Abstract
Cytoplasmic male sterility (CMS) is a widespread phenomenon in higher plants, and several studies have established that this maternally inherited defect is often associated with a mitochondrial mutant. Approximately 10 chimeric genes have been identified as being associated with corresponding CMS systems in the family Brassicaceae, but there is little direct evidence that these genes cause male sterility. In this study, a novel chimeric gene (named orf288) was found to be located downstream of the atp6 gene and co-transcribed with this gene in the hau CMS sterile line. Western blotting analysis showed that this predicted open reading frame (ORF) was translated in the mitochondria of male-sterile plants. Furthermore, the growth of Escherichia coli was significantly repressed in the presence of ORF288, which indicated that this protein is toxic to the E. coli host cells. To confirm further the function of orf288 in male sterility, the gene was fused to a mitochondrial-targeting pre-sequence under the control of the Arabidopsis APETALA3 promoter and introduced into Arabidopsis thaliana. Almost 80% of transgenic plants with orf288 failed to develop anthers. It was also found that the independent expression of orf288 caused male sterility in transgenic plants, even without the transit pre-sequence. Furthermore, transient expression of orf288 and green fluorescent protein (GFP) as a fused protein in A. thaliana protoplasts showed that ORF288 was able to anchor to mitochondria even without the external mitochondrial-targeting peptide. These observations provide important evidence that orf288 is responsible for the male sterility of hau CMS in Brassica juncea.
Keywords: Brassica juncea, cytoplasmic male sterility, cytotoxic protein, mitochondrial-anchored protein, orf288, transgenic plants
Introduction
Cytoplasmic male sterility (CMS) is a maternally inherited defect of higher plants in the production of functional pollen. Several studies have reported that chimeric open reading frames (ORFs) in the mitochondrial genome cause CMS in a variety of higher plant species (Dewey et al., 1986; Chase, 2007). Several CMS-associated aberrant genes are located upstream or downstream of certain known mitochondrial genes and co-transcribed with them. The expression of these novel chimeric ORFs is usually inhibited at different levels by nuclear fertility restorer (Rf) genes (Sarria et al., 1998; Wise et al., 1999; Feng Liu et al., 2001; Koizuka et al., 2003; Kazama et al., 2008; Yamamoto et al., 2008). A survey of the literature shows that the expression of these CMS-associated ORFs in Escherichia coli can inhibit the growth of the host bacteria (Dewey et al., 1988; Duroc et al., 2005; Wang et al., 2006). A series of experiments and observations have led to isolation and characterization of CMS-related genes in various higher plants.
urf13, an aberrant ORF in Texas (T)-cytoplasm maize, is the first identified CMS-related gene in a plant species (Dewey et al., 1987, 1988; Wise et al., 1987). Two nuclear genes, Rf1 and Rf2, restore the male fertility of T-cytoplasm maize (Wise et al., 1999; Feng Liu et al., 2001). The CMS-BT (Boro II) is one of the most widely studied CMS/Rf systems in rice (Oryza sativa L.) (Akagi et al., 2004; Wang et al., 2006; Kazama et al., 2008). It has been reported that orf79, which is a chimeric region located downstream of the atp6 gene, is responsible for this type of CMS in rice (Wang et al., 2006). In the BT-type CMS line, orf79 co-transcribed with the atp6 gene forms a 2.0 kb transcript and encodes a predicted transmembrane protein. In the presence of Rf1a, the 2.0 kb transcript is split into two transcripts of 1.5 kb and 0.5 kb by RNA processing (Wang et al., 2006). A longer orfB transcript may be associated with the CMS-WA (wild abortive) system in rice (Das et al., 2010). The transformation of Arabidopsis with mitochondrion-targeted orf456 causes 45% of the first generation transformed (T1) population to exhibit male sterility (Kim et al., 2007). The expression of orf129 with the assistance of a mitochondrial-targeting pre-sequence results in male sterility in transgenic tobacco plants (Yamamoto et al., 2008). Male sterility in the Helianthus petiolaris (PET1)-CMS system of sunflowers is associated with a novel mitochondrial gene, orfH522, which results in sterile transformed tobacco (Nizampatnam et al., 2009). These well-studied CMS genes have been investigated primarily on the basis of the differences in the DNA sequences of the mitochondrial genomes and in the expression pattern of mitochondrial genes among the CMS, maintainer, and restorer lines. In most cases, these early predictions substantiate further studies of these CMS-associated genes. Furthermore, transgenic plants with these chimeric ORFs failed to produce viable pollen, thereby providing additional important evidence that they are associated with male sterility.
Brassica is one of the most widely used genera for investigating the mechanisms of CMS. Approximately 10 types of CMS have been identified in Brassica, and most of them are due to rearrangements of the mitochondrial genome and expression of chimeric ORFs co-transcribed with functional genes in mitochondria (Landgren et al., 1996; Ashutosh et al., 2008). In the polima CMS system, orf224, which is co-transcribed with the atp6 gene, forms dicistronic 2.2 kb and 1.9 kb transcripts (Singh and Brown, 1993). The CMS-associated atp6/orf224 transcripts were shown to be dramatically reduced in the petals, stamens, and carpals, but not in the sepals of restored line flowers (Li et al., 1998). The ogura CMS system has also been extensively studied owing to its high value in rapeseed breeding (Brown et al., 2003; Gonzalez-Melendi et al., 2008; Duroc et al., 2009). An aberrant region of mitochondrial DNA in the ogura sterile line encodes a peptide of 138 amino acids the accumulation of which can be suppressed by the nuclear Rfo locus (Bellaoui et al., 1999). orf125 may be responsible for Kosena CMS, and the protein levels of the Kosena CMS-associated mitochondrial protein ORF125 were apparently reduced in plants in which male fertility was restored (Koizuka et al., 2003). orf222, which shares 79% sequence similarity with the predicted pol orf224 gene region, may be associated with nap CMS (L'Homme et al., 1997). orf263 co-transcribed with the atp6 gene may be associated with tournefortii CMS and translated into a 32 kDa protein. This 32 kDa protein was only detected in alloplasmic lines (Landgren et al., 1996). In the ‘Tournefortii–Stiewe’ system, orf193, which is co-transcribed with atp9, probably impairs pollen development (Dieterich et al., 2003). orf108, which is co-transcibed with the atpA gene, was found to be associated with Moricandia arvensis CMS, and the long atpA transcript is spliced within orf108 in fertility-restored lines (Ashutosh et al., 2008). A single nucleotide insertion mutation that results in the truncation of atp6 may be associated with DCGMS CMS in radish (Raphanus sativus L.) (Lee et al., 2009). However, these male sterility-associated ORFs are rarely transformed into fertile plants, which further confirm their functions.
