Abstract
The spinach CSP41 protein has been shown to bind and cleave chloroplast RNA in vitro. Arabidopsis thaliana, like other photosynthetic eukaryotes, encodes two copies of this protein. Several functions have been described for CSP41 proteins in Arabidopsis, including roles in chloroplast rRNA metabolism and transcription. CSP41a and CSP41b interact physically, but it is not clear whether they have distinct functions. It is shown here that CSP41b, but not CSP41a, is an essential and major component of a specific subset of RNA-binding complexes that form in the dark and disassemble in the light. RNA immunoprecipitation and hybridization to gene chips (RIP-chip) experiments indicated that CSP41 complexes can contain chloroplast mRNAs coding for photosynthetic proteins and rRNAs (16S and 23S), but no tRNAs or mRNAs for ribosomal proteins. Leaves of plants lacking CSP41b showed decreased steady-state levels of CSP41 target RNAs, as well as decreased plastid transcription and translation rates. Representative target RNAs were less stable when incubated with broken chloroplasts devoid of CSP41 complexes, indicating that CSP41 proteins can stabilize target RNAs. Therefore, it is proposed that (i) CSP41 complexes may serve to stabilize non-translated target mRNAs and precursor rRNAs during the night when the translational machinery is less active in a manner responsive to the redox state of the chloroplast, and (ii) that the defects in translation and transcription in CSP41 protein-less mutants are secondary effects of the decreased transcript stability.
Keywords: Arabidopsis, chloroplast, gene expression, RNA, RNA-binding protein, transcription, translation
Introduction
Chloroplasts are descended from an ancient cyanobacterial endosymbiont and have retained a remnant of the prokaryotic chromosome, which now forms the plastome. Chloroplast genes are still essential for many functions, particularly for plastid gene expression and photosynthesis (Wolfe et al., 1991), but the majority of cyanobacterial genes have been transferred to the host nucleus during evolution. Hence, developmental and metabolic processes in chloroplasts require tight coordination of nuclear and plastid gene expression. Nucleus-encoded chloroplast proteins that act on one or more chloroplast genes or transcripts, or are involved in protein synthesis (Barkan and Goldschmidt-Clermont, 2000), are crucial for chloroplast gene expression. At the RNA level, these regulatory proteins can bind and/or cleave RNA and mediate RNA maturation, editing, or stability (Zerges, 2000; Choquet and Wollman, 2002; Manuell et al., 2004; Marin-Navarro et al., 2007).
All members of the green lineage of photosynthetic eukaryotes have nuclear genes that code for the related proteins CSP41a and CSP41b (chloroplast stem–loop-binding protein of 41 kDa), which are of cyanobacterial origin (Baker et al., 1998; Bollenbach and Stern, 2003b; Yamaguchi et al., 2003). CSP41a was initially isolated as part of a protein complex that binds in vitro to the 3′-terminal stem–loop structure of the petD mRNA (Yang et al., 1995, 1996). Subsequently, CSP41a was shown to exhibit RNase activity, with a preference for 3' stem–loops (Yang and Stern, 1997; Bollenbach and Stern, 2003a, b). CSP41 proteins were also tentatively identified as components of the plastid-encoded RNA polymerase (PEP) in mustard (Pfannschmidt et al., 2000), and an association with the 70S ribosome was suggested in Chlamydomonas reinhardtii (Yamaguchi et al., 2003). Further characterization of the PEP complex failed to identify CSP41a and CSP41b as constituents (Suzuki et al., 2004; Pfalz et al., 2006).
CSP41a and CSP41b are present in three distinct stromal complexes: one that is larger than 0.8 MDa, a 224 kDa complex containing the ribosomal proteins L5 and L31 as potential interaction partners, and a 126 kDa complex that is likely to represent a heterotrimer of CSP41 proteins (Peltier et al., 2006). Further analysis of stromal high molecular weight (HMW) complexes found CSP41b mostly in 0.8–2 MDa fractions together with 30S ribosomal particles, other RNA-binding proteins, and the LTA2 subunit of the pyruvate dehydrogenase (Olinares et al., 2010).
Arabidopsis thaliana mutants lacking both CSP41 proteins were found to be inviable, leading to the proposal that the two CSP41 proteins have redundant functions (Beligni and Mayfield, 2008). However, it was also noted that CSP41a accumulation depended markedly on the presence of CSP41b (Beligni and Mayfield, 2008; Bollenbach et al., 2009). Furthermore, the finding that the two CSP41 proteins physically interact (Bollenbach et al., 2009), as previously suggested by colourless native PAGE analysis (Peltier et al., 2006), implies that the two proteins might act concertedly rather than redundantly.
Multiple functions have been assigned to CSP41 proteins previously. Hassidim et al. (2007) showed that the csp41b mutation affected chloroplast morphology, photosynthetic performance, and circadian rhythms. Moreover, the association of CSP41b with pre-ribosomal particles, and changes in levels of the 23S rRNA precursor seen in csp41 mutants, suggested an involvement of CSP41 proteins in ribosomal biogenesis (Beligni and Mayfield, 2008). Decreases in the transcriptional activity of some chloroplast genes, as well as differential promoter usage, in the csp41b mutant, however, indicated that CSP41b might constitute a component of the plastid transcriptional machinery (Bollenbach et al., 2009). Despite the multiple evidence for an involvement of CSP41 proteins in chloroplast gene expression, CSP41b was also reported to interact in the cytosol with heteroglycans (Fettke et al., 2011).
In the present work, the subcellular localization, function(s), and interactions of the two CSP41 proteins were dissected in detail. It was found that CSP41 proteins reside in the chloroplast and are post-translationally modified, and CSP41b can compensate for the loss of CSP41a, but not vice versa. Both proteins interact in the dark to form a specific subset of riboprotein complexes, which contain chloroplast mRNAs and rRNAs. It is suggested that CSP41 complexes determine the stability of a distinct set of chloroplast transcripts including rRNAs, such that the absence of CSP41b affects both target transcript stability and chloroplast translational activity.
Materials and methods
Plant material, propagation, growth, and chlorophyll a fluorescence measurements
Several mutant lines for CSP41a (At3g63140) and CSP41b (At1g09340) have already been described. Hassidim et al. (2007) reported two mutant alleles of CSP41b, which they designated crb-1 and crb-2. The latter was named csp41b-1 (Beligni and Mayfield, 2008) or csp41b-2 (Bollenbach et al., 2009) by others, and was also used in the present work, under the name csp41b-2. In this study, also a novel insertion line, csp41b-3, which was identified in the GABI-KAT collection, was used. Both csp41b-2 and csp41b-3 are in the Col-0 genetic background. For CSP41a, the mutant alleles csp41a-1 and csp41a-2 have been reported previously; only the latter completely abrogates expression of the CSP41a protein (Beligni and Mayfield, 2008). To obtain additional insertion lines for CSP41a, the T-DNA insertion collection of C. Koncz (Max-Planck-Institute for Plant Breeding Research, Cologne, Germany; Rios et al., 2002) was screened and the csp41a-3 and csp41a-4 mutant alleles (genetic background Col-0) were identified.
Mutant and wild-type [WT; A. thaliana ecotype Columbia (Col-0)] plants were grown on potting soil (A210, Stender AG, Schermbeck, Germany) under controlled greenhouse conditions [daylight supplemented with HQI Powerstar 400W/D lamps (Osram, Munich, Germany) with ∼180 μmol photons m−2 s−1 on the leaf surface from 6:00 to 9:00 and 15:00 to 20:00 h, ∼14 h light/10 h dark photoperiod] except when otherwise stated. Wuxal Super (Manna, Germany) was used as the fertilizer, according to the manufacturer’s instructions. All analyses were performed on 4-week-old mutant and WT plants. Methods used for the measurement of growth have been described previously (Leister et al., 1999).
In vivo chlorophyll a (Chl a) fluorescence of whole plants was recorded using an imaging chlorophyll fluorometer (Walz Imaging PAM, Walz GmbH, Effeltrich, Germany) by exposing plants to actinic light (50 μmol photons m−2 s−1) and after 10 min a saturating light flash (0.8 s, 4000 μmol photons m−2 s−1) to obtain the fluorescence-based effective quantum yield of photosystem II (PSII) in the light (ΦII) (Genty et al., 1989).
Overexpression of CSP41a, CSP41a:eGFP, CSP41b:CFP, and CSP41b:eGFP in planta
For complementation analyses, the CSP41a cDNA was ligated into the plant expression vector pLeela under the control of the 35S promoter from Cauliflower mosaic virus (CaMV). Flowers of csp41b-2 mutant plants were transformed according to Clough and Bent (1998) with the CSP41a overexpression construct and AtCSP41b:CFP-pBA002, which carries the complete CSP41b coding region upstream of a sequence encoding cyan fluorescent protein (CFP) (Raab et al., 2006). In addition, Col-0 was transformed with CSP41a and CSP41b cDNAs ligated into the plant expression vector pB7FWG2 (Karimi et al., 2002), which leads to the expression of a C-terminal enhanced green fluorescent protein (eGFP) fusion protein from the 35S promoter. The plants were then transferred to the greenhouse, and seeds were collected 3 weeks later. Twenty-five independent transgenic plants were selected on the basis of their resistance to Basta. The presence and expression of the transgene was confirmed by PCR, and northern and western analyses.
Nucleic acid analysis
Arabidopsis thaliana DNA was isolated (Ihnatowicz et al., 2004) and T-DNA insertion–junction sites were recovered by PCR using combinations of insertion- and gene-specific primers, and then sequenced. For RNA analysis, total leaf RNA was extracted from fresh tissue using the TRIzol reagent (Invitrogen, Germany). Northern analyses were performed under stringent conditions, according to Sambrook and Russell (2001). Probes complementary to nuclear and chloroplast genes were used for the hybridizations. Primers used to amplify the probes are listed in Supplementary Table S1 available at JXB online. All the probes used were cDNA fragments labelled with 32P. Signals were quantified by using a phosphoimager (Typhoon; GE Healthcare, Munich, Germany) and the respective quantification program ImageQuant.
For quantitative real-time profiling, 7 μg aliquots of total RNA treated with DNase I (Roche Applied Science, Castle Hill, Australia) for at least 1 h were utilized for first-strand cDNA synthesis using Omniscript reverse transcriptase (Qiagen, Doncaster, Australia) and random priming (Sigma, St Louis, MO, USA) according to the supplier’s instructions. The quantitative real-time PCR (qRT-PCR) profiling was carried out on a LightCycler 480 real-time PCR system (Roche Applied Science) using methods and primers as described (de Longevialle et al., 2008). Data were analysed using the Genesis software (http://genome.tugraz.at; Sturn et al., 2002).
In vivo translation assay
Radioactive labelling of thylakoid proteins was performed according to Armbruster et al. (2010). In brief, discs of Arabidopsis leaves were vacuum-infiltrated in a syringe containing 20 μg ml−1 cycloheximide in 10 ml of 10 mM TRIS, 5 mM MgCl2, 20 mM KCl (pH 6.8), and 0.1% (v/v) Tween-20, and incubated for 30 min to block cytosolic translation. Then leaves were again infiltrated with the same solution containing 1 mCi of [35S]methionine, transferred into the light (50 μmol m−2 s−1), and collected after 20 min. Subsequently, thylakoid proteins were prepared and fractionated as described by Armbruster et al. (2010). Signals were detected and quantified using the phosphoimager as described above.
Chloroplast and stroma isolation
Arabidopsis thaliana chloroplasts were isolated according to Kunst (1998) from plants adapted to darkness for 18 h except where otherwise stated. Homogenized leaf tissue was applied to a two-step Percoll gradient and intact chloroplasts were collected from the interphase. Chloroplasts were then lysed in an equal volume of 30 mM HEPES (pH 8.0), 200 mM KOAc, 10 mM MgOAc, and 2 mM dithiothreitol (DTT), and fragmented by passing them 20 times through a 24-gauge syringe. Stromal proteins were separated from the membrane fractions by centrifugation at 16 000 g for 30 min. Stromal protein concentration was determined with the Bradford Protein Assay (Biorad, Munich, Germany).
