunity and the cell differentiation processes that, when dysregulated, lead to endometriosis and SCN. However, the detailed functions and molecular mechanisms whereby miR-196 contributes to the pathophysiology of these diseases remain largely unknown. For example, it is not completely understood how miR-196a is regulated; how miR-196a controls cell proliferation or differentiation; and how miR-196a differentially recognizes its target molecules in a cell- or tissue-specific fashion. Addressing these critical questions will help to establish potential clinical applications of miR-196-directed technology in the diagnosis, treatment, and/or prognosis of development defects, viral infections and different types of human cancers.
Acknowledgments
This study is partially supported by MacDonald General Research Fund Awards (09RDM006 and 09RDM007) from St. Luke’s Episcopal Hospital, Houston, Texas; Duncan Inter and Intra Programmatic Pilot Project (#09–10) from the Dan L. Duncan Cancer Center, Baylor College of Medicine, Houston, Texas; and NIH grant R21CA140828 (Y.Q). L.Z. and S.M.W. were supported by a training grant from NIH (T32HL083774).
Conflict of interest
None.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38:S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–76. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 9.Denli AM, Tops BB, Plasterk RH, et al. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 10.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 11.Lund E, Guttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 12.Hutvágner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 13.Okamura K, Phillips MD, Tyler DM, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3’ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory TI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Nykänen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–21. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 16.Matranga C, Tomari Y, Shin C, et al. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi K, Tsukumo H, Nagami T, et al. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–48. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuschner PJ, Ameres SL, Kueng S, et al. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–20. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–70. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 20.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 22.Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–39. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 23.Mourelatos Z. MicroRNAs: biology and roles in neurodegeneration and brain tumours. Introduction and historical background. Brain Pathol. 2008;18:110–12. doi: 10.1111/j.1750-3639.2007.00118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 25.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 26.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS ONE. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 28.Tanzer A, Amemiya CT, Kim CB, et al. Evolution of microRNAs located within Hox gene clusters. J Exp Zool B Mol Dev Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 29.Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–22. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner GP, Fried C, Prohaska SJ, et al. Divergence of conserved non-coding sequences: rate estimates and relative rate tests. Mol Biol Evol. 2004;21:2116–21. doi: 10.1093/molbev/msh221. [DOI] [PubMed] [Google Scholar]
- 31.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–5. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 32.De Martino I, Visone R, Fedele M, et al. Regulation of microRNA expression by HMGA1 proteins. Oncogene. 2009;28:1432–42. doi: 10.1038/onc.2008.495. [DOI] [PubMed] [Google Scholar]
- 33.Mansfield JH, Harfe BD, Nissen R, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–83. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 34.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–54. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 36.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 37.Hornstein E, Mansfield JH, Yekta S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 38.Kawasaki H, Taira K. MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symp Ser. 2004;48:211–2. doi: 10.1093/nass/48.1.211. [DOI] [PubMed] [Google Scholar]
- 39.Chopra VS, Mishra RK. “Mir”acles in hox gene regulation. Bioessays. 2006;28:445–8. doi: 10.1002/bies.20401. [DOI] [PubMed] [Google Scholar]
- 40.Luthra R, Singh RR, Luthra MG, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–78. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 41.Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–8. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coskun E, von der Heide EK, Schlee C, et al. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leuk Res Jun 4. 2010 doi: 10.1016/j.leukres.2010.05.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 43.Ronshaugen M, Biemar F, Piel J, et al. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–52. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu R, Liu Y, Wu JY, et al. Misexpression of miR-196a induces eye anomaly in Xenopus laevis. Brain Res Bull. 2009;79:26–31. doi: 10.1016/j.brainresbull.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X, Benson KF, Ashar HR, et al. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–4. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 47.Chiappetta G, Avantaggiato V, Visconti R, et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–46. [PubMed] [Google Scholar]
