Summary
Background
Vascular smooth muscle cell (SMC) differentiation is an essential component of vascular repair and tissue engineering. However, currently used cell models for the study of SMC differentiation have several limitations. Multi-lineage progenitor cells (MLPCs) originate from human umbilical cord blood and are cloned from a single cell. The object of this study was to investigate whether MLPCs could differentiate into SMCs in vitro with induction by transforming growth factor β1 (TGF-β1).
Material/Methods
MLPCs were treated without or with TGF-β1 (1 and 5 ng/mL) in mesenchymal stem cell media plus 1% FBS for 7 days. Total RNA was isolated from the MLPCs, and semi-quantitative real-time PCR was performed to test the following mRNA levels: early and late phase SMC-specific markers, two endothelial cell (EC)-specific markers, endothelial progenitor cell (EPC) marker CD34, TGF-β1 accessory protein CD105, and adhesion molecule CD146.
Results
TGF-β1 (1 ng/mL) significantly increased the mRNA levels of SMC-specific markers SM22α, calponin-1, SM α-actin, caldesmon, tropomyosin and MLCK as well as adhesion molecule CD146. The mRNA levels of EC-specific markers VE-cadherin and VEGFR-2, EPC marker CD34 and TGF-β1 accessory protein CD105 were decreased significantly, after MLPC were treated with TGF-β1 (1 ng/mL). TGF-β1 at 5 ng/mL showed similar effect on the expression of these genes.
Conclusions
This study demonstrates that in the presence of TGF-β1, MLPCs undergo SMC lineage differentiation indicating that MLPCs are a promising cell model for SMC lineage differentiation studies, which may contribute to advances in vascular repair and tissue engineering.
Keywords: multi-lineage progenitor cell, transforming growth factor β1, smooth muscle cell, differentiation
Background
Vascular smooth muscle cell (SMC) differentiation is an essential component of vascular development. A variety of human vascular diseases can be traced to a defect in smooth muscle development or proliferation [1–3]. In mice with defective smooth muscle development, embryonic lethality occurs [4]. Strategies that facilitate SMC differentiation should contribute to vascular repair and tissue engineering.
There are several cell models generally used in SMC differentiation studies. They include mouse neural crest stem cell line Monc-1 [5], mouse embryonic fibroblast 10T1/2 [6,7] and mouse embryonic stem cells [8]. Under certain culture conditions, these cells express SMC-specific markers that indicate SMC differentiation, which include contractile apparatus-associated proteins such as calponin-1 and smooth muscle α-actin (SM α-actin). However, several problems arise when using these cell models for the study of SMC differentiation. First, because most of the cells used in SMC differentiation studies are of mouse origin, there may be important interspecies differences in the differentiation environment, intracellular molecules involved in the differentiation process and underlying signaling mechanisms promoting SMC differentiation. This may significantly impair the application of data from mouse to human tissue engineering. Second, some cell lines are not naive cells but immortalized cell lines derived from a primary culture. The regulating mechanisms involved in SMC differentiation may be changed in the immortalized cell lines. As an alternative, Simper et al. [9] have described SMC progenitor cells in human peripheral blood mononuclear cells. However, based on the experience of our lab and several other labs, progenitor cells isolated from human peripheral blood can hardly proliferate under in vitro culture condition and usually deteriorate after 3 weeks, and as a result, it is impossible to maintain the cell line for serial analysis and meaningful comparisons. Also, the cell samples isolated from donors can only be used one time due to the lack of proliferative properties. The variation in blood samples from donors of different ethnicities, ages, genders and medical backgrounds makes the analysis complicated and inaccurate.
Multi-lineage progenitor cell lines (MLPCs) are karyotypically normal multi-potent progenitor cells obtained from post-partum human umbilical cord blood. They have been expanded from a single cell and are clonal. MLPCs are normal, non-transformed, non-immortalized cells. It is possible to continue to expand these cells, and they can differentiate down specific lineages beyond 20 passages. Their high purity and proliferative features make MLPC an ideal tool for the study of SMC differentiation. Transforming growth factor β1 (TGF-β1) is thought to play a key role in SMC differentiation and is known to coordinately upregulate a variety of SMC differentiation markers in cultured SMC from mature blood vessels [10,11] as well as pluri-potential stem cells [5,7,8]. In this study, we investigated the differentiation of MLPC into SMC lineage cells, which was induced by TGF-β1. We found that the mRNA levels of a variety of SMC-specific markers were increased during this process, whereas endothelial cell-specific markers and EPC marker CD34 were decreased in TGF-β1-treated MLPC. This convincingly indicated the cells’ differentiation into SMC lineage.