Two-dimensional PAGE analyses
For two-dimensional (2D) blue native (BN)/SDS–PAGE analysis, samples of stromal proteins equivalent to 30 μg or 160 μg of Chl and treated or untreated with RNase were first fractionated by BN-PAGE (Schägger and von Jagow, 1991) and then by SDS–PAGE (Schägger and von Jagow, 1987). Gradient gels were used in both cases (5–14% acrylamide for the first dimension, 10–16% acrylamide for the second).
For 2D isoelectric focusing (IEF)/SDS–PAGE analysis, stromal proteins were precipitated with acetone and resuspended in 7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 0.5% (v/v) Pharmalyte (GE Healthcare), 0.002% (w/v) bromophenol blue, 18.2 mM DTT, and applied to Immobiline™ Drystrips (gradient pH 3–10 NL, GE Healthcare). After the separation of proteins according to their isoelectric point, the strip was first equilibrated in 6 M urea, 75 mM TRIS-HCl (pH 8.8), 29.3% (v/v) glycerol, 2% (w/v) SDS, 0.002% (w/v) bromophenol blue, 65 mM DTT for 15 min and then in the same buffer supplemented with 135 mM iodoacetamide for another 15 min period. After equilibration, proteins were separated by 10–16% acrylamide gradient SDS–PAGE.
Western analyses
Total leaf proteins were extracted as reported (Martinez-Garcia et al., 1999). For chloroplast fractionation, isolated chloroplasts were re-suspended in TE buffer (10 mM TRIS-HCl, pH 8.0; 1 mM EDTA) at a Chl concentration of 2 mg ml−1 and loaded onto a three-step sucrose gradient consisting of, from the bottom to the top, 1.2, 1, and 0.46 M sucrose in TE. After centrifugation at 30 000 g, 4 °C, the upper phase containing the stroma, the two interphases containing envelopes, and the pellet consisting of thylakoids were collected. Proteins were fractionated on one-dimensional (1D) SDS–PAGE gradient gels (10–16% acrylamide) as described by Schägger and von Jagow (1987). Proteins from 1D and 2D PAGE experiments were transferred to Immobilon-P membranes (Millipore). Replicate filters were incubated with antibodies specific for CSP41a, CSP41b, PsaD, PsbD, PetD, AtpD, Lhcb1, NdhL, phosphothreonine, TIC110, RbcL, or actin. Signals were detected using the Enhanced Chemiluminescence Western Blotting kit (GE Healthcare) and quantified using Imagequant (GE Healthcare).
Cross-linking of stromal proteins with EDC
Stromal extracts (1 mg protein ml−1) of Col-0, csp41b-2, and csp41b-2 plants expressing 35S::CSP41b:CFP were cross-linked with 4 mM 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and 1 mM N-hydroxysulphosuccinimide for 1 h on ice. The reaction was terminated by the addition of 25 mM TRIS.
Co-immunoprecipitation
For co-immunoprecipitation (Co-IP) experiments, 10 μl of anti-GFP antibody (Sigma) was incubated with 100 μl of stromal extract (500 μg of stromal protein) from 4-week-old Arabidopsis plants. The antibody was collected by incubation with Sepharose-coupled protein A (Sigma). For analysis of interaction partners, proteins were eluted by the addition of SDS sample buffer. The sample buffer was then applied to a 15% SDS–polyacrylamide gel and electrophoresis was carried out until all proteins had migrated into the separating gel (as monitored using pre-stained marker proteins). The protein-containing gel slice was excised, washed twice in ddH2O for 10 min, and further processed for mass spectrometry (MS).
Mass spectrometric analysis and database searches
Liquid chromatography (LC)-MS analyses were performed on an LTQ-Orbitrap XL system (Thermo Fisher Scientific Waltham, MA, USA). Trypsin-digested peptides were loaded on a fritless 100 μm capillary, packed in-house with ProntoSIL C18 ace-EPS (Bischoff Analysentechnik und -geräte GmbH, Leonberg, Germany) by using a quaternary HPLC pump (Flux, Basel, Switzerland) including a CTC autosampler. A gradient of 5–80% (v/v) acetonitrile in 0.1% (v/v) formic acid was then passed through the column over a period of 80 min. The eluted peptides were introduced directly into the LTQ orbitrap XL MS at a flow rate of 250 nl min−1 and a spray voltage of 1.3 kV. The LTQ-Orbitrap was operated via Instrument Method files of Xcalibur (Thermo Fisher Scientific) to acquire a full high-resolution MS scan between 400 m/z and 2000 m/z. The SEQUEST algorithm (Thermo Fisher Scientific) was used to interpret MS spectra. Results were interpreted on the basis of a conservative set of criteria: only results with ΔCn (delta normalized correlation) scores >0.2 were accepted, all fragments had to be at least partially tryptic, and the cross-correlation scores (Xcorr) of double-charged or triple-charged ions had to be >2.5 or >3.5, respectively (see Supplementary Table S2 at JXB online). Spectra were manually evaluated to match the following criteria: distinct peaks with signals clearly above noise levels, differences in fragment ion masses in the mass range of amino acids, and fulfilment of consecutive b and y ion series.
RIP-chip analyses
The RIP-chip (RNA immunoprecipitation and hybridization to gene chips) procedures have been described previously (Schmitz-Linneweber et al., 2005). RNA was isolated from pellet and supernatant fractions of the Co-IP by phenol–chloroform extraction, and labelled with Cy3 and Cy5 using the Micromax ASAP RNA labeling kit (Perkin-Elmer Life Sciences, Waltham, MA, USA). Labelled RNA was purified using Qiaquick spin columns (Qiagen, Hilden, Germany) and hybridized to microarrays covering the entire chloroplast chromosome in overlapping DNA fragments (see Supplementary Tables S3 and S4 at JXB online). Slides were scanned with a Scanarray Gx microarray scanner (Perkin-Elmer Life Sciences). Genepix Pro 6.0 software (Molecular Devices, Sunnyvale, CA, USA) was used to identify spots in scanner output files, to calculate local background, and to filter out elements with low signal-to-noise ratios. Background-subtracted data were used to calculate the median of ratios [pellet RNA (F635)/supernatant RNA (F532)]; that is, the enrichment ratio. Normalization was done according to the median log2(F635/F532) value for all above-background spots on each array. Slot-blot analysis of RNA from pellet and supernatant fractions was performed as described previously (Schmitz-Linneweber et al., 2005).
Pigment analysis
Pigments were analysed by reverse-phase HPLC as described previously by Färber et al. (1997). For pigment extraction, leaf discs were frozen in liquid nitrogen and disrupted with beads in microcentrifuge tubes in the presence of acetone. After a short centrifugation, pigment extracts were filtered through a membrane filter (pore size 0.2 μm) and either used directly for HPLC analysis or stored for up to 2 d at –20 °C.
Run-on transcription analysis
Run-on transcription reactions were performed essentially as described by Zoschke et al. (2007). A total of 1.9×107 chloroplasts were used in run-on transcription experiments, which were performed at 25 °C for 10 min in 50 mM TRIS-HCl (pH 8.0), 10 mM MgCl2, 10 mM β-mercaptoethanol, 20 U of RNase inhibitor, and 0.2 mM each of ATP, GTP, and CTP, in the presence of [α-32P]UTP (10 μCi μl−1). RNA was extracted and hybridized overnight at 58 °C to DNA fragments (1 μg and 0.5 μg) dot-blotted onto nylon membranes. Signals were detected and quantified using the phosphoimager as described above.
RNA stability assay
For radioactive labelling of rrn23, psbCD, and psbC RNA species, genes were amplified with specific primers and inserted into pGEM-T easy (Promega) downstream of the T7 promoter. In vitro transcription was performed in the presence of 12.5 μCi of [α-32P]CTP in a final volume of 20 μl. After 60 min incubation at 25 °C, the RNAs were precipitated and 1/10th was applied in each assay. Labelled RNA was incubated in the presence of 1×107 broken chloroplasts in 50 mM TRIS-HCl (pH 8.0), 10 mM MgCl2, 10 mM β-mercaptoethanol for 5 min at 25 °C. The RNAs were recovered by phenol extraction and separated by denaturing 1.5% (w/v) agarose gel electrophoresis.
Statistical analysis
Data were statistically analysed with Student’s t-test using independent one-sample t-tests for mutant/WT ratios and independent two-sample t-tests elsewhere.
Results
CSP41b is required for normal plant development and for maintenance of CSP41a levels
To characterize the functions and interrelationships of CSP41a and CSP41b, T-DNA insertion lines for the genes encoding the two proteins were characterized. For the CSP41a locus, the mutant alleles csp41a-1 and csp41a-2 have already been described; only the latter prevents expression of the CSP41a protein (Beligni and Mayfield, 2008). In the present study, the two novel mutant alleles csp41a-3 and csp41a-4 that carry T-DNA insertions in the 5'-untranslated region and second exon, respectively (Fig. 1A), were used. For CSP41b, two independent mutant alleles were used. The line SALK_021748 was obtained from the SIGnAL collection (Alonso et al., 2003), is identical to crb-1 (Hassidim et al., 2007), csp41b-1 (Beligni and Mayfield, 2008), and csp41b-2 (Bollenbach et al., 2009), and is referred to as csp41b-2 here. In addition, a further mutant allele, csp41b-3, with a T-DNA insertion in the eighth intron of CSP41b, was obtained from the GABI-KAT collection (Li et al., 2003) (Fig. 1A) and used.
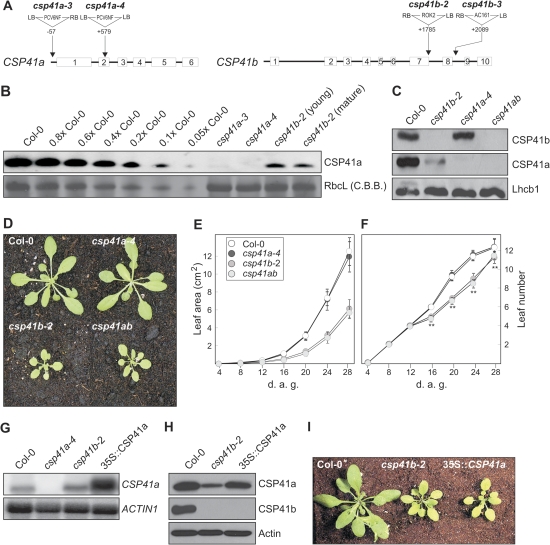
Fig. 1.
CSP41 insertion and overexpression lines: effects on CSP41 levels and plant growth. (A) The translated exons are numbered and shown as white boxes, with introns as black lines. Sites and orientations of T-DNA insertions are indicated. The csp41b-2 allele corresponds to the line SALK_021748 from the SALK T-DNA collection; csp41b-3 originates from the GABI-KAT collection and corresponds to GABI_452H11. The csp41a-3 and -4 alleles were identified in the T-DNA collection of C. Koncz (Max-Planck-Institute for Plant Breeding Research, Cologne, Germany) by PCR screening of lines originally designated as 91.154 and 61.888, respectively. (B) Western analysis of CSP41a in total protein extracts (40 μg) from young (7th–8th true) leaves of WT (Col-0), csp41a-3, csp41a-4, csp41b-2, and mature (3rd–4th true) leaves of csp41b-2. Decreasing amounts of WT extract were added to lanes marked 0.8× to 0.05× WT. The RbcL band visible after staining with Coomassie brilliant blue (C.B.B.) is shown as loading control. (C) Western analysis was performed on total protein extracts (40 μg) obtained from WT (Col-0) and csp41 mutant plants with CSP41a- and CSP41b-specific antibodies, and an antibody against Lhcb1 was used as a loading control. Note that csp41b-2 and csp41b-3 behaved identically (data not shown). (D) Phenotype of WT (Col-0) and csp41 mutant plants grown in the greenhouse. Here too, csp41b-2 and csp41b-3 behaved identically. (E) Growth kinetics of WT (Col-0) and csp41 mutant plants (n ≥10). Leaf area was measured during the period from 4 d to 28 d after germination (d.a.g.). Bars indicate standard deviations. Leaf area was always significantly (P < 0.01) lower for plants lacking CSP41b. (F) Development of WT (Col-0) and csp41 mutant plants (n ≥10). The number of true leaves was determined during the period from 4 to 28 d.a.g. Bars indicate standard deviations. Significant differences between WT/csp41a-4 and plants lacking CSP41b are indicated by two asterisks (P < 0.01). (G) Overexpression of CSP41a mRNA. Northern analysis of CSP41a mRNA from WT (Col-0), csp41a-4, csp41b-2, and 35S::CSP41a csp41b-2 (35S::CSP41a) plants. A CSP41a-specific probe and an ACTIN1-specific probe (as a control) were used. (H) Effects of CSP41a mRNA overexpression on accumulation of the CSP41a protein. Western analysis of WT (Col-0), csp41b-2, and 35S::CSP41a plants. Antibodies specific for CSP41a, CSP41b, and actin (as control) were used. (I) Phenotypes of WT (Col-0), csp41b-2, and 35S::CSP41a plants grown as in D.