- 48.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 49.Szafranska AE, Doleshal M, Edmunds HS, et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54:1716–24. doi: 10.1373/clinchem.2008.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. 2009;2:807–13. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Li M, Wang H, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg. 2009;33:698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hui AB, Shi W, Boutros PC, et al. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 53.Debernardi S, Skoulakis S, Molloy G, et al. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–6. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 54.Thorsteinsdottir U, Sauvageau G, Hough MR, et al. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thorsteinsdottir U, Mamo A, Kroon E, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–9. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 56.Schotte D, Chau JC, Sylvester G, et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–22. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Li Z, He C, et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–7. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–8. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fedele M, Pentimalli F, Baldassarre G, et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene. 2005;24:3427–35. doi: 10.1038/sj.onc.1208501. [DOI] [PubMed] [Google Scholar]
- 60.Xu Y, Sumter TF, Bhattacharya R, et al. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004;64:3371–75. doi: 10.1158/0008-5472.CAN-04-0044. [DOI] [PubMed] [Google Scholar]
- 61.Fedele M, Battista S, Kenyon L, et al. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–8. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- 62.Fedele M, Visone R, De Martino I, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–71. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 63.Kang JS, Calvo BF, Maygarden SJ, et al. Dysregulation of annexin I protein expression in high-grade prostatic intraepithelial neoplasia and prostate cancer. Clin. Cancer Res. 2002;8:117–23. [PubMed] [Google Scholar]
- 64.Shen D, Nooraie F, Elshimali Y, et al. Decreased expression of annexin A1 is correlated with breast cancer development and progression as determined by a tissue microarray analysis. Hum Pathol. 2006;37:1583–91. doi: 10.1016/j.humpath.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 65.Paweletz CP, Ornstein DK, Roth MJ, et al. Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res. 2000;60:6293–7. [PubMed] [Google Scholar]
- 66.Hu N, Flaig MJ, Su H, et al. Comprehensive characterization of annexin I alterations in esophageal squamous cell carcinoma. Clin Cancer Res. 2004;10:6013–22. doi: 10.1158/1078-0432.CCR-04-0317. [DOI] [PubMed] [Google Scholar]
- 67.Vishwanatha JK, Salazar E, Gopalakrishnan VK. Absence of annexin I expression in B-cell non-Hodgkin’s lymphomas and cell lines. BMC Cancer. 2005;4:8. doi: 10.1186/1471-2407-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bai XF, Ni XG, Zhao P, et al. Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J Gastroenterol. 2004;10:1466–70. doi: 10.3748/wjg.v10.i10.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Masaki T, Tokuda M, Ohnishi M, et al. Enhanced expression of the protein kinase substrate annexin in human hepatocellular carcinoma. Hepatology. 1996;24:72–81. doi: 10.1053/jhep.1996.v24.pm0008707286. [DOI] [PubMed] [Google Scholar]
- 70.Sinha P, Hütter G, Köttgen E, et al. Increased expression of annexin I and thioredoxin detected by two-dimensional gel electrophoresis of drug resistant human stomach cancer cells. J Biochem Biophys Methods. 1998;37:105–16. doi: 10.1016/s0165-022x(98)00020-7. [DOI] [PubMed] [Google Scholar]
- 71.Schimanski CC, Frerichs K, Rahman F, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–96. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang YX, Zhang XY, Zhang BF, et al. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–4. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 73.Braig S, Mueller DW, Rothhammer T, et al. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci. 2010;67:3535–48. doi: 10.1007/s00018-010-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y, Zhang M, Chen H, et al. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010;70:7894–904. doi: 10.1158/0008-5472.CAN-10-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 76.Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–31. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 77.Hu Z, Chen F, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 1008;118:2600–8. doi: 10.1172/JCI34934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tian T, Shu Y, Chen J, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–7. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- 79.Peng S, Kuang Z, Sheng C, et al. Association of MicroRNA-196a-2 Gene Polymorphism with Gastric Cancer Risk in a Chinese Population. Dig Dis Sci. 2010;55:2288–93. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 80.Hu Z, Liang J, Wang Z, et al. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum. Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 81.Hoffman AE, Zheng T, Yi C, et al. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dou T, Wu Q, Chen X, et al. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136:1853–9. doi: 10.1007/s00432-010-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–75. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pedersen LM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye L, Wang X, Wang S, et al. CD56+ T cells inhibit hepatitis C virus replication in human hepatocytes. Hepatology. 2009;49:753–62. doi: 10.1002/hep.22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sonkoly E, Stahle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin. Cancer Biol. 2008;18:131–40. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Kim YJ, Bae SW, Yu SS, et al. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–25. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 88.Kannan M, Atreya C. Differential profiling of human red blood cells during storage for 52 selected microRNAs. Transfusion. 2010;50:1581–8. doi: 10.1111/j.1537-2995.2010.02585.x. [DOI] [PubMed] [Google Scholar]