Previously, hau was found to be a novel CMS system in Brassica juncea, and this system has been transferred to Brassica napus. Genetic, morphological, cytological, and molecular analyses showed that hau CMS is different from previously studied CMS systems in Brassica (Wan et al., 2008). In the present work, a chimeric gene, orf288, has been found to be located downstream of the atp6 gene and co-transcribed with it in the hau CMS line. The expression of the putative male sterility-associated gene orf288 in E. coli and Arabidopsis thaliana indicated that orf288 plays an important role in the male sterility of hau CMS in B. juncea.
Materials and methods
Plant materials
The hau CMS (00-6-102A) used in this study was originally discovered as a spontaneous male-sterile mutant in B. juncea in the experimental field of Huazhong Agricultural University in 1999 (Wan et al., 2008). This type of CMS has been successfully transferred into B. napus. The sterile and maintainer lines of this CMS system in B. napus were also used to examine the expression differences of CMS-related genes. The maintainer line of B. napus was used to provide recurrent parents for 10 generations (BC10) of crosses with hau CMS to establish this CMS system in B. napus.
DNA extraction and genome walking
Total genomic DNA samples were prepared from the fresh leaves of plants. To isolate the flanking sequence of the atp6 gene and the whole sequence of orf288, genome walking was performed using the Genome-Walker Universal Kit (Clontech, USA) according to the manufacturer's protocol.
RNA isolation and RT-PCR
Total RNA was isolated from flower buds, roots, and fresh leaves using Trizol (Invitrogen) according to the manufacturer's protocol. Reverse transcription was performed with DNase-treated total RNA samples using the RevertAid™ First-Strand cDNA Synthesis Kit (Fermentas) according to the manufacturer's instructions for cDNA synthesis for reverse transcription-PCR (RT-PCR). Circularized RNA (CR)-RT-PCR (Kuhn and Binder, 2002) was performed to determine the 5' and 3' ends of atp6 transcripts.
Northern blot analysis
The total RNA was fractionated on a 1.5% denaturing agarose gel containing 2% formaldehyde and transferred onto Hybond N+ membranes (Amersham, UK). Hybridizations were carried out at 68 °C overnight in hybridization solution (Toyobo). Filters were washed for 10 min in 2× SSC and 0.1% SDS at 37 °C. The membranes were exposed to a phosphor storage screen for 2–3 h, and the signals were scanned using the Typhoon FLA 9000 (Fujifilm, Japan).
Antibody preparation and western blotting
A peptide antigen corresponding to 17 residues of ORF288 was synthesized by a chemical synthesis method (Neweast, China) and used to immunize rabbits for antibody production. Crude mitochondrial proteins were extracted from the etiolated seedlings of male-sterile and the maintainer lines of hau CMS plants. The proteins were separated by 12% SDS–PAGE and transferred onto a membrane (PVDF type, Millipore). The membrane blots were incubated in blocking buffer (1% bovine serum albumin, 0.05% Tween-20, 20 mM TRIS-HCl, and 500 mM NaCl, pH 7.5) for 1.5 h and then incubated with primary antibody serum (1:500 dilution) for 3 h at room temperature. After three rinses (10 min each) with TBST, the blots were incubated in secondary antibody solution (affinity-purified phosphatase-labelled goat anti-rabbit IgG[HþL], 1:3000 dilution) for 1.5 h at room temperature and finally washed three times (10 min each) with TBST.
Expression of orf288 in E. coli
The whole fragment of orf288 was amplified from the cDNA of flower buds using the primer pairs KpnI288STR and BamHI288ED; in Supplementary Table S1, available at JXB online, the KpnI and BamHI sites in the primer sequences are underlined. The corresponding enzymes were then used to clone this fragment into the bacterial expression vector PET32a (Novagen). The expression of ORF288 in E. coli BL21(DE3)plysS (Promega) was induced by adding 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) when the optical density (OD) of the samples reached ∼0.6. Over the next 2.5 h, the OD of the samples was measured every 30 min at 600 nm using a UV-1601 spectrophotometer (Shimadzu, Japan)
Vector construction and expression of orf288 in A. thaliana
The AP3 promoter fragment (base pairs –1 to –867 of the AP3 gene) was amplified from A. thaliana using the oligonucleotide primer pairs AP3BR and AP3F; in Supplementary Table S1 at JXB online, the KpnI and BamHI sites in the primer sequences are underlined. The Cauliflower mosaic virus (CaMV) 35S promoter was amplified from pCAMBIA2301 (CAMBIA) using the primers VNOF and VNOL. The pre-sequence of coxIV (mitochondrial transit peptide of the cytochrome oxidase subunit IV from yeast) (Köhler et al., 1997) and the coding frame of orf288 were also amplified using coxVIF, T-288L, T-288F, and 288L and fused by overlap extension PCR. In the absence of the mitochondrial-targeting peptide, an additional primer, VNO2, was used. The NOS sequence was amplified from pCAMBIA2301 using the primer pairs NOSR and NOSF and was cloned to the 3' end of the expression fragment. All of the PCR products were inserted into the pCAMBIA2300 vector (CAMBIA), and the AP3 promoter fragment was also inserted into pBI101 (Clontech) as a control. Agrobacterium GV3101 and A. thaliana Columbia were used for the floral dipping, and the Agrobacterium-mediated transformation was performed according to the modified floral-dip method (Clough and Bent, 1998).