The csp41a-4 mutant lacked the full-length CSP41a protein, whereas the csp41a-3 mutant accumulated ∼5% of WT CSP41a levels (Fig. 1B); therefore, csp41a-4 was used in all subsequent experiments. The csp41a-4 mutant still expressed the CSP41b protein in WT-like amounts and grew like the WT in the greenhouse (Fig. 1C–F), as described previously for the csp41a-2 mutant allele (Beligni and Mayfield, 2008). The existing literature shows variation in the phenotypes reported for the csp41b-2 mutant, possibly due to the different growth conditions employed. Hassidim et al. (2007) observed a pronounced phenotype when plants were grown on Murashige and Skoog medium supplemented with 1% sucrose in a 16 h light/8 h dark photoperiod of 125 μmol photons m−2 s−1. Beligni and Mayfield (2008) did not detect any visually discernible phenotype when plants were grown on 0.5% sucrose-supplemented media at a light intensity of 40 μmol photons m−2 s−1 and a 12 h light/12 h dark photoperiod. Bollenbach et al. (2009) described a developmental phenotype with paling mature leaves (≥15 d) when plants were grown in climate chambers at a light intensity of 100 μmol photons m−2 s−1 and a photoperiod of 16 h light/8 h dark. In the present experiments, greenhouse-grown csp41b-2 mutants (light intensity: ∼180 μmol photons m−2 s−1, 14 h light/10 h dark photoperiod) had in all developmental stages far smaller and paler leaves than the WT (Fig. 1D, E), and leaf formation was delayed from the second week after germination onwards (Fig. 1F). In the climate chamber at a light intensity of 80 μmol photons m−2 s−1 and a 12 h light/12 h dark photoperiod, csp41b-2 plants were only slightly paler, but not notably smaller than the WT (Supplementary Fig. S1A at JXB online). Increasing growth light intensities led to a stronger visually discernible phenotype of csp41b-2 mutant plants (Supplementary Fig. S1B at JXB online). When grown at 500 μmol photons m−2 s−1, but not at 80 μmol photons m−2 s−1, these mutants exhibited a significant reduction (P < 0.01) in total Chl content (Chl a+b), measured as nmol g−1 fresh weight (WT500, 2993±452; csp41b-2500, 1629±213; WT80, 2571±157; csp41b-280, 2509±84). Moreover, the size of the violaxanthin+antheraxanthin+zeaxanthin (VAZ) pool—an indicator for oxidative stress—was significantly increased (P < 0.01) in csp41b-2 plants grown at 500 μmol m−2 s−1 compared with the WT (WT500, 61.2±9.8; csp41b-2500, 139.4±15.2; WT80, 31.0±1.2; csp41b-280, 39.0±2). These data indicate that the intensity of incident light is critical for the effects of the csp41b mutation on plant performance. The previously reported developmental phenotype (Bollenbach et al., 2009) of csp41b plants was corroborated by analysing the photosynthetic performance, determined as ΦII, of whole WT and mutant plants. Thus, a decrease in ΦII was found with increasing leaf age of csp41b-2 mutants grown in the greenhouse or at 80 μmol m−2 s−1 in the climate chamber (Supplementary Fig. S1B at JXB online).
To obtain plants which entirely lacked both CSP41 proteins, the csp41a-4 csp41b-2 (csp41ab) double mutant was generated by crossing. Strikingly, csp41ab plants were viable and displayed a csp41b-like phenotype (Fig. 1D–F). This finding is at variance with the observation of Beligni and Mayfield (2008), who reported embryo lethality for the double mutant, but is compatible with the observation that already the csp41b mutant itself, which is viable, is severely depleted of CSP41a (Beligni and Mayfield, 2008; Bollenbach et al., 2009; Fig. 1B, C).
The marked drop in CSP41a protein levels in the absence of CSP41b could be caused either by a decrease in CSP41a mRNA accumulation owing to plastid-to-nucleus (retrograde) signalling or by a post-translational effect, for instance if the two proteins physically interact and CSP41b stabilizes CSP41a. In csp41b-2 plants, CSP41a RNA levels were increased relative to the WT (Fig. 1G), arguing against regulation of CSP41a abundance at the transcriptional level. Overexpression of CSP41a under the control of the CaMV 35S promoter in the csp41b-2 mutant background (35S::CSP41a plants) was found to boost the steady-state level of the CSP41a transcript several-fold (Fig. 1G). However, although CSP41a protein levels increased in 35S::CSP41a plants relative to those in csp41b-2 plants, they remained below WT concentrations (Fig. 1H). Moreover, the increase in levels of CSP41a in 35S::CSP41a plants did not attenuate the csp41b-2 mutant phenotype (Fig. 1I).
In consequence, CSP41b seems to be critical for the stability of CSP41a (see Beligni and Mayfield, 2008; Bollenbach et al., 2009), but not vice versa. Clearly, therefore, CSP41b, but not CSP41a, is required for normal plant growth and leaf coloration, and simultaneous absence of both proteins results in a csp41b-like phenotype—but not in lethality as reported earlier (Beligni and Mayfield, 2008).
Lack of CSP41b is associated with a fall in levels of thylakoid proteins and decreased plastid translational activity
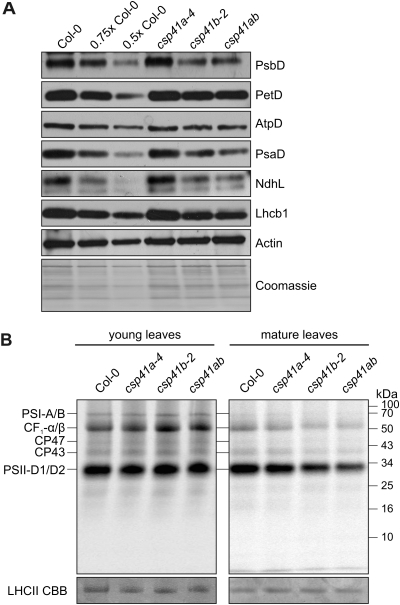
To obtain an overview of the alterations in thylakoid composition associated with loss of CSP41 proteins, total proteins from whole plants were fractionated by SDS–PAGE and analysed with antibodies raised against representative subunits of the major thylakoid multiprotein complexes (Fig. 2A). As expected from their essentially WT appearance (see Fig. 1D), no aberrations were detected in csp41a-4 mutant plants (Fig. 2A). In plants lacking CSP41b, levels of all thylakoid multiprotein complexes were markedly lower (Table 1). The greatest reduction (∼30%) was observed for PsaD of PSI, PsbD of PSII, and AtpD of the ATP synthase. NdhL of the NAD(P)H dehydrogenase (NDH) complex, Lhcb1 of light-harvesting complex II (LHCII), and PetD of the cytochrome (Cyt) b6/f complex showed reductions of ∼20%. Again, the csp41ab double mutant and csp41b-2 plants behaved similarly. A decrease in amounts of the chloroplast ATPase (cpATPase) and Cyt b6/f complexes has been noted previously in csp41b mutants grown in climate chambers (100 μmol photons m−2 s−1, 16/8 h light/dark cycles; Bollenbach et al., 2009). These observations indicate that, under the greenhouse conditions used here, absence of CSP41b, but not of CSP41a, also affects the accumulation of subunits of PSI, PSII, and NDH.
Fig. 2.
Composition and synthesis of thylakoid proteins in greenhouse-grown csp41 mutants and the WT. (A) Western analysis was performed on total protein extracts (40 μg) obtained from WT (Col-0) and mutant plants. Decreasing amounts of WT extract were added to lanes marked 0.75× Col-0 and 0.5× Col-0. Protein complexes were visualized and quantified using antibodies specific for representative subunits. Actin was used as a loading control. As a further control for loading, a replicate gel was stained with Coomassie brillant blue (C.B.B). One representative of three independently prepared immunoblots is shown. (B) Pulse labelling of thylakoid membrane proteins in young (7th–8th true) leaves and mature (3rd–4th true) leaves of WT and csp41 mutants. After inhibition of cytosolic translation with cycloheximide, plastid protein synthesis was monitored by pulse-labelling with [35S]methionine for 20 min; thylakoid membrane proteins were isolated from WT and csp41 mutant leaves, fractionated by SDS–PAGE, and analysed by autoradiography. Prior to drying, the gel was stained with C.B.B. and the stained LHCII bands are indicated. The experiment was performed three times with similar results.
Table 1.
Thylakoid protein levels
| csp41a–4 | csp41b–2 | csp41ab | |
| PsbD | 1.10 | 0.68 | 0.67 |
| Lhcb1 | 1.03 | 0.79 | 0.78 |
| PsaD | 0.98 | 0.71 | 0.69 |
| AtpD | 1.00 | 0.70 | 0.67 |
| PetD | 1.02 | 0.81 | 0.79 |
| NdhL | 1.04 | 0.78 | 0.76 |
Values for proteins are average signals derived from three independent western analyses (PsbD, Lhcb1, PsaD, AtpD, PetD, and NdhL; see also Fig. 2A), expressed relative to WT levels (=1). Standard deviations were <5% and values were significantly different from the WT with P < 0.05 for csp41b-2 and csp41ab.
To clarify whether the decrease in thylakoid protein levels is due to changes in the rate of translation in the plastid, leaves from plants (as in Fig. 1D) were treated with an inhibitor of cytosolic translation and fed with radioactively labelled methionine. Because the phenotype of plants lacking CSP41b is development dependent (Bollenbach et al., 2009; this study (Supplementary Fig. S1B at JXB online)], mature (3rd–4th true) and young (7th–8th true) leaves of 4-week-old plants were analysed. After pulse labelling, thylakoid proteins were prepared and fractionated by SDS–PAGE (Fig. 2B). In csp41b-2 and csp41ab plants, the plastid translation rate was markedly decreased in mature but not in younger leaves, as shown by the reduction in the amounts of D1, CP47, PsaA/B, and CF1-α/β synthesized de novo (total label incorporated relative to the WT (=1.0), csp41a-4, 1.1±0.2; csp41b-2, 0.7±0.1; csp41ab, 0.7±0.1; results for both csp41b-2 and csp41ab were significantly different from the WT with P < 0.05).
Taken together, these results indicate that in mature leaves, lack of CSP41b (in csp41b-2 and csp41ab) leads to a decrease in the chloroplast translation rate, resulting in reduced levels of thylakoid proteins (see also Bollenbach et al., 2009) and a drop in photosynthetic activity (Supplementary Fig. S1B at JXB online).
CSP41 proteins form several stromal multiprotein complexes
Recently, a cytosolic location of CSP41 was reported by Fettke et al. (2011), a finding which is at variance with the study of Raab et al. (2006), in which CSP41b was unambiguously identified in chloroplasts, and also in contradiction to the general consensus that CSP41 proteins are involved in chloroplast gene expression (Yang et al., 1995; Bollenbach et al., 2003, 2009; Beligni and Mayfield, 2008; this study). In order to test whether a substantial proportion of CSP41b is indeed present outside of the chloroplast, the amount of CSP41 proteins detected by immunoblot analysis in protein preparations from entire leaves and isolated chloroplasts, which were normalized according to their Chl content, was compared. Like the large subunit of RubisCO (RbcL), both CSP41a and CSP41b showed equal signal strengths in the two samples (Fig. 3A), indicating that the vast majority of CSP41 proteins should be located in chloroplasts. In proteomic analyses of chloroplast subfractions, both CSP41 proteins were detected in the stroma, as well as in thylakoid and envelope subfractions (PPDB database: http://ppdb.tc.cornell.edu; Sun et al., 2009). To clarify the suborganellar localization of the CSP41 proteins, chloroplasts were fractionated into thylakoids, stroma, and envelopes. Specific proteins were detected with antibodies raised against marker proteins for the individual fractions, as well as against CSP41a and CSP41b (Fig. 3B). Most CSP41a was detected in the stroma, although—like for RbcL—traces were also present in the two envelope fractions. CSP41b was exclusively found in the stroma (Fig. 3B).