Material and Methods
Chemicals and reagents
Recombinant human TGF-β1 was obtained from R&D systems (Minneapolis, MN, USA). MLPCs were purchased from BioE company (St. Paul, MN, USA). The mesenchymal stem cell medium Bulletkit was obtained from Cambrex-walker (Walkersville, MD, USA). RNAquous-4PCR kit was purchased from Ambion (Austin, TX, USA). IQ SYBR Green super-mix kit was obtained from Bio-Rad (Hercules, CA, USA). All of the primers were synthesized by Sigma Genosys (The Woodlands, TX, USA).
Cell culture
MLPCs were cultured in mesenchymal stem cell basic medium to maintain their undifferentiated status and were then subcultured by using trypsin-EDTA reagent in the regular way. MLPCs at passage 4 to passage 6 were used in the experiment. To induce cell differentiation, MLPCs were seeded at 1.5×105 in each well on a 6 well plate containing mesenchymal stem cell basic medium plus 1% FBS with or without TGF-β1 (1 ng/mL or 5 ng/mL). MLPCs were cultured for 7 days, and cell RNA was then harvested for PCR analysis.
Real-time PCR
Total cellular RNA was isolated using the RNAquous-4PCR kit. The genomic DNA contamination in RNA preparation was removed by using the DNA-free kit (Ambion, Austin, TX), and a lack of detectable genomic DNA was confirmed by PCR. Total RNA (0.5 μg) was reverse-transcribed into cDNA using the iScipt cDNA synthesis kit (Bio-Rad) following the manufacturer’s instruction. Primers for all tested genes were designed using the Beacon Designer 2.1 software (Bio-Rad). The sequences of primers are shown in Table 1. The quality of individual pairs of primers was confirmed by running conventional PCR before real-time PCR to make sure there were no detectable primer dimers or non-specific products yielded. The real-time PCR reaction mixture included the following: 250 nM primers, 50 ng cDNA, and iQ SYBR Green supermix (0.2 mM of each dNTP, 25 U/mL iTaq DNA polymerase, SYBR Green I, 10 nM fluorescein, 3 mM MgCl2, 50 mM KCl, and 20 mM Tris-HCl). Using the iCycler iQ Real-time PCR detection system (Bio-Rad), PCR cycling conditions were set as follows: 95°C for 90 seconds, 40 cycles at 95°C for 20 seconds, and 60°C for 1 minunte. Melting curve analysis was performed on the iCycler over the range 55–95°C by monitoring iQ SYBR green fluorescence with increasing temperature (0.5°C increment changes at 10 seconds intervals). Specific products were determined as clear single peaks at their melting curves. All sample measurements were performed in triplicate. Sample cycle threshold (Ct) values were determined from plots of relative fluorescence units (RFU) versus PCR cycle number during exponential amplification so that sample measurement comparison was possible. Standard curves for all primer amplifications were generated by plotting average Ct values against the logarithm starting quantity of target template molecules (series dilution of cDNA template: 50, 10, 2, 0.4, and 0.08 ng), followed by a sum of least squares regression analysis. The correlation coefficiency and PCR efficiency of all primers were above 90%, respectively. The gene expression in each sample was normalized to GAPDH expression as [2^(CtGAPDH−Ctgene)].
Table 1.
Primers Sequence for Real-Time PCR*.
| Gene | Gene bank No. | Forward primer | Reverse primer |
|---|---|---|---|
| SM22α | D17409 | AGATCATCAGTTAGAGCGGAGAGG | GTGTGGGTGAGGCAGGCTAAG |
| calponin-1 | BC036307 | CAACCACCACGCACACAACTAC | GGTCCAGCCAAGAGCAGCAG |
| SM α-actin | NM_001613 | GTGTTGCCCCTGAAGAGCAT | GCTGGGACATTGAAAGTCTCA |
| caldesmon | NM_033140 | CTGGCTTGAAGGTAGGGGTTT | TTGGGAGCAGGTGACTTGTTT |
| tropomyosin | M19713 | CGAACAACTTGAAGTCACTGGA | CGTACAGCTCGTCTTCTAAGTCA |
| MLCK | NM_053026 | GCAAGGCTGCTAACAGGAGAA | GGCAAGCCCTTCACATCTGA |
| VE-cadherin | NM_001795 | GATCAAGTCAAGCGTGAGTCG | AGCCTCTCAATGGCGAACAC |
| VEGFR-2 | AF063658 | GCAGGGGACAGAGGGACTTG | GAGGCCATCGCTGCACTCA |
| CD34 | S53910 | CAACACCTAGTACCCTTGGAAGT | ACTGTCGTTTCTGTGATGTTTGT |
| CD146 | NM_006500 | TCCAGCTCCGCGTCTACAA | CTACACAGGTAGCGACCTCC |
| CD105 | NM_000118 | AGCCCCACAAGTCTTGCAG | GCTAGTGGTATATGTCACCTCGC |
Primers for all tested genes were designed via the Beacon Designer 2.1 software (Bio-Rad Inc., Hercules, CA).