Transient expression and subcellular localization of ORF288
The full-length cDNA of orf288 was PCR-amplified using primers HF and HL (Supplementary Table S1 at JXB online) and inserted into the pM999GFP (Supplementary Fig. S1) vector, that was provided by Dr Jian Xu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China), to generate an N-terminal green fluorescent protein (GFP) fusion product. This vector allows for the in planta expression of proteins under the control of the constitutive CaMV double 35S promoter. The mitochondria of protoplasts were marked using Mitotracker Red CMX-Ros staining solution (Molecular Probes, Invitrogen). The fusion construct was introduced into Arabidopsis protoplasts that were prepared from whole seedlings by polyethylene glycol (PEG)/calcium-mediated transformation (Yoo et al., 2007). Fluorescence microscopy was performed using a confocal laser microscope.
Sequence analysis
Gene sequences were analysed using the BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and DNAStar software. Multiple alignments of DNA sequences were performed with ClustalX 2.0 (Larkin et al., 2007) and GeneDoc (Nicholas and Nicholas, 1997), and the output was manually modified. The ORF288 transmembrane structure was predicted by the software TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM). The mitochondrial transit peptide was predicted by PredSL (http://hannibal.biol.uoa.gr /PredSL/) and SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/).
Results
A specific sequence located downstream of the atp6 gene in hau CMS cytoplasm
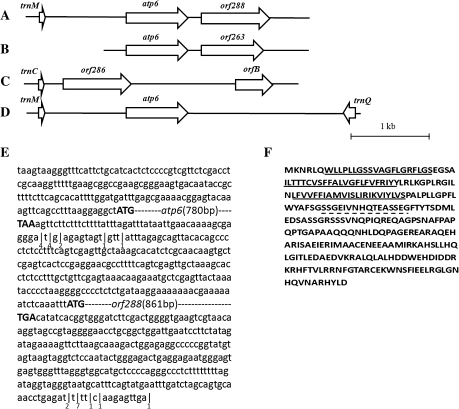
Previously, restriction fragment length polymorphisms (RFLPs) were detected for atp1, atp6, and atp9 genes in the sterile and the maintainer lines (Wan et al., 2008). Here, the flanking sequences of these genes in the male-sterile line were compared with those in the maintainer line. A chimeric fragment was found to be located downstream of the atp6 gene (Fig. 1A). BLAST searches indicated that this fragment shares high similarity with orf263 (Fig. 1B) in alloplasmic male-sterile Brassica lines (Landgren et al., 1996). To identify the organization of this fragment, genome walking was carried out based on the previous results. The BLAST searches with a product of 629 nucleotides confirmed that the sequence similarity between orf263 and the region downstream of atp6 in the hau CMS mitotype was very high (99.5%). Four point mutations were detected: three were present in the coding region and one was located downstream of orf263 (Supplementary Fig. S2 at JXB online). PCR amplification of atp6–orf288 with the primers atp6288UP2 and atp6288DOWN2 (shown in Supplementary Table S1) also showed that the chimeric fragment was located downstream of the atp6 gene. It was predicted that a deletion in these four point mutations would cause the length of the orf263 product to increase to 288 amino acids. Therefore, this chimeric fragment was designated as orf288. Multiple alignments of orf286, orf288, and orf263 showed that orf286 (Fig. 1C) from the mitochondrial genome of B. napus (GenBank accession no. AP006444) shares 93% nucleotide sequence identity with orf288 (Supplementary Fig. S2). orf286 was identical to orf288 at the sites of four single nucleotide differences between orf288 and orf263 (Supplementary Fig. S2). Sequence analysis showed that this orf288 was a chimeric fragment located downstream of the atp6 gene in the hau mitochondrial genome, but was not detected downstream of the atp6 gene in the hau CMS maintainer line (Fig. 1D).
Fig. 1.
(A–D) The organization of mitochondrial genome regions associated with the orf288 gene for four different mitotypes. (A) The trnM–atp6–orf288 region of the hau mitotype. (B) The atp6–orf263 region of tour CMS: orf263 is associated with this type of CMS (Landgren et al., 1996). (C) The trnC–orf286–orfB region of the nap mitotype: orf286 is an unidentified ORF in nap CMS (Handa, 2003). (D) The trnM–atp6–trnQ region of the hau CMS maintainer line. (E) Complete cDNA sequence of the atp6/orf288 transcripts of the male-sterile line. The initiation and termination codon are highlighted in bold. The different 3' ends of transcripts are indicated by vertical bars, and the number under the bar indicates the number of the ends sequenced. (F) Amino acid sequence of ORF288: the transmembrane regions are underlined, and the dotted underlining indicates that the peptide was synthesized and used for antibody production. Arrows indicate the direction of transcription.
Two different transcripts of the atp6 gene in the hau CMS sterile line
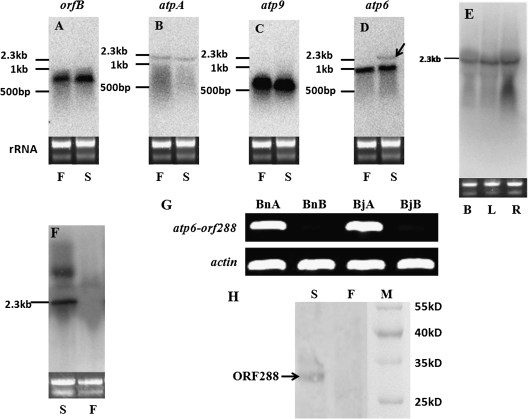
Four mitochondrial gene-specific probes, atpA, atp6, atp9, and orfB, were used to analyse the expression differences of these gene transcripts between the male-sterile and the maintainer lines of hau CMS in B. juncea. Northern blotting results revealed no difference for the orfB, atpA, and atp9 probes (Fig. 2A–C), but the atp6 transcripts showed different banding patterns (Fig. 2D). There are two bands in the male-sterile line, and between them the shorter one was observed in the maintainer line (Fig. 2D). Therefore, the atp6 gene and its flanking region were suggested together to be a strong candidate region of CMS-associated fragments in the hau CMS line. Because a chimeric fragment had been previously identified, northern blotting experiments with the orf288 probe confirmed that the downstream region of atp6 was expressed constitutively in the male-sterile line (Fig. 2E). The 2.3 kb transcript was also found in the hau CMS line of B. napus (Fig. 2F). The RT-PCR analysis also revealed that the atp6 gene was co-transcribed with the orf288 region in the hau CMS sterile line (Fig. 2G). To identify the RNA editing sites in orf288, the nucleotide sequences of the RT-PCR were compared with PCR products, and the alignment exhibited no evidence of RNA editing in the orf288 coding region.