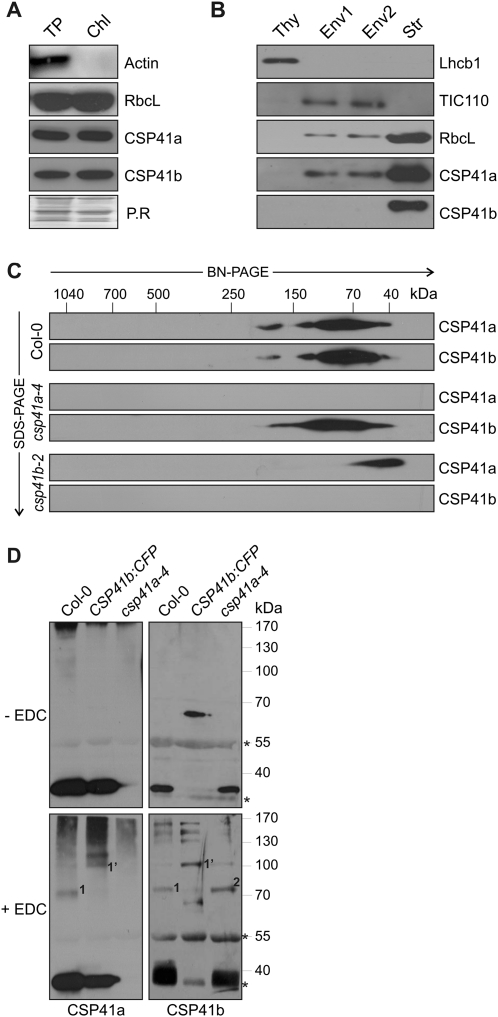
Fig. 3.
Characterization of complexes containing CSP41 proteins. (A) Total protein (TP) and chloroplast protein (Chl) extracts corresponding to 3 μg of Chl were subjected to SDS–PAGE, followed by immunoblot analysis using antibodies raised against CSP41 proteins. As controls, RbcL for chloroplast and actin for non-chloroplast proteins were used. As control, Ponceau Red stain (P.R.) of the membrane prior to antibody immunodecoration is shown. (B) Chloroplasts from WT plants were subfractionated into thylakoids (Thy), two envelope fractions (Env1 and Env2), and stroma (Str). Aliquots (40 μg) of proteins from each fraction were subjected to SDS–PAGE, followed by immunoblot analysis using antibodies raised against CSP41 proteins. As controls for the purity of the different fractions, antibodies recognizing Lhcb1, TIC110, and RbcL, which are located in thylakoids, envelope, and stroma, respectively, were used. (C) Stromal proteins from chloroplast samples corresponding to 30 μg of Chl were fractionated by BN-PAGE in the first dimension and by SDS–PAGE in the second. CSP41a and CSP41b were each detected using specific antibodies. The approximate molecular masses of the labelled protein complexes were estimated from the mobilities of other complexes with known molecular masses (Peltier et al., 2006). (D) Stromal proteins (70 μg) from WT (Col-0), csp41a-4 mutants, and a csp41b-2 line expressing CSP41b:CFP were cross-linked with EDC and fractionated by SDS–PAGE. Protein detection was carried out with specific antibodies as in A–C. Asterisks indicate unspecific bands detected by the CSP41b antibody.
The fact that CSP41a depends on CSP41b for its own stability (Beligni and Mayfield, 2008; Bollenbach et al., 2009; this study), together with the Co-IP of CSP41a with a FLAG-tagged CSP41b variant (Bollenbach et al., 2009), suggests that CSP41a and CSP41b physically interact. To investigate further the nature of the CSP41-containing complexes, stromal protein extracts equivalent to 60 μg of Chl were fractionated by 2D BN/SDS–PAGE, blotted onto nylon membrane, and treated with antibodies specific for CSP41a or CSP41b. In WT plants, CSP41a and CSP41b were detected at several positions in the BN gel, but always together; the signals corresponded to molecular species with masses of ∼40, ∼80, ∼120, and ∼200 kDa (Fig. 3C), with the ∼40 kDa spot representing the monomeric forms (CSP41a, 36 kDa; CSP41b, 37 kDa). In the absence of CSP41a (in the csp41a-4 mutant), CSP41b still formed complexes resembling those in WT plants, but CSP41a migrated as a monomer in csp41b-2 mutant plants (Fig. 3C).
In order to analyse the composition of CSP41-containing complexes further, stromal proteins were cross-linked with EDC. Cross-linking experiments were performed on stromal extracts from WT, csp41a-4 mutant, and csp41b-2 mutant plants expressing a CSP41b:CFP construct under control of the 35S promoter (35S::CSP41b:CFP). The latter line contains CSP41a and the 64 kDa CSP41b:CFP construct, but no CSP41b. Because the CSP41b:CFP fusion can complement the csp41b-2 mutant phenotype (see Supplementary Fig. S2 at JXB online), the tagged protein is functional. Cross-linking of WT proteins, followed by SDS–PAGE and immunoblotting, resulted in detection of a product of ∼75 kDa by both CSP41a- and CSP41b-specific antibodies (marked ‘1’ in Fig. 3D). Because the 75 kDa band was replaced by one of ∼100 kDa (marked ‘1’) in plants expressing the larger CSP41b:CFP construct, these two species presumably result from cross-linking of the constituents of the heterodimers CSP41a/CSP41b and CSP41a/CSP41b:CFP, respectively. In the csp41a-4 line, a cross-linking product of ∼70 kDa containing CSP41b was detectable (marked ‘2’). This product must result either from cross-linking of two CSP41b monomers, or from interaction of CSP41b with another protein of similar size.
Taken together, it can be concluded that (i) CSP41 proteins are predominantly localized in the stroma; (ii) higher order complexes containing both CSP41 proteins exist; (iii) formation of these complexes requires CSP41b; (iv) in the absence of CSP41b (in csp41b-2 plants), CSP41a appears in monomeric form; and (v) CSP41b can form complexes in the absence of CSP41a also with itself or with another protein of similar size.
Stoichiometry and post-translational modifications of CSP41 proteins
For CSP41 proteins several post-translational modifications, including phosphorylation (CSP41a; Reiland et al., 2009, 2011; CSP41b, Reiland et al., 2011) and lysine acetylation (CSP41a; Finkemeier et al., 2011), have been reported. Moreover, both CSP41 proteins contain redox active cysteines (Ströher and Dietz 2008); in particular, CSP41a has been identified as a putative thioredoxin target (Marchand et al., 2006). Because both phosphorylation and acetylation of amino acid residues add negative charges to the molecule and therefore shift the pI of the modified proteins towards decreased pH, 2D IEF/SDS–PAGE analyses were perfomed to characterize further the extent of post-translational modifications of CSP41a and CSP41b. Unmodified CSP41a and CSP41b have predicted pIs of 6.04 and 6.36 respectively, whereas unmodified RbcL (which in vivo also becomes phosphorylated and acetylated; Lohrig et al., 2009; Finkemeier et al., 2011) with a predicted pI of 5.88 was employed as control. As expected, CSP41a protein species migrated at a more basic pH relative to the main RbcL species and, in turn, CSP41b proteins at a more basic pH than CSP41a (Fig. 4A, B). Comparison of Coomassie-stained 2D IEF/SDS–polyacrylamide gels from WT and csp41b-2 mutant plants allowed identification of spots tentatively representing CSP41a and CSP41b, which were subsequently unambiguously assigned to CSP41a and CSP41b by immunoblot analysis employing antibodies specific for the two proteins (Fig. 4B). In total, six different protein species could be observed for CSP41a and five for CSP41b, indicating that in vivo both CSP41 proteins occur with post-translational modifications.
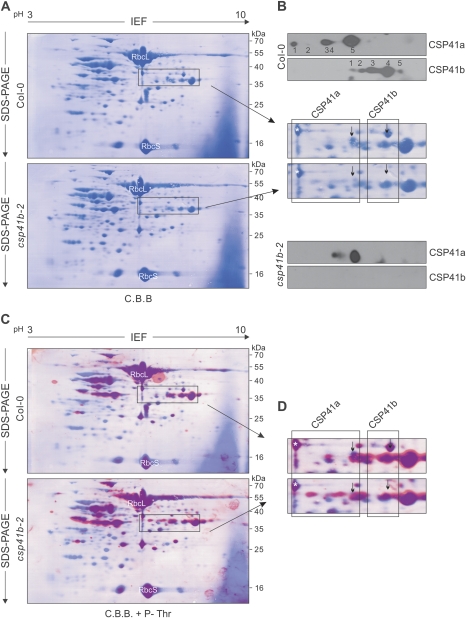
Fig. 4.
CSP41 proteins are post-translationally modified. (A) Stromal proteins (500 μg) from WT (Col-0) and csp41b-3 plants were fractionated by IEF in the first dimension and by SDS–PAGE in the second. Proteins were transferred to PVDF membranes and stained with Coomassie brilliant blue (C.B.B.). (B) Sections of A containing CSP41 proteins stained with C.B.B. or immunodecorated with specific antibodies raised against CSP41a or CSP41b are shown. Arrows indicate the main CSP41a and CSP41b protein species. (C) The membranes from A were immunodecorated with antibodies against phosphorylated threonine residues; the detected signals were converted to red and merged with the image shown in A. Phosphorylated proteins are shown in purple, and highly phosphorylated proteins in red. (D) Sections of C containing CSP41 proteins. The asterisk marks glyceraldehyde-3-phosphate dehydrogenase B (Goulas et al., 2006). This protein, together with RbcL and RbcS, has been previously found to be phosphorylated (Lohrig et al., 2009; Reiland et al., 2009). Arrows indicate the main CSP41a and CSP41b protein species. (A–D) At least two independent experiments were performed with similar results.
To corroborate that CSP41a and CSP41b undergo post-translational modification by phosphorylation, protein blot analyses of IEF/SDS–polyacrylamide gels were carried out with phosphothreonine-specific antibodies. The images derived from Coomassie staining (Fig. 4A, blue) and from the immunodecoration of phosphorylated threonines (red) were merged, such that phosphorylated proteins are indicated by purple spots and highly phosphorylated proteins by red ones (Fig. 4C). The pattern of stromal phosphoproteins is in accordance with previous studies (Goulas et al., 2006; Reiland et al., 2009, 2011; Lohrig et al., 2009) and, indeed, for both CSP41a and CSP41b, phosphorylated variants were detected (Fig. 4D). Moreover, it was found that lack of CSP41b changes the phosphorylation state of several other stromal proteins.
In addition, the results of the 2D PAGE analyses allowed estimation of the relative abundance of CSP41a and CSP41b. To this end, the main spot intensities of CSP41a and CSP41b relative to the complete signals were determined by protein blot analysis (Fig. 4B), quantified from the Coomassie gel (Fig. 4B, see arrow), and their relative ratio was calculated. The results indicate that CSP41b is ∼2.6-fold more abundant than CSP41a.
HMW CSP41 complexes are associated with RNAs in the dark
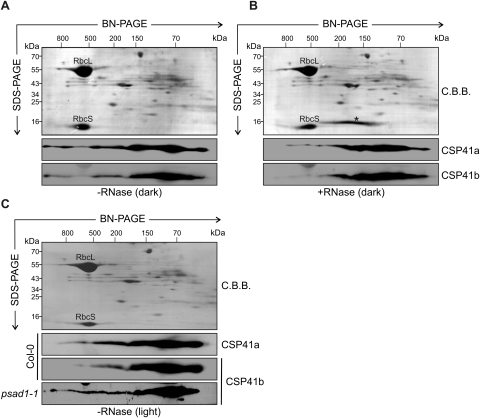
Because CSP41 proteins have been found to bind and cleave RNA in vitro (Yang et al., 1995, 1996; Yang and Stern, 1997; Bollenbach and Stern, 2003a, b), investigations were carried out to determine whether stromal CSP41 complexes are associated with chloroplast RNAs. To this end, stromal extracts of WT plants (from chloroplasts containing 160 μg of Chl) adapted to the dark (to avoid formation of starch granules which might disrupt chloroplasts during their isolation) were treated with RNase A or left untreated, separated by 2D BN/SDS–PAGE, and subjected to immunoblot analysis with CSP41a- and CSP41b-specific antibodies. In untreated samples, multiple HMW complexes could be detected, which largely disappeared in RNase-treated samples (Fig. 5A, B). This implies that the formation of HMW CSP41 complexes depends on the interaction with RNA.