Statistical analysis
Data from the control and TGF-β1-treated groups were analyzed using a paired Student’s t test (one tail, Minitab software, Sigma Breakthrough Technologies, Inc., San Marcos, TX). Statistics are reported as mean ± the standard deviation (SD). P value <0.05 was considered statistically significant.
Results
TGF-β1 increased the expression of SMC markers in MLPCs
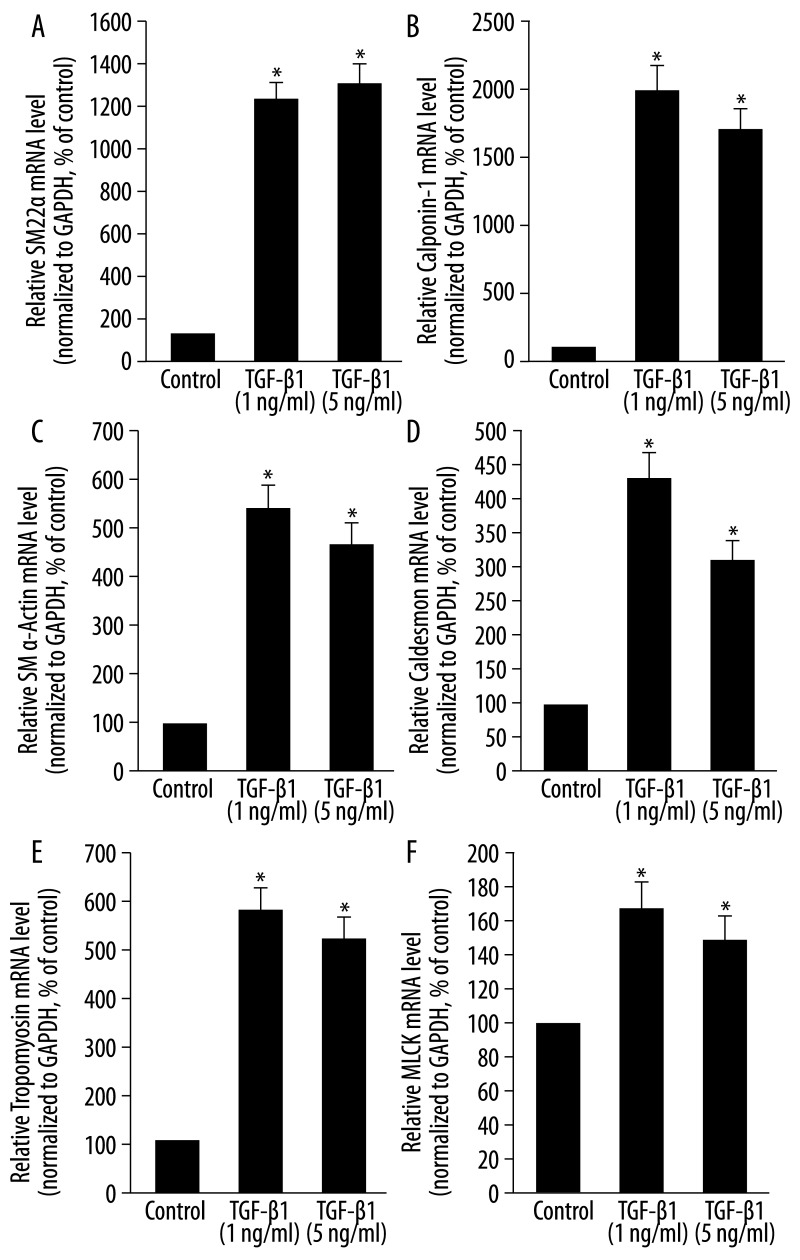
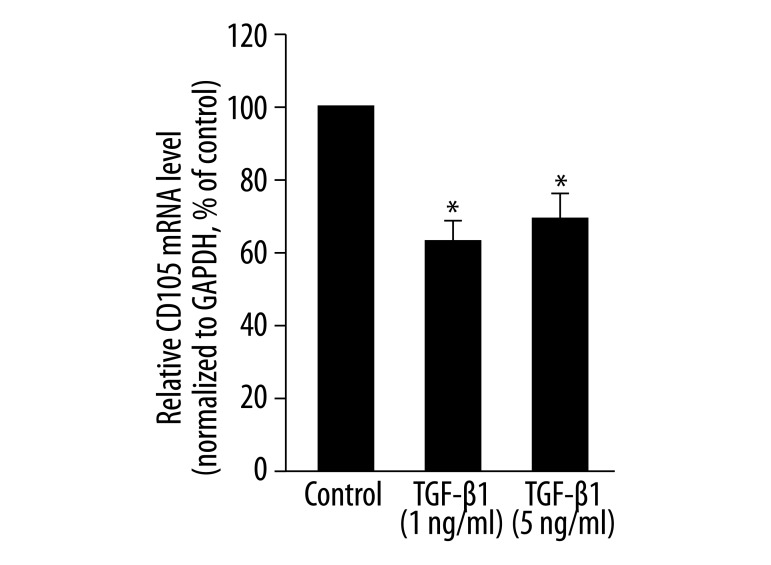
After 7 days of exposure to TGF-β1, several SMC-specific markers were dramatically increased. The addition of TGF-β1 (1 ng/mL) to mesenchymal stem cell medium significantly increased the mRNA levels of SM22α, calponin-1, SM α-actin, caldesmon, tropomyosin and myosin light chain kinase (MLCK) to 1215.5%, 1974.6%, 567%, 429.7%, 567% and 162.8%, respectively, when compared to controls (medium only) (P<0.05, Figure 1A–F). TGF-β1 (5 ng/mL) also increased the mRNA levels of these markers significantly (P<0.05), but the effect was less than that of the TGF-β1 treatment at 1 ng/mL (Figure 1A–F). In addition, we investigated the expression level of CD105 (a TGF-β accessory receptor) in MLPCs. We found that CD105 is expressed at a high level in MLPCs under control conditions (relative mRNA level is 0.156, data not shown). After 7 days, TGF-β1 at 1 ng/mL and 5 ng/mL led to a significant downregulation of CD105 mRNA levels to 61.7% and 70.8% of that of controls, respectively (P<0.05, Figure 2).
Figure 1.