Fig. 2.
(A–D) Northern blotting analysis of total RNA from buds of the male-sterile line (S) and the maintainer line (F) for four mitochondrial probes (orfB, atpA, atp9, and atp6). The atp6 gene showed polymorphic band patterns of RNA transcripts (indicated by the black arrow). (E) Total RNA from the floral buds (B), leaf (L), and roots (R) of the hau CMS sterile line in Brassica juncea was blotted with the orf288 probe. (F) The total RNA from the male-sterile line (S) and the maintainer line (F) of hau CMS in Brassica napus was detected using the orf288 probe. (G) RT-PCR to demonstrate the expression of the atp6–orf288 region in the hau mitotype, but not in its maintainer line. BnA, the male-sterile line in Brassica napus; BnB, the maintainer line in Brassica napus; BjA, the male-sterile line in Brassica juncea; BjB, the maintainer line in Brassica juncea. (H) Identification of ORF288 unique to the sterile line of hau CMS. Mitochondrial proteins were extracted from etiolated seedlings of male-sterile (S) and maintainer (F) lines and then were separated by 12% SDS–PAGE. The protein blots were then probed with an antibody to ORF288. The specific band (lane S) for ORF288 is indicated with an arrow. A pre-stained marker (M) was used for the detection of the transfer process and an evaluation of the molecular mass of the detected protein.
The 5' and 3' ends of the two different orf288-related transcripts
CR-RT-PCR was performed to identify the 5′ and 3′ ends of the atp6 gene transcripts. The identified 5′ ends of the two atp6 mRNAs were –165 nucleotides from the initiation codon (Fig. 1E). It was also observed that the mRNA ends of atp6 could self-ligate without treating the RNA with tobacco alkaline phosphatase, which indicated that the 5' end of the atp6 mRNA was generated by post-transcriptional processing. One atp6 3′ end was detected from 48 to 61 nucleotides downstream of the stop codon, and another was located from 276 to 289 nucleotides downstream of the orf288 stop codon (Fig. 1E).
The chimeric gene orf288 encodes a 32 kDa peptide in male-sterile mitochondria
The northern blotting and CR-RT-PCR results showed that orf288 was completely co-transcribed with the atp6 gene in the male-sterile line. This gene was predicted to encode a 32 kDa protein with a triple transmembrane region (Fig. 1F). To confirm that this gene was actually translated into a protein, a peptide antigen corresponding to 17 residues (Fig. 1F) of ORF288 was synthesized and used to produce a polyclonal antibody. The mitochondrial proteins extracted from the etiolated seedling tissue of the fertile and sterile lines were detected using the above-mentioned antibody. A band of ∼32 kDa was detected exclusively for the male-sterile sample (Fig. 2H).
The expression of orf288 represses the growth of E. coli
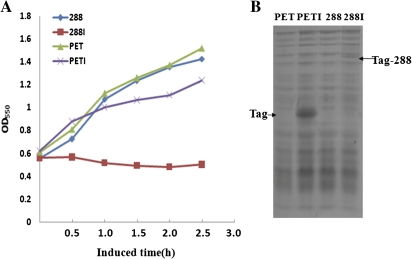
To examine the function of orf288, its coding sequence was cloned into the expression region of a PET32a vector, and IPTG was used to induce its expression in E. coli. The growth of the host bacteria was repressed significantly with the expression of ORF288 (Fig. 3A). The growth of E. coli was normal when the tag peptide was highly expressed in the vector but was repressed when low levels of ORF288 fused with the tag peptide were expressed (Fig. 3B). This finding indicated that orf288 encodes a peptide that is cytotoxic to host bacteria.
Fig. 3.
(A) The effect of orf288 expression on the growth of E. coli cells in liquid cultures with or without IPTG. IPTG was added when the cell growth reached OD550=0.6. The expression vector PET32a was set as a control. 288, orf288-containing vector not induced by IPTG, 288I, induced with IPTG; PET, the control expression vector not induced by IPTG; PETI, the control expression vector induced by IPTG. (B) The expressed recombinant protein. Tag, the tag peptide in pet32a; Tag-288, the fusion peptide of tag and ORF288. (This figure is available in colour at JXB online.)
Expression of orf288 in Arabidopsis significantly impairs the development of anthers
To investigate the association of this chimeric gene with the male abortion of the hau CMS sterile line, four constructs were prepared and transformed into A. thaliana (Fig. 4A). The VNO construct contained a 35S promoter and orf288, which was fused to the mitochondrial transit peptide sequence of the nuclear coxIV gene of yeast for mitochondrial targeting. The fused ORF that was driven by the AP3 promoter was specifically designed to investigate its effects on floral organs. To determine whether the chimeric gene still functioned without a mitochondrial-targeting peptide, the VNII vector was constructed, in which orf288 was driven by the AP3 promoter but lacked the coxIV pre-sequence. A construct with the β-glucuronidase (GUS) gene driven by the AP3 promoter was transformed into the plants as a control.
Fig. 4.
(A) Schematic illustration of constructs used to transform Arabidopsis. The mitochondrial-targeting pre-sequence is the N-terminal 57 amino acids of the coxIV gene in yeast (grey boxes). ORFs are indicated by open boxes. Arrows indicate the CaMV35S or the promoter of the Arabidopsis AP3 gene. The fragments were cloned to the expression sites of pCAMBIA2300 (constructs: VN3, VNO, and VNII). and the AP3 promoter fragment used was inserted into pBI101 as a control. The direction of transcription is from left to right. (B) Expression analysis of orf288 in transgenic plants with the VN3 construct. The transgenic plants showed male sterility (S) and full fertility (F). Wild buds (WT) were used as a control. (C–H) Phenotype analyses of male-sterile transformants with the VN3 construct. (C) Feature of a wild silique (black arrow); (D) feature of a male-sterile transgenic non-pollinated pistil (black arrow); (E) a wild flower with normal petals (white arrow); (F) a male-sterile transgenic flower without white petals; (G) wild anthers (white arrow); (H) no anther on top of a filament (white arrow).