Fig. 5.
CSP41-containing complexes interact with RNA. (A) Stromal proteins from dark-adapted plants corresponding to 160 μg of Chl were fractionated by BN-PAGE in the first dimension and by SDS–PAGE in the second. CSP41a and CSP41b were each detected using specific antibodies. As control, the Coomassie brilliant blue (C.B.B.) staining of the membrane prior to immunodecoration is shown. (B) Stromal proteins as in A, but treated with RNase. The asterisk indicates RNase A. (C) Stromal proteins from light-adapted WT and psad1-1 plants detected as in A. (A–C) The approximate molecular masses of the labelled protein complexes were estimated from the mobility of other complexes with known molecular masses (Peltier et al., 2006).
In order to determine whether the association of CSP41 proteins in HMW complexes is regulated by light, the formation of HMW CSP41 complexes in samples taken from light-adapted plants was investigated (Fig. 5C). In fact, in light-adapted plants, the accumulation of HMW CSP41 complexes was similar to that of dark-adapted plants treated with RNase A. This suggests that the formation of CSP41 riboprotein complexes is negatively regulated by light. Because the stromal redox state is shifted in the light to reducing conditions, it is tempting to speculate that the formation of CSP41–RNA complexes is redox regulated. This hypothesis was experimentally addressed by analysing the formation of CSP41 complexes in light-adapted psad1-1 mutants (Fig. 5C). This mutant displays a strong decrease in linear photosynthetic electron flow and consequently possesses a more oxidized stroma (Ihnatowicz et al., 2004; DalCorso et al., 2008). Because in light-adapted psad1-1 plants an increase in the formation of HMW complexes was noted, it can be concluded that CSP41 riboprotein complex assembly is indeed regulated by the redox state of the chloroplast, with more reducing stromal conditions (light versus dark, WT versus psad1-1), resulting in the dissociation of HMW CSP41–RNA complexes. A tentative cause for this behaviour could be the redox-dependent post-translational modification of CSP41 proteins, and, in fact, the pIs of CSP41b species were found to be shifted towards higher pH values in the light (see Supplementary Fig. S3 at JXB online).
Target transcripts of CSP41 complexes encode photosynthetic core proteins, and 16S and 23S rRNAs
CSP41a has previously been identified as a protein bound to the 3' stem–loop of the petD mRNA (Yang et al., 1995), and putative protein interactions have mostly been proposed because of co-migration behaviour in sucrose gradients and colourless native PAGE (Peltier et al., 2006; Beligni and Mayfield, 2008). In order to determine systematically the composition of CSP41–RNA complexes, Co-IP experiments in combination with MS analysis, as well as RIP-chip, were carried out. Because the CSP41a- and CSP41b-specific antibodies proved to be unsuitable for Co-IP experiments, the analyses were performed on stromal extracts from the lines that express CSP41b:CFP in the csp41b-2 mutant background using a GFP antibody (see Fig. 3D; see Supplementary Fig. S2 at JXB online). As control, Co-IP was also performed on stroma from plants expressing eGFP that is targeted to the chloroplast by the N-terminal chloroplast transit peptide from ferredoxin-NADP(+) oxidoreductase (FNR) which is cleaved off after chloroplast import (cp-eGFP; Marques et al., 2004). After analysing the immunoprecipitations by western analysis employing a GFP antibody, which also recognizes CFP and cp-eGFP (see Supplementary Fig. S4A at JXB online), the precipitate was subjected to MS. This experiment was performed in duplicate and only proteins detected in both experiments in the CSP41b:CFP sample but not in the cp-eGFP control were considered as tentative components of the CSP41 complex (Table 2; Supplementary Table S2). As expected, CSP41b and CSP41a were identified and exhibited the highest total sequence coverage by detected tryptic peptides. In addition, LTA2, the E2 subunit of the plastid pyruvate carboxylase, which was shown to co-migrate with CSP41b in HMW complexes (Olinares et al., 2010), was also detected (Table 2; Supplementary Table S2). These results suggest that CSP41b is not stably associated with PEP or ribosomes, as previously suggested (Pfannschmidt et al., 2000; Yamaguchi et al., 2003), but might interact to some extent with LTA2, which co-migrates with CSP41b in HMW complexes (Olinares et al., 2010).
Table 2.
MS analysis of co-immunoprecipitates of CSP41b:CFP Data are given for two independent experiments. Only proteins for which tryptic peptides were found in CSP41b:CFP, but not in cp-eGFP Co-IP analyses are considered. Peptides were assigned to MS/MS spectra by the SEQUEST searching algorithm. Identified peptides are listed in Supplementary Table S2 at JXB online.
| Gene | Protein | No. of matched peptides |
| At1g09340 | CSP41b | 29/86 |
| At3g63140 | CSP41a | 5/20 |
| At3g25860 | LTA2 | 2/3 |
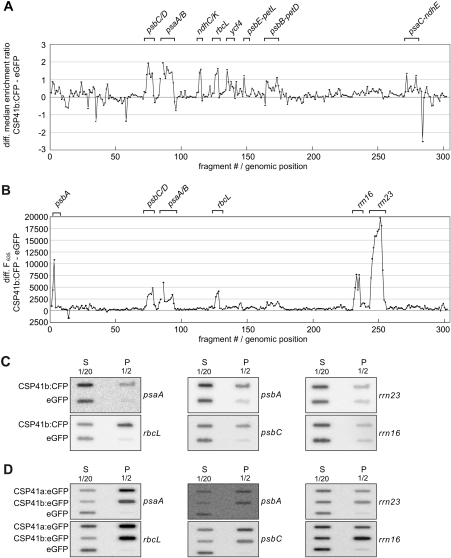
For RIP-chip assay (Schmitz-Linneweber et al., 2005), RNAs from pellet and supernatant fractions of the immunoprecipitate were purified and labelled with red (Cy5) and green (Cy3) fluorescing dyes, respectively. Both RNA fractions were then competitively hybridized to an A. thaliana chloroplast genome tiling microarray. The ratio of red (F635) to green (F532) fluorescence (F635/F532 or enrichment ratio) for each spot reflects the degree to which the corresponding RNA sequence is enriched in the pelleted immunoprecipitate. CSP41b:CFP-associated RNAs were identified as RNAs that were enriched to a higher degree in immunoprecipitates from CSP41b:CFP stroma than in immunoprecipitates from cp-eGFP stroma. Data from two replicate experiments were normalized and used to calculate median enrichment ratios for each DNA fragment among the replicate spots (six per array) (see Supplementary Table S3 at JXB online). To visualize sequences that were preferentially enriched from the CSP41b:CFP extract, the difference between CSP41b:CFP and cp-eGFP samples with respect to the median normalized values for each DNA fragment was plotted as a function of chromosomal position (Fig. 6A; Supplementary Table S3). Only those peaks were considered to represent true RNA targets that (i) showed >2-fold enrichment; (ii) hybridized with more than one genomic fragment on the array; and (iii) for which a t-test indicated significant enrichment (P < 0.01). Based on these criteria, enrichment of RNAs for the genes psbD/C, psaA/B, and rbcL was observed. For psbD/C, five contiguous genomic fragments were co-enriched, eight for psaA/B and three for rbcL. If only two of the three criteria were employed, additional putative targets could be identified— transcripts of the genes ycf4, psbE-petL, psbB-petD, ndhD, ndhG/I, and ndhK/C. Strikingly, enrichment was observed only for mRNAs which code for proteins involved in photosynthesis, but not for tRNAs.
Fig. 6.
Identification of RNAs associated with CSP41b by RIP-chip analysis. (A) Differential enrichment ratios. The enrichment ratios (F635/F532) obtained from an assay involving CSP41b:CFP stroma were normalized with respect to a control assay using cp-eGFP stroma (both assays were performed in duplicate). The median normalized values for replicate spots from the cp-eGFP data were subtracted from the CSP41b data, log2 transformed, and plotted according to fragment number. Fragments are numbered according to their chromosomal positions. The data used to generate this figure are provided in Supplementary Table S3 at JXB online. Prominent peaks are labelled with gene names. (B) Differential pellet signals. The data from the experiments described in the upper panel were used to subtract normalized mean pellet signals (F635) obtained in cp-eGFP assays from those calculated for CSP41b:CFP assays. The corresponding data can also be found in Supplementary Table S4. (C) Slot-blot hybridization of RNAs that co-immunoprecipitate with CSP41b:CFP. One-tenth of the RNA recovered from each supernatant (sn) or half of the RNA recovered from each immunoprecipitation pellet (p) was applied to slot blots and hybridized with probes specific for rrn23, rrn16, psbA, rbcL, psaA, or psbC. (D) Slot-blot hybridization of RNAs that co-immunoprecipitate with CSP41a:eGFP and CSP41b:eGFP. One-tenth of the RNA recovered from each supernatant (sn) or half of the RNA recovered from each immunoprecipitation pellet (p) was applied to slot blots and hybridized with probes specific for rrn23, rrn16, psbA, rbcL, psaA, or psbC.
Previous experiments had suggested that CSP41b might interact with 23S rRNA precursors (Beligni and Mayfield, 2008). Because of their high abundance, rRNAs give by far the strongest signal in the supernatant in RIP-chip experiments. rRNA precursors are far less abundant, and if CSP41b were to interact with such precursors, the minimal amounts of this subpopulation of rRNA precipitated might not produce a sufficiently large shift in enrichment ratios between experiments and controls to be scored as significant. To exclude the dominant impact of excessive amounts of mature rRNA in the supernatant, only the pellet signals (F635) between CSP41b:CFP and cp-eGFP experiments were compared. This comparison resulted in the identification of psbD/C, psaA/B, and rbcL, which had already been recognized based on the ratio comparisons described above (see Fig. 6A). In addition, very prominent peaks for rrn16, rrn23, and psbA, which are known to correspond to abundant chloroplast RNAs, were found (Fig. 6B; Supplementary Table S4 at JXB online). Because none of the highly abundant tRNAs gave a signal in this type of analysis, the appearance of peaks for rrn16, rrn23, and psbA is unlikely to be attributable to higher overall background contamination by abundant RNAs. Moreover, the relative abundances of the different RNAs in the supernatants of CSP41b:CFP and cp-eGFP immunoprecipitations were very similar (see Supplementary Fig. S5 at JXB online). No bias suggesting a general excess of RNAs or an excess of specific RNAs in CSP41b:CFP experiments was observed. This implies that one-channel comparisons in RIP-chip experiments can uncover RNA ligands that represent just a minor fraction of all RNAs corresponding to a specific gene (Prikryl et al., 2008). One drawback of the differential pellet analysis may lie in the difficulty in visualizing differences for transcripts that are of low abundance.
To confirm the chip hybridization data, slot-blot analyses with gene-specific probes were carried out employing RNA from the supernatant and pellet of immunoprecipitates from CSP41b:CFP and cp-eGFP stroma (see Supplementary Fig. S4 at JXB online), demonstrating enrichment of psaA, rbcL, psbA, and psbC in the pellet fraction of CSP41b:CFP immunoprecipitates (Fig. 6C). In order to verify that the RNA is bound by both CSP41a and CSP41b or by a complex containing both proteins (see Figs 3C, D, 4A; Table 2; Supplementary Table S2), RNA Co-IP was performed on stroma from plants expressing eGFP-tagged versions of CSP41a and CSP41b, and the immunoprecipitation of the proteins was confirmed by protein gel analysis (Supplementary Fig. S4B). In fact, all tested RNAs were highly enriched in the pellet of both CSP41a:eGFP and CSP41b:eGFP immunoprecipitates with respect to immunoprecipitates from the line expressing eGFP alone (Fig. 6D).