Effects of TGF-β1 on the mRNA levels of SMC-specific markers in MLPCs. MLPCs were treated without or with TGF-β1 (1 ng/mL and 5 ng/mL) for 7 days. TGF-β1 significantly increased the mRNA levels of SM22α and calponin-1 (A, B), SM α-actin and caldesmon (C, D) and tropomyosin and MLCK (E, F). Total RNA was extracted and then reverse-transcribed to cDNA. Fifty nanograms of cDNA of each sample were used in real-time PCR analysis to detect gene expression. The mRNA level of each gene in each sample was normalized to that of GAPDH. The relative mRNA level of each gene in TGF-β1-treated cells was presented as the percentage of that in control cells. Data are expressed as means ± SD of triplicate values from three separate experiments. *P<0.05 compared with control group (Student’s t test).
Figure 2.
Effects of TGF-β1 on the mRNA level of CD105 in MLPCs. MLPCs were treated without or with TGF-β1 (1 ng/mL and 5 ng/mL) for 7 days. Real-time PCR was performed to detect the mRNA level of CD105. TGF-β1 decreased the mRNA levels of CD105. Data are expressed as means ± SD of triplicate values from three separate experiments. * P<0.05 compared with control group (Student’s t test).
TGF-β1 decreased the expression of endothelial cell-specific markers in MLPCs
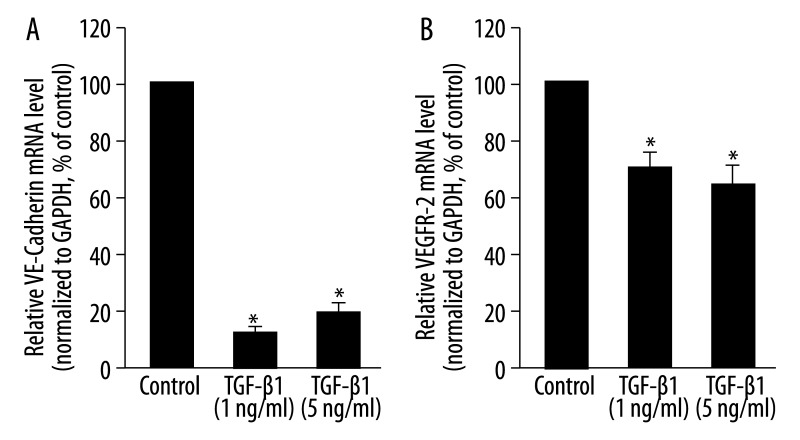
Two EC-specific markers, VE-cadherin and VEGFR-2, were detected in MLPCs under control conditions. After 7 days of exposure to TGF-β1 at 1 ng/mL, there was a significant reduction in VE-cadherin and VEGFR-2 mRNA levels to 15.4% and 70.8% of control levels, respectively. TGF-β1 at 5 ng/mL also significantly decreased VE-cadherin and VEGFR-2; mRNA levels were 21.8% and 66.1% of controls, respectively (P<0.05, Figure 3A and B).
Figure 3.