The statistical data showed that 80% of the T1 plants with the VN3 construct developed abnormal flowers (Table 1; Fig. 4C–H). The pistil was surrounded by sepals but lacked stamens and petals (Fig. 4D, F, H; compare with the wild type, Fig. 4C, E, G). The stamens were instead represented only by one or two filaments with absent anthers for each flower (Fig. 4H). Surprisingly, ∼80% of the transgenic plants with the VNII fragment showed phenotypes similar to those with the mitochondrial-targeting pre-sequence (Table 1). The transgenic plants that were transformed using the construct with the CaMV35S promoter were as fertile as the wild-type plants (Table 1). The control transformation with the AGUS construct did not change the male fertility of the transgenic plants. These transformation results indicated that the expression of orf288 in Arabidopsis could lead to male sterility in the absence or presence of an external mitochondrial-targeting peptide, and an appropriate promoter is pivotal for this male sterility-associated gene to affect plant function significantly.
Table 1.
Number of A. thaliana transgenic plants obtained with each construct
| Constructs | No. of PCR positive transgenic plants | No. of fertile plants | No. of sterile plants |
| VNII | 20 | 4 | 16 |
| VNO | 22 | 22 | 0 |
| VN3 | 15 | 3 | 12 |
| AGUS | 6 | 6 | 0 |
RT-PCR analysis was performed to examine the expression of the chimeric gene in the transgenic plants. Male-sterile T1 plants were found to express the orf288 gene strongly, but it was not expressed in fertile transgenic plants (Fig. 4B). To confirm that the male-sterile phenotype was transmitted to the second generation of transformed (T2) plants, three male-sterile T1 transformants were selected at random to assess the association of the orf288 fragment with male sterility. These selected plants were cross-pollinated with wild-type pollen, and then the T2 progeny were selected on kanamycin-containing medium and detected by the PCR amplification of orf288 using specific primers. Each of the 158 T2 plants with the transgenic fragments showed similar male-sterile phenotypes to their parents.
ORF288 is a mitochondrial-anchored protein
The above transformation experiments showed that the expression of orf288 in Arabidopsis disrupted anther development even without a mitochondrial-targeting pre-sequence, and the morphologies of the transgenic plants were similar to those that possessed the external signal. This might suggest that orf288 is functional in the cytoplasmic matrix or is targeted to the mitochondria without the assistance of an external transit peptide. PredSL (Petsalaki et al., 2006) was used to predict the subcellular localization of ORF288, and the result showed that the N-terminal sequence could have its own signal function, because the mTP score was nearly 0.95126. The SignalP 3.0 results also revealed that the N-terminus of ORF288 may be a signal anchor peptide (signal anchor probability: 0.954) (Nielsen et al., 1997; Nielsen and Krogh, 1998; Bendtsen et al., 2004). This suggested that ORF288 is a mitochondrial-anchored protein that is originally expressed in the mitochondria of hau CMS cytoplasm and is capable of disrupting floral organ development.
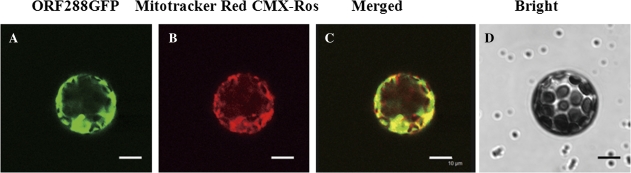
To confirm that ORF288 is able to anchor to the mitochondria, the entire ORF was fused with GFP, and the fusion protein was expressed transiently in Arabidopsis protoplasts using the PEG transfection method (Fig. 5A–D). Mitotracker Red CMX-Ros dye was used to stain the mitochondria. The GFP fluorescence (Fig. 5A) and Mitotracker dye (Fig. 5B) images matched perfectly (Fig. 5C). These results demonstrated that without the external signal pre-sequence, ORF288 that is expressed in the cellular matrix additionally targets itself to the mitochondria.
Fig. 5.
Subcellular localization of ORF288. (A) The protoplast showed a green fluorescent signal at 488 nm; (B) the same protoplast showed a red fluorescent signal (stained by Mitotracker Red CMX-Ros) at 561 nm; (C) merged image of the green and red signals; (D) bright-field image. Data are representative of the transformed protoplasts. Green fluorescent signals were examined 16 h after transformation. Scale bars=10 μm.
Discussion
orf288, a specific chimeric fragment located downstream of the atp6 gene
The male-sterile hau CMS line was found to be a spontaneous male-sterile mutant in B. juncea. As a chimeric gene in the mitochondrial genome, the nucleotide sequence of orf288 shared 99.5% similarity with the corresponding fragment of male-sterile Brassica lines containing B. tournefortii mitochondria (Landgren et al., 1996). There were four point mutations: one deletion at base pair 764 and three nucleotide changes at base pairs 485, 785, and 802 in orf288 (Supplementary Fig. S2 at JXB online). At these four positions, the nucleotides of orf288 and orf286 (located upstream of orfB) were identical. These observations might indicate that this region of hau CMS is evolutionarily closer to the homologous segment of the nap mitotype (Handa, 2003) than that of B. tournefortii CMS (Landgren et al., 1996). Tournefortii–Stiewe CMS (B. napus) originated from a donor–recipient protoplast fusion of B. tournefortii and B. napus (Stiewe and Röbbelen, 1994). However, in this B. tournefortii-related male sterility system, the expression of the atp6 gene is similar between the male-sterile line and its maintainer line (Dieterich et al., 2003). Brassica tour CMS and Tournefortii–Stiewe CMS share a 1.58 kb atp9 gene transcript (Dieterich et al., 2003), which was not detected in the hau CMS mitotype and could contribute to the male sterility of the latter system. These observations indicated that the organization and expression of the CMS-associated genes were different in these three CMS systems.