In summary, the RIP-chip analysis points to several RNAs as putative ligands of CSP41 complexes. These include mRNAs for the large subunit of Rubisco (RbcL), core proteins of PSI (PsaA and PsaB) and PSII (D1, D2, CP43, C47, Cyt b559, and PsbH), and subunits of the NDH (NdhK, NdhC, NdhD, NdhI, NdhG) and Cyt b6/f complexes (Cyt b6, SubIV, and PetL), as well as 16S and 23S rRNAs.
Lack of CSP41b leads to a decrease in levels of 16S and 23S rRNAs and of target mRNAs
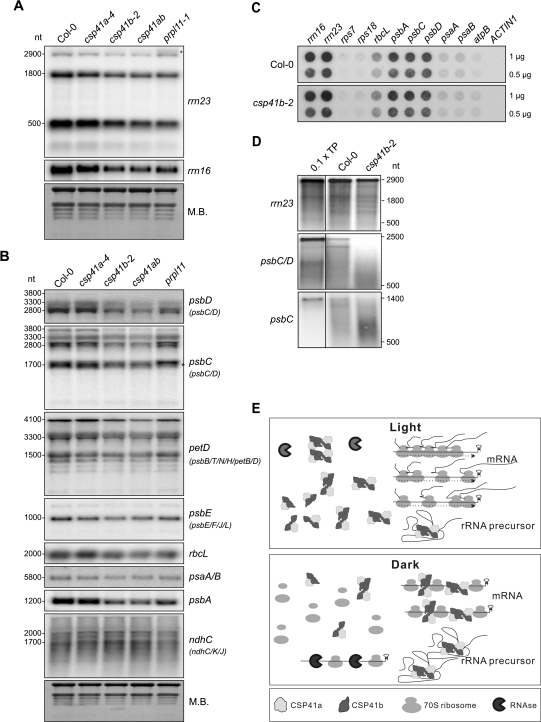
The decrease in translational activity in mature leaves of the csp41b-2 mutant, together with the interaction of CSP41b with 16S and 23S rRNAs, suggests that the influence of CSP41b on translation is mediated by its interaction with these rRNAs. In this context, Beligni and Mayfield (2008) have previously shown that lack of CSP41 proteins alters 23S rRNA accumulation: in particular, there is an increase in relative levels of unprocessed 23S rRNA in csp41a and csp41b mutants. To reinvestigate the accumulation of 16S and 23S rRNAs in the absence of CSP41 proteins, RNA was isolated from leaves of whole Col-0 and csp41 mutant plants. As a control, total leaf RNA was also isolated from prpl11-1, a mutant that is specifically affected in plastid translation due to lack of the L11 protein of the plastid 50S ribosomal subunit (Pesaresi et al., 2001). Northern analysis with a probe recognizing the 5' end (500 nucleotides) of 23S rRNA indicated that in plants lacking CSP41b, levels of all 23S rRNA species (the precursor and ribosome-incorporated forms, processed at hidden breaks) and 16S rRNA were reduced (Fig. 7A, Table 3). In prpl11-1, the unprocessed 23S rRNA species accumulated to WT levels. Therefore, it appears likely that the decrease in unprocessed 23S rRNA in csp41b-2 and csp41ab mutants is directly associated with the absence of CSP41b (and therefore CSP41 complexes) and is not the consequence of a general drop in translational activity.
Fig. 7.
Lack of CSP41b affects the accumulation of chloroplast rRNAs and specific mRNAs. (A) Samples (20 μg) of total RNA from light-adapted WT (Col-0), csp41, and prpl11-1 mutant plants grown in the greenhouse (14 h/10 h light/dark regime at ∼180 μmol photons m−2 s−1) were size-fractionated by agarose gel electrophoresis, transferred to nitrocellulose filters, and probed with cDNA fragments specific for the 5′ end of rrn23 and rrn16. Methylene blue (M.B.) staining of total leaf RNA served as loading control. The asterisk marks unprocessed rrn23. Quantification relative to WT can be found in Table 3. (B) Northern analyses of samples as in A with cDNA fragments specific for the coding regions of psbC, psbD, petD, psbE, rbcL, psaA/B, psbA, and ndhC. M.B. staining of total leaf RNA served as a loading control. (C) Run-on transcription in mature leaf chloroplasts isolated from WT and csp41b-2 plants. Duplicate nylon membranes were spotted with 1 μg or 0.5 μg of PCR products corresponding to the genes indicated at the top. Isolated chloroplasts were incubated in the presence of radiolabelled UTP. RNA from chloroplasts that had been pulse-labelled for 10 min was used to probe the membranes shown. The nuclear ACTIN1 gene was included as negative control. Quantification relative to rps18 can be found in Table 4. (D) Radiolabelled transcription products (TPs) from rrn23, psbC/D, and psbC were incubated with broken chloroplasts from WT and csp41b-2 mutants followed by re-isolation of RNA, separation on a denaturing 1.5% agarose gel, and transfer to nylon membranes. One representative of three independent experiments is shown. Note that in lane TP only one-tenth of the experimentally employed amount of transcription product was loaded. (A–D) Signals were analysed by autoradiography. (E) Working model for action of CSP41 protein complexes: in the light, newly synthesized rRNA precursors are rapidly incorporated into functional ribosomes, which in turn stabilize mRNAs during the translational process (arrows on ribosomes indicate translationally active ribosomes); CSP41 protein complexes do not bind to RNA. In the dark, CSP41 protein complexes associate with RNAs and protect them from nucleolytic cleavage. Untranslated RNA not stabilized by CSP41 protein complexes (i.e. in the csp41b and csp41ab double mutant) are degraded by RNases.
Table 3.
Differential accumulation of 23S and 16S rRNA species in greenhouse-grown csp41 mutants relative to WT (=1) Signals from two (16S rRNA) or three (23S rRNA) independent northern analyses were quantified and normalized with respect to the 25S rRNA methylene blue signal. Mean values ±SD are shown. Asterisks indicate that results were significantly different from the WT with Student’s t-test P-value <0.03 (*) and <0.02 (**).
| 23S rRNA |
16S rRNA | |||
| 2900 nt | 1800 nt | 500 nt | ||
| csp41a-4 | 0.92±0.07 | 0.98±0.20 | 1.15±0.12 | 0.90±0.02* |
| csp41b-2 | 0.60±0.19 | 0.53±0.13* | 0.59±0.09** | 0.45±0.02** |
| csp41ab | 0.57±0.18 | 0.50±0.14* | 0.56±0.10** | 0.44±0.01** |
| prpl11-1 | 1.38±0.19 | 0.74±0.13 | 0.56±0.08** | 0.53±0.03* |
The decrease in all 23S rRNA species observed here in all genotypes lacking CSP41b is at variance with the findings of Beligni and Mayfield (2008), and might be attributable to the different growth conditions employed in the two studies (Murashige and Skoog medium and a 12 h/12 h light/dark cycle of 40 μmol photons m−2 s−1 in the earlier work; 14 h/10 h light/dark cycle in the greenhouse, natural light supplemented with ∼180 μmol photons m−2 s−1 in the present study). Alternatively, a different developmental state of plants [4-week-old plants in the present study; unknown age in Beligni and Mayfield (2008)] might have caused the different results.
Northern analyses were also performed with probes for the putative target mRNAs psbD, psbC, petD, psbE, rbcL, psaA, psbA, and ndhC. In csp41b-2 and csp41ab plants, steady-state levels of the majority of transcripts analysed decreased relative to those in csp41a-4 plants, which behaved similarly to the WT (Fig. 7B). Only the level of the polycistronic ndhC/K/J transcripts was slightly increased. An intermediate behaviour was observed for prpl11-1 leaves, in which some transcripts were decreased in their abundance compared with the WT and csp41a-4 plants. In order to study the effects of loss of CSP41 proteins on the levels of all chloroplast transcripts, the entire plastid transcriptomes of csp41b-2 and csp41a-4 plants grown in the climate chamber were analysed by qRT-PCR (Supplementary Table S5 at JXB online). These analyses established that deviations from WT were slight, and more prominent in csp41b-2 than in csp41a-4 mutants. A comparison with other mutants defective in chloroplast gene expression (Okuda et al., 2007; Chateigner-Boutin et al., 2008; de Longevialle et al., 2008; Delannoy et al., 2009; Pfalz et al., 2009; Tillich et al., 2009) revealed that the profiles for csp41a-4 and csp41b-2 plants were related to those of crr (Okuda et al., 2007) and cp31 (Tillich et al., 2009) mutants (see Supplementary Fig. S6 at JXB online). However, RNA analysed by (qRT)-PCR was isolated from plants grown in the climate chamber at 80 μmol photons m−2 s−1 (see Supplementary Fig. S1A) and therefore the results cannot be directly compared with the RNA blot analysis.
At the level of the transcript pattern recognized by individual probes, it was interesting to note that the psbC transcript pattern varied in WT (Col-0), CSP41-less plants and prpl11-1. In the WT, two psbC-specific transcripts of ∼1700 nucleotides (marked by an asterisk) were present, which were not detected by the specific psbD probe used (Fig.7B). The lower transcript was more prominent in the WT and hardly detectable in prpl11-1 with increased intensity of the higher transcript, while both transcript forms had approximately the same intensity in csp41b-2 and csp41ab mutants. Because the psbC gene contains a promoter specific for PEP (Ishizaki et al., 2005), the larger psbC-specific transcript might be due to preferential transcription from an alternative and more distal nuclear-encoded RNA polymerase (NEP) promoter as the result of a drop in amounts of PEP owing to the translational defect in prpl11-1 (Pesaresi et al., 2001) and csp41b (Fig. 2B) mutants. In fact, upstream of the psbC PEP promoter several NEP promoter consensus sequences (YRTA) (Liere et al., 2004) can be found. Such a PEP-to-NEP promoter switch has already been described for the atpBE transcript in the absence of Sig6 (Schweer et al., 2006) and in the csp41b mutant (Bollenbach et al., 2009).
Taken together, the data suggest that CSP41b controls the accumulation of target rRNAs and mRNAs. Because these effects are less pronounced in the prpl11-1 mutant with a specific defect in plastid translation, CSP41b should have a characteristic function in transcript accumulation. Moreover, absence of CSP41a from CSP41 complexes also does not have any major effect at the level of putative RNA ligands.
RNA stability is decreased in the absence of CSP41b
To analyse whether the decrease in transcript accumulation observed above was due to changes in the transcription rate, a run-on transcription assay was performed with broken chloroplasts from WT and csp41b-2 plants (Fig. 7C). The newly synthesized labelled transcripts were hybridized to probes against the putative CSP41 complex targets rrn16, rrn23, rbcL, psbA, psaA, psaB, psbC, and psbD, and against probes representing the non-target transcripts rps7, rps18, and atpB as controls. When the signal intensities of the de novo synthesized transcripts were compared with those of the NEP promoter transcribed non-target rps7 and rps18 transcripts (Hajdukiewicz et al., 1997), it appeared that the transcription rates of all analysed putative RNA targets were lower in plants lacking CSP41b (Table 4). For the RNAs that are primarily transcribed from a PEP promoter (rbcL, psbA, psbC/D, and psaA/B; Allison et al., 1996; Hajdukiewicz et al., 1997) this could be due to lower levels of the PEP as a consequence of the decrease in plastid translation (see Fig. 2B). Indeed, the transcription rate of atpB, for which Bollenbach et al. (2009) have shown a PEP-to-NEP promoter switch in csp41b plants, was not changed in csp41b-2 mutants. The two putative rRNA targets, rrn16 and rrn23, which can be transcribed from both PEP and NEP promoters, also displayed a decrease in de novo syntheses, with 0.69 and 0.79 of WT, respectively (Table 4). This could indicate that for these two transcripts a PEP-to-NEP promoter switch does not occur as efficiently as for atpBE. The fact that the steady-state accumulation of putative RNA targets was lower than the decrease in transcriptional rate indicated that the direct role of CSP41 complexes might be the stabilization of target RNAs.
Table 4.