Effects of TGF-β1 on the mRNA levels of endothelial cell-specific markers in MLPCs. MLPCs were treated without or with TGF-β1 (1 ng/mL and 5 ng/mL) for 7 days. Real-time PCR was performed to detect VE-cadherin and VEGFR-2 mRNA levels. TGF-β1 decreased the mRNA levels of VE-cadherin (A) and VEGFR-2 (B). Data are expressed as means ±SD of triplicate values from three separate experiments. *P<0.05 compared with control group (Student’s t test).
L2 TGF-β1 decreased the expression of endothelial progenitor cell (EPC) marker CD34 in MLPCs
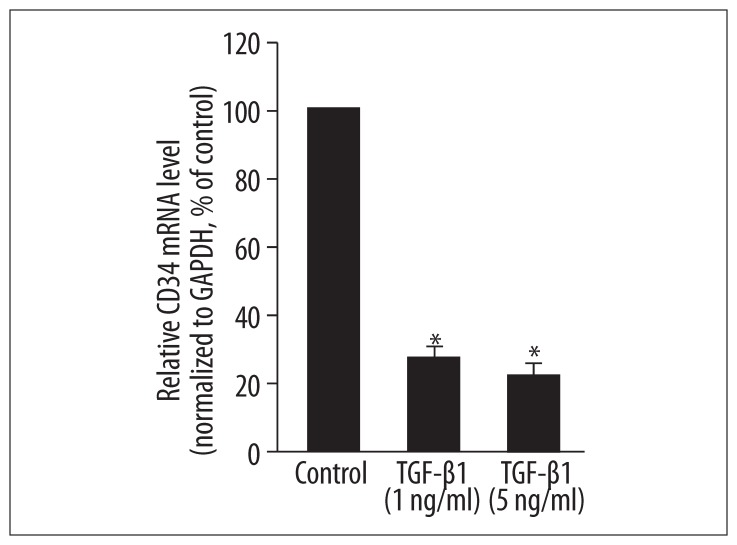
We investigated the expression of CD34, a marker of endothelial progenitor cells, in MLPCs. CD34 was expressed at a low level in MLPCs under control conditions (relative mRNA level is 2.34E-05, data not shown). After 7 days, the addition of TGF-β1 at 1 ng/mL and 5 ng/mL to the mesenchymal stem cell culture media significantly decreased the mRNA level of CD34 to 28.7% and 25.1% of control levels, respectively (P<0.05, Figure 4).
Figure 4.
Effects of TGF-β1 on the mRNA level of EPC marker CD34 in MLPCs. MLPCs were treated without or with TGF-β1 (1 ng/mL and 5 ng/mL) for 7 days. Real-time PCR was performed to detect the mRNA level of CD34. TGF-β1 decreased the mRNA levels of CD34. Data are expressed as means ±SD of triplicate values from three separate experiments. * P<0.05 compared with control group (Student’s t test).
TGF-β1 increased the expression of adhesion molecule CD146 in MLPCs
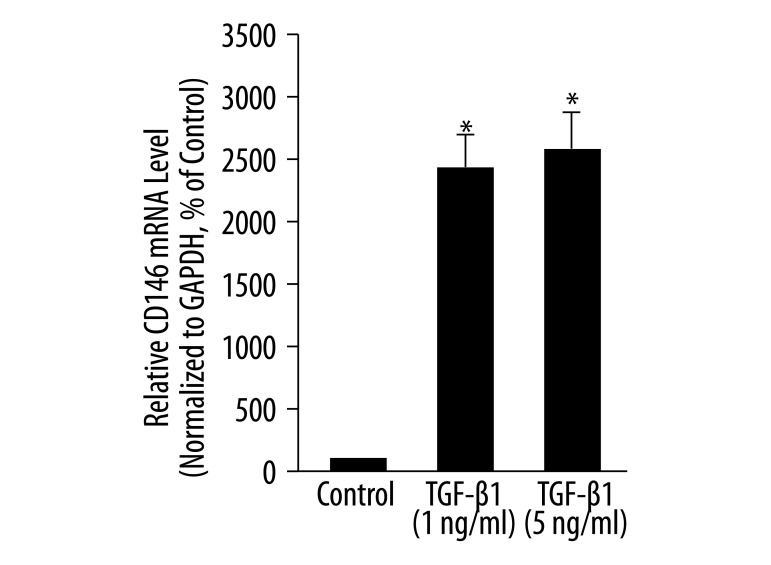
We also tested the expression of CD146 in MLPCs. The mRNA level of CD146 was low in MLPCs under control conditions (relative mRNA level is 3.8E-05, data not shown). After 7 days, TGF-β1 at 1 ng/mL and 5 ng/mL significantly increased the mRNA level of CD146 to 2430% and 2605% of that of the controls, respectively (P<0.05, Figure 5).