The characterization of the transcriptional pattern of orf288
In most of the CMS systems that have been investigated, there is an association with mitochondrial genome rearrangement, and several CMS-associated genes are co-transcribed with functional mitochondrial genes (Dewey et al., 1986; Hanson, 1991). Therefore, previous studies have routinely employed Southern and northern blotting analyses (Stahl et al., 1994; Kim et al., 2007). It was reported that three mitochondrial genes possessed RFLPs in the CMS and male-fertile lines (Wan et al., 2008). In this study, northern blotting and RT-PCR analyses were used to detect transcriptional differences between fertile and sterile lines. A longer atp6 transcript was found only in the male-sterile line, and this type of co-transcription was also found in several other CMS systems with known mitochondrial gene probes (Bonhomme et al., 1992; Krishnasamy and Makaroff, 1993; Wang et al., 2006).
Previous Southern blotting results indicated that the atp6 gene is located at a single locus in the hau CMS mitochondrial genome (Wan et al., 2008). Northern blotting with the atp6 probe showed that two different transcripts were present in the sterile line. Furthermore, 12 independent CR-RT-PCR products of each of the above transcripts were sequenced, and two slightly scattering 3' ends were found (Fig. 1E). These observations suggested that the two atp6 transcripts with different products originated from the same locus, and the insertion of the chimeric fragment orf288 disrupted the transcriptional termination of the atp6 gene in certain nuclear backgrounds.
ORF288 is a cytotoxic protein
BLASTP analysis showed that the N-terminus of ORF288 was identical to the NADH dehydrogenase subunit 5 and shared 68% identity with an uncharacterized ATP synthase C chain-like protein in Arabidopsis. The TMHMM result revealed that ORF288 is a multiple transmembrane protein with three transmembrane regions at its N-terminus. It was previously reported that ORF79 in Boro II cytoplasm and ORF129 in wild sugar beet are mitochondrial transmembrane proteins (Wang et al., 2006; Yamamoto et al., 2008), and the expression of several male-sterile genes is toxic or lethal to host bacterial cells (Dewey et al., 1988; Duroc et al., 2005; Wang et al., 2006). The data showed that the expression of orf288 in E. coli significantly repressed host cell growth (Fig. 3A). Therefore, it was suggested that ORF288 might also affect the development of floral organs by weakening mitochondria, as has been predicted by other researchers for several other CMS-associated genes (Wang et al., 2006).
The multiple effects of ORF288 in transgenic plants
The Arabidopsis APETALA3 gene is expressed specifically in stamens and petals during most floral development (Hill et al., 1998), and its promoter has been successfully used to identify the CMS-associated gene orf129 in sugar beet (Yamamoto et al., 2008). The pre-sequence of coxIV (partial or full) in yeast is also efficient for mitochondrial localization (Köhler et al., 1997; Kim et al., 2007; Nizampatnam et al., 2009). Therefore, in this study, the pre-sequence of coxIV was fused to the 5' end of orf288 driven by the AP3 gene promoter. For this promoter, ORF288 disrupted the development of the anther, and the male-sterile transformants with the VN3 and VNII constructs could not develop white petals. These phenotypes suggested that ORF288 not only strongly disturbs the differentiation of stamens but also affects the development of the petals in the transgenic plants. Previous experiments have shown that the anthers of the hau CMS line are transformed into thickened petal-like structures that lack anthers and filaments. The stamen primordia deviated from normal polarization and formed petal primordia, which developed into petal-like structures where the stamen would have been located (Wan et al., 2008). Although the flower morphologies of the male-sterile transgenic plants were different from those of the male-sterile hau CMS line, both reached their male abortion stages earlier than those of other previously reported CMS types in Brassica. The male-sterile phenotype was also found to be co-segregated with a transgenic fragment in the T2 progeny of male-sterile plants that were pollinated by wild pollen.
It was initially hypothesized that the transgenic plants with orf288 driven by CaMV35S would be male sterile, but the result showed that the male fertility was not affected. In fact, several other researchers also found the same phenomenon, when they used CaMV35S to drive their CMS-associated genes (Chaumont et al., 1995; Duroc et al., 2006; Nizampatnam et al., 2009). According to present knowledge, it was suggested that this might be due to the weak promotion of the CaMV35S promoter in anther/floral tissues and the male-sterile genes were not expressed in sufficient amounts in the appropriate anther/floral cells. In addition, several CMS-associated genes were also detected in vegetative tissues (Dewey et al., 1987; Bellaoui et al., 1999), but the growth of most vegetative tissues is not being notably affected. This may be due to the vulnerability of anther/floral tissues or specific interaction of CMS-associated genes with some factors in floral organs.
The subcellular localization of ORF288 expressed in the cytoplasmic matrix
For the first time, a CMS-associated gene expressed in the cellular matrix of transgenic plants was found to be targeted to the mitochondria without the assistance of an external transit peptide. In chili pepper and wild beet, similar transgenic experiments with the CMS-associated gene did not result in male-sterile plants when a targeting pre-sequence was not used to assist in the mitochondrial targeting process (Kim et al., 2007; Yamamoto et al., 2008). However, the expression of the common bean CMS-associated gene (orf239) caused male sterility in transgenic tobacco plants even when ORF239 was not targeted to the mitochondria (He et al., 1996). Subsequently, it was found that ORF239 accumulated within the callose layer of the pre-meiotic pollen mother cell wall and the primary cell wall (Abad et al., 1995; He et al., 1996; Sarria et al., 1998). In this study, the male-sterile transgenic plants obtained with the VNII construct showed similar phenotypes to those with the mitochondrial-targeting peptide. Therefore, considering the targeting function and subcellular localization of these CMS-associated genes, the function of orf288 in male sterility in the hau CMS system may be distinct from that of orf239 in the common bean or orf129 in wild beets.
Restoring genes are imperative for the production of hybrid seeds in breeding and are useful for the characterization of cytoplasmic sterile genes. orf288 has been detected in the mitochondrial genome of a wild B. tournefortii plant that is male fertile. Therefore, it was predicted that this plant may possess hau CMS-restoring genes. This work provides convincing evidence that orf288 disrupts stamen development in transgenic plants, but the mechanism that results in this phenotype currently is not clear. Therefore, the nuclear gene expression profiles of flowers of the hau CMS line are now being compared with those of the maintainer line using microarray analysis to investigate further the mechanism of male sterility in hau CMS.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Map of the pM999GFP vector.
Figure S2. Multiple alignment of DNA sequences of orf288, orf286, and orf263
Table S1. Primers used in the study.