Transcription rates compared with rps18 signals
| rrn16 | rrn23* | rps7 | rps18 | rbcL** | psbA | psbC** | psbD* | psaA | psaB | atpB | |
| Col-0 | 17.3±4.7 | 24.0±2.1 | 0.7±0.2 | 1.0±0.0 | 12.0±1.5 | 23.7±7.0 | 20.1±2.1 | 20.0±3.2 | 5.8±2.4 | 5.3±1.9 | 1.4±0.3 |
| csp41b-2 | 12.4±1.4 | 18.9±1.3 | 0.7±0.1 | 1.0±0.0 | 5.3±1.1 | 13.9±2.9 | 10.1±1.5 | 10.1±2.6 | 2.9±0.3 | 2.6±0.4 | 1.2±0.2 |
Values for RNAs are average signals derived from three independent run-on transcription assays (rrn16, rrn23, rps7, rbcL, psbA, psbC, psbD, psaA, psaB, and atpB, see also Fig 7C), expressed relative to the rps18 signal (=1). Mean values ±SD are shown. Asterisks indicate that results were significantly different between the WT and csp41b-2 with Student’s t-test P-value <0.05 (*) or <0.01 (**).
In order to assess directly the effect of the CSP41 complexes on the 23S rRNA, an in organello RNA stability assay was employed. Because CSP41a and CSP41b form HMW complexes, broken chloroplasts from Col-0 and csp41b-2 leaves were used for the assay, essentially as in the run-on transcription assay, to provide conditions in which multiprotein complexes are maintained. After addition of radioactively labelled precursor rrn23 RNA, RNA was re-isolated after 5 min, separated on a denaturing agarose gel, blotted onto nylon membranes, and analysed by autoradiography. In both genotypes, the majority of RNA molecules were degraded after incubation with broken chloroplasts, and the rrn23 RNA appeared to be partially processed into distinct bands, possibly representing rRNA segments protected by ribosomal proteins or stable secondary structures (Fig. 7D). Precursor rrn23 species were significantly (P < 0.05) less stable in csp41b-2 samples compared with the WT (0.84±0.04, WT=1). The same assay was also performed on the psbC/D full-length transcript, and here also the transcript was less stable in csp41b-2 broken chloroplasts. To investigate whether the decrease in transcript stability was specific for full-length transcripts, the psbC coding sequence, lacking 5′ and 3′ stability determinants, was also analysed. Again, the stability of the mRNA was found to be decreased in csp41b-2 chloroplasts. This indicates that CSP41 complexes stabilize target transcripts, possibly by unspecifically shielding sites of nucleolytic action.
Discussion
Do CSP41a and CSP41b have redundant functions?
The present data provide multiple lines of evidence for the notion that CSP41a and CSP41b do not have entirely redundant functions and that CSP41b appears to be functionally more important than its homologue under the conditions used. Loss of CSP41a has no obvious phenotypic effect, while lack of CSP41b reduces chloroplast RNA levels and impairs plant performance and, accordingly, the csp41ab double mutant exhibits the same phenotype as csp41b plants. Secondly, although CSP41 protein complexes seem to contain both proteins in WT plants, CSP41b, which is also present at higher levels, but not CSP41a, is essential for their formation. This also implies that CSP41b can form at least both homo- and heterodimers. Thirdly, the A. thaliana CSP41b and its C. reinhardtii orthologue RAP38 are more closely related to their cyanobacterial CSP41 counterparts (slr1540 and alr_4831) than are the CSP41a/RAP41 pair from A. thaliana and C. reinhardtii (Yamaguchi et al., 2003) (see Supplementary Fig. S7A at JXB online). Additionally, the predicted three-dimensional structure and surface charge distribution of the cyanobacterial CSP41 (slr1540) is more similar to Arabidopsis CSP41b than to Arabidopsis CSP41a (see Supplementary Fig. S7B). This, together with the lower level of divergence between RAP38 and CSP41b compared with RAP41 and CSP41a, suggests that CSP41a might be less constrained evolutionarily, either because it is intrinsically less important for chloroplast gene expression than CSP41b, or because CSP41b can supply all its functions.
What types of complexes do CSP41 proteins form?
CSP41a and CSP41b proteins have been found to be associated with the ribosome in C. reinhardtii (Yamaguchi et al., 2003), and both proteins were further shown to co-migrate in 2D gels with the L5 and L31 proteins of the 50S subunit of the plastid ribosome in A. thaliana (Peltier et al., 2006). Moreover, CSP41b has been co-purified with PEP from mustard (Pfannschmidt et al., 2000). The present Co-IP experiments with tagged CSP41b identified CSP41a as its major interaction partner. Because of the co-migration of the majority of both CSP41 proteins in several distinct spots during 2D BN/SDS–PAGE, it can be assumed that they are present in multimeric protein complexes mainly comprised of these two subunits. In consequence, it can be concluded that the function of CSP41 proteins in chloroplast gene expression does not depend on a tight association with PEP or the 70S ribosome, as proposed earlier (Pfannschmidt et al., 2000; Yamaguchi et al., 2003). Indeed, the only other protein—besides CSP41a—found in immunoprecipitates of CSP41b:CFP was LTA2, the E2 subunit of the plastid pyruvate decarboxylase. The physiological significance of such an interaction, however, remains to be investigated.
The finding that HMW CSP41 complexes are disrupted by treatment with RNase shows that they are associated with RNAs. RIP-chip analyses indicated that these RNAs include mRNAs for subunits of photosynthetic complexes, as well as 16S and 23S rRNAs. No tRNAs, or mRNAs for ribosomal proteins, were found to be targeted by CSP41.
At which step(s) in plastid RNA metabolism does the CSP41 complex act?
Various roles for the interaction of CSP41 proteins with RNAs have already been suggested, including functions in the binding and stability of RNA (Yang et al., 1996; Yang and Stern, 1997; Bollenbach and Stern, 2003a, b), in the biogenesis of functional ribosomes (Beligni and Mayfield, 2008), and in transcription (Bollenbach et al., 2009). The present results demonstrate that CSP41 complexes bind various chloroplast RNAs, the levels of which decrease in their absence. Because no direct evidence for an interaction of CSP41 proteins with plastid ribosomes or the PEP complex could be obtained in the present study (see above), it can be assumed that CSP41 complexes might serve to stabilize RNAs. Indeed, the in organello assay showed that the stability of two putative target RNAs including the 23S rRNA precursor was decreased in mutants lacking CSP41b. Therefore, the lowered chloroplast translation rate in CSP41-less mutants might be due to the destabilization of 23S and 16S rRNA precursors, resulting in fewer (∼50%) functional ribosomes. Moreover, the decreased levels of target mRNAs might further suppress translation of distinct transcripts. In accordance with this model, absence of CSP41b proteins is expected to lead to (i) a drop in steady-state levels of their target transcripts including 23S and 16S rRNA; (ii) a concomitant decrease in translation; and (iii) a fall in levels of PEP synthesis and, consequently, of transcription. However, it cannot be entirely excluded that binding of target transcripts also directly influences their transcription or translation.
Do CSP41 proteins act as RNases in planta?
It has been shown previously that CSP41 proteins display endonuclease activity in vitro (Yang et al., 1996; Yang and Stern 1997; Bollenbach and Stern 2003a, b). However, the present data indicate that CSP41 protein complexes stabilize target RNAs in vivo. These results do not have to be mutually exclusive, because the endoribonuclease activity of CSP41 might be highly regulated in vivo, for example by post-translational modifications (see Fig. 4; Supplementary Fig. S3 at JXB online; Marchand et al., 2006; Ströher and Dietz, 2008; Reiland et al., 2009, 2011; Finkemeier et al., 2011). In consequence, CSP41 complexes might stabilize RNAs by binding in an inactive state (without endonuclease activity) to sites that otherwise could be targeted by other RNases. Specific cues (not present under laboratory growth conditions) could then lead to the activation of ribonucleolytic activity and degradation of target transcripts.
How can the development-dependent phenotype of plants lacking CSP41b be explained?
CSP41 proteins are expressed in green tissue at all plant stages (Genevestigator, www.genevestigator.com; Hruz et al., 2008), and CSP41a accumulates equally in young and mature leaves (Bollenbach et al., 2009). However, the function of CSP41 complexes seems to be especially required in mature leaves. One explanation is that in younger leaves processes operate which compensate for the lack of CSP41b. Interestingly, mutants with reduced levels of the plastid-encoded polymerase show an inverse developmental phenotype compared with csp41b mutant plants with pale and photosynthesis-compromised younger leaves but normal mature leaves (Chi et al., 2008; Chateigner-Boutin et al., 2011). The set of transcripts targeted by CSP41 complexes and transcribed by the PEP overlap. Therefore, it is plausible to conclude that the RNA-stabilizing function of CSP41 complexes is not required in young leaves where sufficient transcripts are synthesized by the PEP. In older leaves, however, CSP41-dependent transcript stabilization processes might become crucial to maintain chloroplast gene expression. Accordingly, it has been shown that chloroplast transcript stability increases with leaf age (Klaff and Gruissem 1991). Alternatively, or additionally, the post-translational modification of CSP41 proteins might regulate their function in a development-dependent manner.
What is the physiological role of CSP41 complexes?
CSP41 proteins do not form HMW complexes to a detectable extent in the light, when the translational machinery of the chloroplast is most active (reviewed by Marin-Navarro et al., 2007). In the dark, however, CSP41 proteins are found in HMW complexes that can be disrupted by RNase treatment (see Fig. 5). Provided that increased ribosomal and polysomal association during translation contribute to stabilize chloroplast RNAs in the light (Klein et al., 1988; Berry et al., 1990; Yohn et al., 1996), a working model for the function of CSP41 complexes as a complementary RNA-stabilizing element in the dark can be proposed (Fig. 7E). Thus, non-translated mRNAs and non-ribosome-incorporated rRNA precursors are bound by CSP41 complexes in the dark and thereby protected from nucleolytic cleavage. This would allow rapid translation initiation and elongation from these stabilized transcripts as well as de novo ribosome assembly, when the translational machinery is boosted in the light (reviewed by Marin-Navarro et al., 2007). In mutants lacking CSP41 complexes, untranslated target transcripts and rRNA precursors would be more rapidly degraded. The association of CSP41 complexes with RNA ligands in the dark and the dissociation in the light seem to be controlled by the redox state of the chloroplast, because HMW complexes which only assemble in the dark can be found in the psad1-1 mutant with reduced linear electron transport and therefore more oxidizing conditions (Ihnatowicz et al., 2004; DalCorso et al., 2008) also in the light (Fig. 5C). Changes in the pIs of CSP41b species between dark and light conditions (Supplementary Fig. S3 at JXB online) indicate that the redox state of the stroma might be signalled to CSP41b via post-translational modifications and thereby regulate the RNA-binding properties of CSP41 complexes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phenotypic characterization of csp41 insertion lines.
Figure S2. CFP-tagged CSP41b can complement the csp41b-2 mutation.
Figure S3. IEF analysis of CSP41b proteins from light- and dark-adapted plants.
Figure S4. Analyses of immunoprecipitated CSP41b:CFP, chloroplast-targeted eGFP (cp-eGFP), CSP41a:eGFP, and CSP41b:eGFP.
Figure S5. RIP-chip analysis: differential supernatant signals and validation of data.
Figure S6. Comparative plastid transcriptome analysis in mutants affecting plastid gene expression.
Figure S7. Unrooted phylogram of the CSP41 family of proteins and predicted protein structures.
Table S1. Overview of primers used for amplification of probes for northern, slot blot, and transcription run-on assays
Table S2. MS data of proteins from co-immunoprecipitates of CSP41b:CFP.
Table S3. RIP-chip data from differential enrichment ratios.
Table S4. RIP-chip data from differential pellet signals.
Table S5. Log2 values of mRNA expression (relative to WT) measured by qRT-PCR.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB-TR1 TP B8 to DL and SFB429 TP A14 to CSL) and by the Australian Research Council (grant CE0561495 to IS). We thank Paul Hardy, Jörg Nickelsen, Martin Jonikas, and Mia Terashima for critical comments on the manuscript, Reik Modrozynski for excellent technical support, Rob Sharwood and David Stern for providing antibodies against CSP41a and CSP41b, Toshiharu Shikanai for antibodies against NdhL, Ute Vothknecht for antibodies against TIC110, Stefan Hoth for the CSP41b:CFP fusion construct, Bernd Weisshaar and colleagues for providing the GABI-KAT line, Csaba Koncz for allowing us to screen his T-DNA insertion library, and Ralf Bernd Kloesgen for providing cTPFNR:eGFP seeds. Additionally, we acknowledge Youlia Davidova and Mathias Pribil for MS analysis.