Figure 5.
Effects of TGF-β1 on the mRNA level of adhesion molecule CD146 in MLPCs. MLPCs were treated without or with TGF-β1 (1 ng/mL and 5 ng/mL) for 7 days. Real-time PCR was performed to detect the mRNA level of CD146. TGF-β1 increased the mRNA levels of CD146. Data are expressed as means ±SD of triplicate values from three separate experiments. * P< 0.05 compared with control group (Student’s t test).
Discussion
MLPCs originate from human umbilical cord blood, are karyotypically normal, demonstrate proliferative features, and are highly pure. In the present study, we observed the differentiation of MLPCs to SMC lineage in the presence of TGF-β1 as evidenced by an upregulation of the mRNA levels of both early and late phase SMC-specific markers and adhesion molecule CD146. Two EC markers, as well as EPC marker CD34 and TGF-β1 cell surface accessory protein CD105, were decreased in TGF-β1 treated MLPCs. As such, these data suggest that MLPCs represent a promising tool for the study of SMC differentiation.
Quiescent vascular SMCs exhibit a phenotype characterized by the expression of several contractile apparatus-associated proteins; these are used as markers to identify SMC lineage differentiation. We checked both early phase markers (SM α-actin, SM22α) and late phase markers (MLCK, calponin-1, caldesmon, and tropomyosin) for SMC differentiation in MLPCs. We found that treatment with TGF-β1 led to a dramatic increase in the mRNA levels of all the tested markers in MLPCs after 7 days of culture; this strongly indicates the differentiation of MLPCs into SMC lineage in the presence of TGF-β1. Chen S et al. [5] reported that TGF-β1 (5 ng/mL) increases the expression of SM α-actin, SM22α, calponin-1 and myosin in the neural crest stem cell line Monc-1 in vitro. Lien SC et al. [7] also found that TGF-β1 (2 ng/mL) induces SM α-actin, SM22α and smooth muscle myosin heavy chain (SMMHC) in 10T1/2 mesenchymal cells in vitro. In our study, the mRNA levels of all the tested SMC markers in MLPCs were upregulated to a comparable level with TGF-β1 treatment when compared to those found in neural crest stem cell line (Monc-1) or 10T1/2 mesenchymal cells.
The signaling pathway underlying TGF-β1-induced SMC-specific marker expression in MLPCs was not investigated in this study. Sinha et al. [8] investigated differential TGF-β-Smad signaling for early versus late SMC marker expression; SM α-actin promoter activity was found to be dependent on both Smad 2 and Smad 3 whereas smooth muscle myosin heavy chain (SMMHC) activity is Smad2 dependent in mouse embryonic stem cell. Chen S et al. [5] also found that TGF-β increased SM α-actin and SM22α in the neural crest stem cell line through activation of Smad2 and Smad3. In addition, RhoA was reported to be essential in Smad signaling during TGF-β-induced SM a-actin, SM22α and calponin expression in the neural crest stem cell line Monc-1 [12]. Meanwhile, the PI3 kinase/Akt pathway was found to be involved in TGF-β1-induced SM α-actin, SM22α and SMMHC expression in 10T1/2 mesenchymal cells [7].
We determined the expression of CD105 (endoglin) in the current study. CD105 is a transmembrane accessory receptor for TGF-β. By forming a heteromeric complex with distinct TGF-β receptors, it modulates the access of TGF-β to the signaling complex and the postponed cellular responses to TGF-β [13,14]. We found that CD105 is expressed at a high level in MLPCs in the control condition (relative mRNA level is 0.156), which may facilitate TGF-β1/TGF-β receptor signaling transduction. Over a long duration of TGF-β1 treatment (7 days), expression of CD105 was decreased, which may be due to a negative feedback mechanism. The specific signaling mechanism involved in TGF-β1-induced differentiation of MLPCs into SMC will need to be further elucidated.
Progenitor cells are primitive cells that possess the capacity to differentiate into multiple lineages under different conditions [15,16]. Evidence has shown that progenitor cells from human peripheral blood can differentiate into EC when stimulated by shear stress [17] and into SMCs in conditions of cyclic strain [18]. Hence, we investigated the expression of two EC specific markers to confirm that the potential for EC lineage differentiation by the MLPCs was suppressed under the designated conditions of this study. We found that the mRNA levels of VE-cadherin and VEGFR-2 were significantly decreased by TGF-β1 on day 7 of culture. VEGFR-2 is the pivotal receptor mediating the mitogenic action of VEGF; it plays an essential role in angiogenesis, neovascularization and EPC differentiation [19]. VE-cadherin is another EC-specific marker which has been shown to play important role in vasculogenesis and angiogenesis. The downregulated VE-cadherin and VEGFR-2 observed in the MLPCs provides further confirmation that TGF-β1 induced SMC lineage differentiation in the MLPCs. Similarly, Chen S et al. [5] found that TGF-β1 reduced epithelial markers while inducing SMC-specific markers in the neural crest stem cell line Monc-1. The signaling pathways underlying TGF-β1-decreased EC markers remain unclear. Watabe et al. [20] reported that TGF-β inhibited proliferation and sheet formation of embryonic stem cell (ESC)-derived ECs. Stimulation of ESC-derived ECs with TGF-β resulted in phosphorylation of both Smad2 and Smadl/5. The specific signaling mechanism underlying the TGF-β1-induced reduction in EC markers in MLPC need to be further studied.