Acknowledgments
We greatly acknowledge the efforts of Mayank Gautam in reading through the manuscript. We also appreciate the anonymous reviewers for their valuable comments and suggestions on the manuscript. This work was supported by the National Key Basic Research Special Foundation of China ‘973’ Project (grant no. 2007CB109005) and High-Tech Program ‘863’ (grant no. 2011AA10A104).
Glossary
Abbreviations
- AP3
APETALA3
- BT
Boro II
- CaMV
Cauliflower mosaic virus
- CMS
cytoplasmic male sterility
- coxIV
cytochrome oxidase subunit IV
- CR
circularized RNA
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- IPTG
isopropyl-β-D-thiogalactopyranoside
- OD
optical density
- ORF
open reading frame
- PET1
Helianthus petiolaris cytoplasm based male-sterile line in sunflower
- Rf
fertility restorer
- RFLP
restriction fragment length polymorphism
- RT-PCR
reverse transcription-PCR
- WA
wild abortive
References
- Abad AR, Mehrtens BJ, Mackenzie SA. Specific expression in reproductive tissues and fate of a mitochondrial sterility-associated protein in cytoplasmic male-sterile bean. The Plant Cell. 1995;7:271–285. doi: 10.1105/tpc.7.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi H, Nakamura A, Yokozeki-Misono Y, Inagaki A, Takahashi H, Mori K, Fujimura T. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theoretical and Applied Genetics. 2004;108:1449–1457. doi: 10.1007/s00122-004-1591-2. [DOI] [PubMed] [Google Scholar]
- Ashutosh Kumar P, Dinesh Kumar V, Sharma PC, Prakash S, Bhat SR. A novel orf108 co-transcribed with the atpA gene is associated with cytoplasmic male sterility in Brassica juncea carrying Moricandia arvensis cytoplasm. Plant and Cell Physiology. 2008;49:284–289. doi: 10.1093/pcp/pcm182. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Grelon M, Pelletier G, Budar F. The restorer Rfo gene acts post-translationally on the stability of the ORF138 Ogura CMS-associated protein in reproductive tissues of rapeseed cybrids. Plant Molecular Biology. 1999;40:893–902. doi: 10.1023/a:1006223908044. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bonhomme S, Budar F, Lancelin D, Small I, Defrance MC, Pelletier G. Sequence and transcript analysis of the Nco2.5 Ogura-specific fragment correlated with cytoplasmic male sterility in Brassica cybrids. Molecular and General Genetics. 1992;235:340–348. doi: 10.1007/BF00279379. [DOI] [PubMed] [Google Scholar]
- Brown GG, Formanová N, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. The Plant Journal. 2003;35:262–272. doi: 10.1046/j.1365-313x.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends in Genetics. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Chaumont F, Bernier B, Buxant R, Williams ME, Levings CS, 3rd, Boutry M. Targeting the maize T-urf13 product into tobacco mitochondria confers methomyl sensitivity to mitochondrial respiration. Proceedings of the National Academy of Sciences, USA. 1995;92:1167–1171. doi: 10.1073/pnas.92.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Das S, Sen S, Chakraborty A, Chakraborti P, Maiti MK, Basu A, Basu D, Sen SK. An unedited 1.1 kb mitochondrial orfB gene transcript in the wild abortive cytoplasmic male sterility (WA-CMS) system of Oryza sativa L. subsp. indica. BMC Plant Biology. 2010;10:39. doi: 10.1186/1471-2229-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey RE, Levings CS, 3rd, Timothy DH. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell. 1986;44:439–449. doi: 10.1016/0092-8674(86)90465-4. [DOI] [PubMed] [Google Scholar]
- Dewey RE, Siedow JN, Timothy DH, Levings CS., 3rd A 13-kilodalton maize mitochondrial protein in E.coli confers sensitivity to Bipolaris maydis toxin. Science. 1988;239:293–295. doi: 10.1126/science.3276005. [DOI] [PubMed] [Google Scholar]
- Dewey RE, Timothy DH, Levings CS. A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proceedings of the National Academy of Sciences, USA. 1987;84:5374–5378. doi: 10.1073/pnas.84.15.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich JH, Braun HP, Schmitz UK. Alloplasmic male sterility in Brassica napus (CMS ‘ Tournefortii-Stiewe’) is associated with a special gene arrangement around a novel atp9 gene. Molecular Genetics and Genomics. 2003;269:723–731. doi: 10.1007/s00438-003-0886-3. [DOI] [PubMed] [Google Scholar]
- Duroc Y, Gaillard C, Hiard S, Defrance MC, Pelletier G, Budar F. Biochemical and functional characterization of ORF138, a mitochondrial protein responsible for Ogura cytoplasmic male sterility in Brassiceae. Biochimie. 2005;87:1089–1100. doi: 10.1016/j.biochi.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Duroc Y, Gaillard C, Hiard S, Tinchant C, Berthom R, Pelletier G, Budar F. Nuclear expression of a cytoplasmic male sterility gene modifies mitochondrial morphology in yeast and plant cells. Plant Science. 2006;170:755–767. [Google Scholar]
- Duroc Y, Hiard S, Vrielynck N, Ragu S, Budar F. The Ogura sterility-inducing protein forms a large complex without interfering with the oxidative phosphorylation components in rapeseed mitochondria. Plant Molecular Biology. 2009;70:123–137. doi: 10.1007/s11103-009-9461-6. [DOI] [PubMed] [Google Scholar]
- Feng Liu, Xiangqin Cui, Harry T. Horner, Henry Weiner, Schnable PS. Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. The Plant Cell. 2001;13:1063–1078. doi: 10.1105/tpc.13.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Melendi P, Uyttewaal M, Morcillo CN, Hernandez Mora JR, Fajardo S, Budar F, Lucas MM. A light and electron microscopy analysis of the events leading to male sterility in Ogu-INRA CMS of rapeseed (Brassica napus) Journal of Experimental Botany. 2008;59:827–838. doi: 10.1093/jxb/erm365. [DOI] [PubMed] [Google Scholar]
- Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Research. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR. Plant mitochondrial mutations and male sterility. Annual Review of Genetics. 1991;25:461–486. doi: 10.1146/annurev.ge.25.120191.002333. [DOI] [PubMed] [Google Scholar]