Glossary
Abbreviations
- BN-PAGE
blue native PAGE
- Chl
chlorophyll
- CSP41
chloroplast stem–loop-binding protein of 41 kDa
- HMW
high molecular weight
- NEP
nuclear-encoded RNA polymerase
- PEP
plastid-encoded RNA polymerase
- RIP-Chip
RNA immunoprecipitation and hybridization to gene chips
References
- Allison LA, Simon LD, Maliga P. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO Journal. 1996;15:2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Armbruster U, Zühlke J, Rengstl B, et al. The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. The Plant Cell. 2010;22:3439–3460. doi: 10.1105/tpc.110.077453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ME, Grundy WN, Elkan CP. Spinach CSP41, an mRNA-binding protein and ribonuclease, is homologous to nucleotide-sugar epimerases and hydroxysteroid dehydrogenases. Biochemical and Biophysical Research Communications. 1998;248:250–254. doi: 10.1006/bbrc.1998.8951. [DOI] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- Beligni MV, Mayfield SP. Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism. Plant Molecular Biology. 2008;67:389–401. doi: 10.1007/s11103-008-9328-2. [DOI] [PubMed] [Google Scholar]
- Berry JO, Breiding DE, Klessig DF. Light-mediated control of translational initiation of ribulose-1, 5-bisphosphate carboxylase in amaranth cotyledons. The Plant Cell. 1990;2:795–803. doi: 10.1105/tpc.2.8.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB. The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Molecular Biology. 2009;69:541–552. doi: 10.1007/s11103-008-9436-z. [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Stern DB. Divalent metal-dependent catalysis and cleavage specificity of CSP41, a chloroplast endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. Nucleic Acids Research. 2003a;31:4317–4325. doi: 10.1093/nar/gkg640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Stern DB. Secondary structures common to chloroplast mRNA 3′-untranslated regions direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. Journal of Biological Chemistry. 2003b;278:25832–25838. doi: 10.1074/jbc.M303559200. [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Tatman DA, Stern DB. CSP41a, a multifunctional RNA-binding protein, initiates mRNA turnover in tobacco chloroplasts. The Plant Journal. 2003;36:842–852. doi: 10.1046/j.1365-313x.2003.01935.x. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. The Plant Journal. 2011;65:532–542. doi: 10.1111/j.1365-313X.2010.04441.x. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-Garcia A, et al. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. The Plant Journal. 2008;56:590–602. doi: 10.1111/j.1365-313X.2008.03634.x. [DOI] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiology. 2008;147:573–584. doi: 10.1104/pp.108.116194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Wollman FA. Translational regulations as specific traits of chloroplast gene expression. FEBS Letters. 2002;529:39–42. doi: 10.1016/s0014-5793(02)03260-x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dipa: simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008;132:273–285. doi: 10.1016/j.cell.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Delannoy E, Le Ret M, Faivre-Nitschke E, Estavillo GM, Bergdoll M, Taylor NL, Pogson BJ, Small I, Imbault P, Gualberto JM. Arabidopsis tRNA adenosine deaminase arginine edits the wobble nucleotide of chloroplast tRNAArg(ACG) and is essential for efficient chloroplast translation. The Plant Cell. 2009;21:2058–2071. doi: 10.1105/tpc.109.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle FA, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. The Plant Journal. 2008;56:157–168. doi: 10.1111/j.1365-313X.2008.03581.x. [DOI] [PubMed] [Google Scholar]
- Färber A, Young AJ, Ruban AV, Horton P, Jahns P. Dynamics of xanthophyll-cycle activity in different antenna subcomplexes in the photosynthetic membranes of higher plants (the relationship between zeaxanthin conversion and nonphotochemical fluorescence quenching) Plant Physiology. 1997;115:1609–1618. doi: 10.1104/pp.115.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J, Nunes-Nesi A, Fernie AR, Steup M. Identification of a novel heteroglycan-interacting protein, HIP 1.3, from Arabidopsis thaliana. Journal of Plant Physiology. 2011;168:1415–1425. doi: 10.1016/j.jplph.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Finkemeier I, Laxa M, Miguet L, Howden AJ, Sweetlove LJ. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiology. 2011;155:1779–1790. doi: 10.1104/pp.110.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B, Briantais JM, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochimica et Biophysica Acta. 1989;990:87–92. [Google Scholar]
- Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardestrom P, Schröder W, Hurry V. The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. The Plant Journal. 2006;47:720–734. doi: 10.1111/j.1365-313X.2006.02821.x. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO Journal. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim M, Yakir E, Fradkin D, Hilman D, Kron I, Keren N, Harir Y, Yerushalmi S, Green RM. Mutations in CHLOROPLAST RNA BINDING provide evidence for the involvement of the chloroplast in the regulation of the circadian clock in Arabidopsis. The Plant Journal. 2007;51:551–562. doi: 10.1111/j.1365-313X.2007.03160.x. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihnatowicz A, Pesaresi P, Varotto C, Richly E, Schneider A, Jahns P, Salamini F, Leister D. Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. The Plant Journal. 2004;37:839–852. doi: 10.1111/j.1365-313x.2004.02011.x. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. The Plant Journal. 2005;42:133–144. doi: 10.1111/j.1365-313X.2005.02362.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Sciences. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Klaff P, Gruissem W. Changes in chloroplast mRNA stability during leaf development. The Plant Cell. 1991;3:517–529. doi: 10.1105/tpc.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Mason HS, Mullet JE. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. Journal of Cell Biology. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. Preparation of physiologically active chloroplasts from Arabidopsis. Methods in Molecular Biology. 1998;82:43–48. doi: 10.1385/0-89603-391-0:43. [DOI] [PubMed] [Google Scholar]
- Leister D, Varotto C, Pesaresi P, Niwergall A, Salamini F. Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiology and Biochemistry. 1999;37:671–678. [Google Scholar]
- Li Y, Rosso MG, Strizhov N, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: a flanking sequence tag (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics. 2003;19:1441–1442. doi: 10.1093/bioinformatics/btg170. [DOI] [PubMed] [Google Scholar]
- Liere K, Kaden D, Maliga P, Börner T. Overexpression of phage-type RNA polymerase RpoTp in tobacco demonstrates its role in chloroplast transcription by recognizing a distinct promoter type. Nucleic Acids Research. 2004;32:1159–1165. doi: 10.1093/nar/gkh285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrig K, Müller B, Davydova J, Leister D, Wolters DA. Phosphorylation site mapping of soluble proteins: bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana. Planta. 2009;229:1123–1134. doi: 10.1007/s00425-009-0901-y. [DOI] [PubMed] [Google Scholar]
- Manuell A, Beligni MV, Yamaguchi K, Mayfield SP. Regulation of chloroplast translation: interactions of RNA elements, RNA-binding proteins and the plastid ribosome. Biochemical Society Transactions. 2004;32:601–605. doi: 10.1042/BST0320601. [DOI] [PubMed] [Google Scholar]
- Marchand C, Le Marechal P, Meyer Y, Decottignies P. Comparative proteomic approaches for the isolation of proteins interacting with thioredoxin. Proteomics. 2006;6:6528–6537. doi: 10.1002/pmic.200600443. [DOI] [PubMed] [Google Scholar]
- Marin-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynthesis Research. 2007;94:359–374. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- Marques JP, Schattat MH, Hause G, Dudeck I, Klosgen RB. In vivotransport of folded EGFP by the ΔpH/TAT-dependent pathway in chloroplasts of Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:1697–1706. doi: 10.1093/jxb/erh191. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Monte E, Quail PH. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. The Plant Journal. 1999;20:251–257. doi: 10.1046/j.1365-313x.1999.00579.x. [DOI] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proceedings of the National Academy of Sciences, USA. 2007;104:8178–8183. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PD, Ponnola L, van Wijk KJ. Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry and hierarchical clustering. Molecular and Cellular Proteomics. 2010;9:1594–1615. doi: 10.1074/mcp.M000038-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, van Wijk KJ. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Molecular and Cellular Proteomics. 2006;5:114–133. doi: 10.1074/mcp.M500180-MCP200. [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D. Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. The Plant Journal. 2001;27:179–189. doi: 10.1046/j.1365-313x.2001.01076.x. [DOI] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO Journal. 2009;28:2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. The Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G. The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle-specific functions. European Journal of Biochemistry. 2000;267:253–261. doi: 10.1046/j.1432-1327.2000.00991.x. [DOI] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Research. 2008;36:5152–5165. doi: 10.1093/nar/gkn492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab S, Toth Z, de Groot C, Stamminger T, Hoth S. ABA-responsive RNA-binding proteins are involved in chloroplast and stromule function in Arabidopsis seedlings. Planta. 2006;224:900–914. doi: 10.1007/s00425-006-0282-4. [DOI] [PubMed] [Google Scholar]
- Reiland S, Finazzi G, Endler A, et al. Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF) Proceedings of the National Academy of Sciences, USA. 2011;108:12955–12960. doi: 10.1073/pnas.1104734108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiology. 2009;150:889–903. doi: 10.1104/pp.109.138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios G, Lossow A, Hertel B, et al. Rapid identification of Arabidopsis insertion mutants by non-radioactive detection of T-DNA tagged genes. The Plant Journal. 2002;32:243–253. doi: 10.1046/j.1365-313x.2002.01416.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schägger H, von Jagow G. Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Analytical Biochemistry. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. The Plant Cell. 2005;17:2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweer J, Loschelder H, Link G. A promoter switch that can rescue a plant sigma factor mutant. FEBS Letters. 2006;580:6617–6622. doi: 10.1016/j.febslet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Ströher E, Dietz KJ. The dynamic thiol–disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiologica Plantarum. 2008;133:566–583. doi: 10.1111/j.1399-3054.2008.01103.x. [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Research. 2009;37:D969–D974. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki JY, Ytterberg AJ, Beardslee TA, Allison LA, Wijk KJ, Maliga P. Affinity purification of the tobacco plastid RNA polymerase and in vitro reconstitution of the holoenzyme. The Plant Journal. 2004;40:164–172. doi: 10.1111/j.1365-313X.2004.02195.x. [DOI] [PubMed] [Google Scholar]
- Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz-Linneweber C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proceedings of the National Academy of Sciences, USA. 2009;106:6002–6007. doi: 10.1073/pnas.0808529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. Ins and outs of plastid genome evolution. Current Opinion in Genetics and Development. 1991;1:523–529. doi: 10.1016/s0959-437x(05)80202-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Beligni MV, Prieto S, Haynes PA, McDonald WH, Yates JR, 3rd, Mayfield SP. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70S ribosome. Journal of Biological Chemistry. 2003;278:33774–33785. doi: 10.1074/jbc.M301934200. [DOI] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB. CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. The Plant Cell. 1996;8:1409–1420. doi: 10.1105/tpc.8.8.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Stern DB. The spinach chloroplast endoribonuclease CSP41 cleaves the 3′-untranslated region of petD mRNA primarily within its terminal stem–loop structure. Journal of Biological Chemistry. 1997;272:12874–12880. doi: 10.1074/jbc.272.19.12874. [DOI] [PubMed] [Google Scholar]
- Yang J, Usack L, Monde RA, Stern DB. The 41 kDa protein component of the spinach chloroplast petD mRNA 3′ stem–loop:protein complex is a nuclear encoded chloroplast RNA-binding protein. Nucleic Acids Symposium Series. 1995;33:237–239. [PubMed] [Google Scholar]
- Yohn CB, Cohen A, Danon A, Mayfield SP. Altered mRNA binding activity and decreased translational initiation in a nuclear mutant lacking translation of the chloroplast psbA mRNA. Molecular and Cellular Biology. 1996;16:3560–3566. doi: 10.1128/mcb.16.7.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- Zoschke R, Liere K, Börner T. From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. The Plant Journal. 2007;50:710–722. doi: 10.1111/j.1365-313X.2007.03084.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.