In addition to EC markers, we tested the EPC marker CD34 in MLPCs. CD34 has been used as a hematopoietic stem cell marker; endothelial progenitor cells are characterized by the expression of CD34, CD133 and VEGFR-2 [21]. In this study, CD34 was expressed at a low level in MLPC under the control conditions, and expression was further reduced by TGF-β1 treatment; this suggests that the tendency for EC lineage differentiation by the MLCPs was suppressed under the designated conditions. This in accordance with a previous finding that CD34 expression was downregulated during TGF-β1 induced myofibroblast differentiation [22]. Our data provide further confirmation for SMC lineage differentiation in MLPCs. The specific mechanisms whereby TGF-β1 mediates this phenomenon remain unclear.
CD146, melanoma cell adhesion molecule, is also expressed in SMC and plays an important role in vasculogenesis and embryo development [23]. CD146, when activated, induces association of p59fyn with CD146, resulting in the phosphorylation of p125FAK and its binding with paxillin; this finding suggests that CD146 is involved in outside-in signaling and may contribute to focal adhesion assembly, reorganization of the cytoskeleton, intercellular interaction, maintenance of cell shape, and control of cell migration and proliferation [24]. In this study, CD146 was significantly increased by TGF-β1 treatment in MLPCs, which may facilitate signaling transduction by TGF-β1 and contribute to the differentiation of MLPCs into mature SMC.
Conclusions
In conclusion, a variety of SMC-specific markers, including early and late phase markers, were dramatically increased in MLCP treated with TGF-β1. Meanwhile, two EC-specific markers as well as the EPC marker CD34 were significantly decreased. These data strongly indicate that MLCPS differentiate into SMC lineage cells in the presence of TGF-β1. MLPCs are karyotypically normal, non-transformed, non-immortalized cells that are obtained from post-partum human umbilical cord blood. Because they have been expanded from a single cell and have the capacity to differentiate into multiple lineages, they are highly pure and proliferative. MLPCs offer significant advantages over other currently used cell models, such as 10T1/2 cells, the neural crest stem cell line Monc-1 and SMC progenitor cells from human peripheral blood for the study of SMC differentiation. This study demonstrates a novel cell model for SMC lineage differentiation analysis, which may increase our understanding of SMC differentiation and help contribute to the field of vascular repair and tissue engineering.
Footnotes
Source of support: This work was partially supported by research grants from the National Institutes of Health (R01 EB002436 and R01 HL083471 to C.C.) and by a grant from the Alliance for NanoHealth (W81XWH-09-2-0139) awarded from the U.S. Army Research and Materiel Command’s Telemedicine and Advanced Technology Research Medicine program (TATRC). L.Z and S.M.W. were supported by a training grant from NIH (T32HL083774)
References
- 1.Milewicz DM, Kwartler CS, Papke CL, et al. Genetic variants promoting smooth muscle cell proliferation can result in diffuse and diverse vascular diseases: evidence for a hyperplastic vasculomyopathy. Genet Med. 2010;12:196–203. doi: 10.1097/GIM.0b013e3181cdd687. [DOI] [PubMed] [Google Scholar]
- 2.Meyrick B. The pathology of pulmonary artery hypertension. Clin Chest Med. 2001;22:393–404. doi: 10.1016/s0272-5231(05)70279-3. [DOI] [PubMed] [Google Scholar]
- 3.Caglayan E, Romeo GR, Kappert K, et al. Profilin-1 is expressed in human atherosclerotic plaques and induces atherogenic effects on vascular smooth muscle cells. PLoS One. 2010;5:e13608. doi: 10.1371/journal.pone.0013608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmeliet P, Collen D. Transgenic mouse models in angiogenesis and cardiovascular disease. J Pathol. 2000;190:387–405. doi: 10.1002/(SICI)1096-9896(200002)190:3<387::AID-PATH595>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Lechleider RJ. Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circ Res. 2004;94:1195–202. doi: 10.1161/01.RES.0000126897.41658.81. [DOI] [PubMed] [Google Scholar]