- He S, Abad AR, Gelvin SB, Mackenzie SA. A cytoplasmic male sterility-associated mitochondrial protein causes pollen disruption in transgenic tobacco. Proceedings of the National Academy of Sciences, USA. 1996;93:11763–11768. doi: 10.1073/pnas.93.21.11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development. 1998;125:1711–1721. doi: 10.1242/dev.125.9.1711. [DOI] [PubMed] [Google Scholar]
- Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. The Plant Journal. 2008;55:619–628. doi: 10.1111/j.1365-313X.2008.03529.x. [DOI] [PubMed] [Google Scholar]
- Kim DH, Kang JG, Kim BD. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.) Plant Molecular Biology. 2007;63:519–532. doi: 10.1007/s11103-006-9106-y. [DOI] [PubMed] [Google Scholar]
- Köhler RH, Zipfel WR, Webb WW, Hanson MR. The green fluorescent protein as a marker to visualize plant mitochondria in vivo. The Plant Journal. 1997;11:613–621. doi: 10.1046/j.1365-313x.1997.11030613.x. [DOI] [PubMed] [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. The Plant Journal. 2003;34:407–415. doi: 10.1046/j.1365-313x.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- Krishnasamy S, Makaroff CA. Characterization of the radish mitochondrial orfB locus: possible relationship with male sterility in Ogura radish. Current Genetics. 1993;24:156–163. doi: 10.1007/BF00324680. [DOI] [PubMed] [Google Scholar]
- Kuhn J, Binder S. RT-PCR analysis of 5' to 3'-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Research. 2002;30:439–446. doi: 10.1093/nar/30.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Homme Y, Stahl RJ, Li XQ, Hameed A, Brown GG. Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing a chimeric gene similar to the pol CMS-associated orf224 gene. Current Genetics. 1997;31:325–335. doi: 10.1007/s002940050212. [DOI] [PubMed] [Google Scholar]
- Landgren M, Zetterstrand M, Sundberg E, Glimelius K. Alloplasmic male-sterile Brassica lines containing B. tournefortii mitochondria express an ORF 3' of the atp6 gene and a 32 kDa protein. Plant Molecular Biology. 1996;32:879–890. doi: 10.1007/BF00020485. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee YP, Kim S, Lim H, Ahn Y, Sung SK. Identification of mitochondrial genome rearrangements unique to novel cytoplasmic male sterility in radish (Raphanus sativus L.) Theoretical and Applied Genetics. 2009;118:719–728. doi: 10.1007/s00122-008-0932-y. [DOI] [PubMed] [Google Scholar]
- Li XQ, Jean M, Landry BS, Brown GG. Restorer genes for different forms of Brassica cytoplasmic male sterility map to a single nuclear locus that modifies transcripts of several mitochondrial genes. Proceedings of the National Academy of Sciences, USA. 1998;95:10032–10037. doi: 10.1073/pnas.95.17.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K, Nicholas HB., Jr GeneDoc: a tool for editing and annotating multiple sequence alignments. 1997 http://www.nrbsc.org/gfx/genedoc/ [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proceedings of the International Conference on Intelligent Systems for Molecular Biology. 1998;6:122–130. [PubMed] [Google Scholar]
- Nizampatnam NR, Doodhi H, Narasimhan YK, Mulpuri S, Viswanathaswamy DK. Expression of sunflower cytoplasmic male sterility-associated open reading frame, orfH522 induces male sterility in transgenic tobacco plants. Planta. 2009;229:987–1001. doi: 10.1007/s00425-009-0888-4. [DOI] [PubMed] [Google Scholar]
- Petsalaki E, Bagos P, Litou Z, Hamodrakas S. PredSL: a tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics, Proteomics and Bioinformatics. 2006;4:48–55. doi: 10.1016/S1672-0229(06)60016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarria R, Lyznik A, Vallejos CE, Mackenzie SA. A cytoplasmic male sterility-associated mitochondrial peptide in common bean is post-translationally regulated. The Plant Cell. 1998;10:1217–1228. doi: 10.1105/tpc.10.7.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Brown GG. Characterization of expression of a mitochondrial gene region associated with the Brassica ‘Polima’ CMS: developmental influences. Current Genetics. 1993;24:316–322. doi: 10.1007/BF00336783. [DOI] [PubMed] [Google Scholar]
- Stahl R, Sun S, L'Homme Y, Ketela T, Brown GG. RNA editing of transcripts of a chimeric mitochondrial gene associated with cytoplasmic male-sterility in Brassica. Nucleic Acids Research. 1994;22:2109–2113. doi: 10.1093/nar/22.11.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiewe G, Röbbelen G. Establishing cytoplasmic male sterility in Brassica napus by mitochondrial recombination with B. tournefortii. Plant Breeding. 1994;113:294–304. [Google Scholar]
- Wan ZJ, Jing B, Tu JX, Ma CZ, Shen JX, Yi B, Wen J, Huang T, Wang XJ, Fu TD. Genetic characterization of a new cytoplasmic male sterility system (hau) in Brassica juncea and its transfer to B. napus. Theoretical and Applied Genetics. 2008;116:355–362. doi: 10.1007/s00122-007-0673-3. [DOI] [PubMed] [Google Scholar]
- Wang ZH, Zou Y, Li X, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. The Plant Cell. 2006;18:676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Bronson CR, Schnable PS, Horner HT. The genetics, pathology, and molecular biology of T-cytoplasm male sterility in maize. Advances in Agronomy. 1999;65:79–130. [Google Scholar]
- Wise RP, Pring DR, Gengenbach BG. Mutation to male fertility and toxin insensitivity in Texas (T)-cytoplasm maize is associated with a frameshift in a mitochondrial open reading frame. Proceedings of the National Academy of Sciences, USA. 1987;84:2858–2862. doi: 10.1073/pnas.84.9.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto MP, Shinada H, Onodera Y, Komaki C, Mikami T, Kubo T. A male sterility-associated mitochondrial protein in wild beets causes pollen disruption in transgenic plants. The Plant Journal. 2008;54:1027–1036. doi: 10.1111/j.1365-313X.2008.03473.x. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.