- 6.Hirschi KK, Rohovsky SA, D’Amore PA. PDGF, TGF-beta, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J Cell Biol. 1998;141:805–14. doi: 10.1083/jcb.141.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lien SC, Usami S, Chien S, Chiu JJ. Phosphatidylinositol 3-kinase/Akt pathway is involved in transforming growth factor-beta1-induced phenotypic modulation of 10T1/2 cells to smooth muscle cells. Cell Signal. 2006;18:1270–78. doi: 10.1016/j.cellsig.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Sinha S, Hoofnagle MH, Kingston PA, et al. Transforming growth factor-beta1 signaling contributes to development of smooth muscle cells from embryonic stem cells. Am J Physiol Cell Physiol. 2004;287:C1560–68. doi: 10.1152/ajpcell.00221.2004. [DOI] [PubMed] [Google Scholar]
- 9.Simper D, Stalboerger PG, Panetta CJ, et al. Smooth muscle progenitor cells in human blood. Circulation. 2002;106:1199–204. doi: 10.1161/01.cir.0000031525.61826.a8. [DOI] [PubMed] [Google Scholar]
- 10.Becker C, Laeufer T, Arikkat J, Jakse G. TGFbeta-1 and epithelial-mesenchymal interactions promote smooth muscle gene expression in bone marrow stromal cells: possible application in therapies for urological defects. Int J Artif Organs. 2008;31:951–59. doi: 10.1177/039139880803101105. [DOI] [PubMed] [Google Scholar]
- 11.Kennard S, Liu H, Lilly B. Transforming growth factor-beta (TGF-1) down-regulates Notch3 in fibroblasts to promote smooth muscle gene expression. J Biol Chem. 2008;283:1324–33. doi: 10.1074/jbc.M706651200. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Crawford M, Day RM, et al. RhoA modulates Smad signaling during transforming growth factor-beta-induced smooth muscle differentiation. J Biol Chem. 2006;281:1765–70. doi: 10.1074/jbc.M507771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derynck R, Feng XH. TGF-beta receptor signaling. Biochim Biophys Acta. 1997;1333:F105–50. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- 14.Wrana JL, Attisano L, Wieser R, et al. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–47. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 15.Campard D, Lysy PA, Najimi M, Sokal EM. Native umbilical cord matrix stem cells express hepatic markers and differentiate into hepatocyte-like cells. Gastroenterology. 2008;134:833–48. doi: 10.1053/j.gastro.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 16.Wu KH, Zhou B, Lu SH, et al. In vitro and in vivo differentiation of human umbilical cord derived stem cells into endothelial cells. J Cell Biochem. 2007;100:608–16. doi: 10.1002/jcb.21078. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto K, Takahashi T, Asahara T, et al. Proliferation, differentiation, and tube formation by endothelial progenitor cells in response to shear stress. J Appl Physiol. 2003;95:2081–88. doi: 10.1152/japplphysiol.00232.2003. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton DW, Maul TM, Vorp DA. Characterization of the response of bone marrow-derived progenitor cells to cyclic strain: implications for vascular tissue-engineering applications. Tissue Eng. 2004;10:361–69. doi: 10.1089/107632704323061726. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita J, Itoh H, Hirashima M, et al. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 20.Watabe T, Yamashita JK, Mishima K, Miyazono K. TGF-beta signaling in embryonic stem cell-derived endothelial cells. Methods Mol Biol. 2006;330:341–51. doi: 10.1385/1-59745-036-7:341. [DOI] [PubMed] [Google Scholar]
- 21.Hristov M, Erl W, Weber PC. Endothelial progenitor cells: mobilization, differentiation, and homing. Arterioscler Thromb Vasc Biol. 2003;23:1185–89. doi: 10.1161/01.ATV.0000073832.49290.B5. [DOI] [PubMed] [Google Scholar]
- 22.Espana EM, Kawakita T, Liu CY, Tseng SC. CD-34 expression by cultured human keratocytes is downregulated during myofibroblast differentiation induced by TGF-beta1. Invest Ophthalmol Vis Sci. 2004;45:2985–91. doi: 10.1167/iovs.04-0201. [DOI] [PubMed] [Google Scholar]
- 23.Ouhtit A, Gaur RL, Abd Elmageed ZY, et al. Towards understanding the mode of action of the multifaceted cell adhesion receptor CD146. Biochim Biophys Acta. 2009;1795:130–36. doi: 10.1016/j.bbcan.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Anfosso F, Bardin N, Frances V, et al. Activation of human endothelial cells via S-endo-1 antigen (CD146) stimulates the tyrosine phosphorylation of focal adhesion kinase p125(FAK) J Biol Chem. 1998;273:26852–56. doi: 10.1074/jbc.273.41.26852. [DOI] [PubMed] [Google Scholar]