Abstract
The inflorescence of flowering plants is a highly organized structure, not only contributing to plant reproductive processes, but also constituting an important part of the entire plant morphology. Previous studies have revealed that the class-I KNOTTED1-like homeobox (KNOX) genes BREVIPEDICELLUS (BP or KNAT1), KNAT2, and KNAT6 play essential roles in inflorescence architecture. Pedicel morphology is known to contribute greatly to inflorescence architecture, and BP negatively regulates KNAT2 and KNAT6 to ensure that pedicels have a normal upward-pointing orientation. These findings indicate that a genetic network exists in controlling pedicel orientation, but how this network functions in the developmental process remains elusive. Here it is reported that the ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) gene, which belongs to the BELL1-like homeodomain gene family, is a new member participating in regulating pedicel orientation in the class-I KNOX network. In a genetic screening for suppressors of isoginchaku-2D, a gain-of-function ASYMMETRIC LEAVES2 mutant that displays downward-pointing pedicels, a suppressor mutant was obtained. Characterization of this mutant revealed that the mutation corresponds to ATH1. Genetic analysis indicated that ATH1 acts mainly in the KNAT2 pathway. Yeast two-hybrid and bimolecular fluorescence complementation assays demonstrated that ATH1 physically interacts with KNAT2. The data indicate that the ATH1–KNAT2 complex acts redundantly with KNAT6, both of which are negatively regulated by BP during pedicel development.
Keywords: Arabidopsis, ATH1, BP, inflorescence architecture, KNAT2
Introduction
In flowering plants, the inflorescence is a highly organized structure bearing flowers connected by pedicels. The pedicel characteristics are one of the key contributors to the display of the whole inflorescence architecture, which is highly diverse among flowering plant species (Douglas et al., 2002; Venglat et al., 2002). In recent years, inflorescence architecture, especially pedicel development, has been studied extensively. In Arabidopsis, several members in the class-I KNOTTED1-like homeobox (KNOX) gene family were reported to play central roles in regulating pedicel development (Douglas et al., 2002; Venglat et al., 2002; Ragni et al., 2008).
The class-I KNOX genes in Arabidopsis comprise four members, namely SHOOT MERISTEMLESS (STM), BREVIPEDICELLUS (BP, also called KNAT1), KNAT2, and KNAT6 (Hake et al., 2004). A loss-of-function bp mutant exhibits defective inflorescence architecture, with downward-pointing pedicels (Douglas et al., 2002; Venglat et al., 2002). Consistent with these abnormal bp phenotypes, BP is strongly expressed in pedicels, as well as in other inflorescence parts such as young flowers (Lincoln et al., 1994; Douglas et al., 2002; Alonso-Cantabrana et al., 2007). A genetic study revealed that the pedicel phenotype of bp is caused by increased expression of two additional class-I KNOX genes, KNAT2 and KNAT6, in pedicels, and double mutations in these two genes in the bp background fully rescue the pedicel phenotype caused by the bp single mutation (Ragni et al., 2008). BP and KNAT2 are negatively regulated by two leaf development-controlling genes, ASYMMETRIC LEAVES1 (AS1) and AS2. These two genes encode transcription factors that form a protein complex (Xu et al., 2003) to regulate BP and KNAT2 directly in leaves (Guo et al., 2008). Overexpression of AS2 strongly represses BP in the inflorescence, resulting in downward-pointing pedicels, mimicking those in the bp mutant (Lin et al., 2003; Xu et al., 2003). Although recent progress has greatly improved our understanding of pedicel development, a detailed network for such regulation remains unresolved. In the current work it is reported that the ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) gene, which plays an important role in the KNAT2 pathway, regulates pedicel development.
ATH1 is a member of the BELL1-like (BELL) transcription factor subfamily, which together with the KNOX subfamily belongs to the three-amino-acid-loop-extension (TALE) superfamily (Hake et al., 2004). In Arabidopsis, the BELL subfamily comprises 13 members (Hamant and Pautot, 2010). It was reported that two other members, PENNYWISE (PNY, also called BELLRINGER and REPLUMLESS) and POUND-FOOLISH (PNF), act redundantly in flowering initiation and inflorescence architecture. Both PNY and PNF proteins are able to form heterodimers with class-I KNOX proteins (Byrne et al., 2003; Smith and Hake, 2003; Smith et al., 2004). The ATH1 gene was first identified as a target of the photomorphogenic genes CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and DEETIOLATED1 (DET1), as ATH1 was derepressed in cop1 and det1 mutants (Quaedvlieg et al., 1995). In addition, the ath1 mutant displayed elongated rosette internodes, similar to those of the photoreceptor mutants phytochromeA (phyA), phyB, and cryptochrome1 (Devlin et al., 1996; Mazzella et al., 2000). Further studies revealed that ATH1 functions in multiple developmental processes. For example, ATH1 is required for development of the basal boundaries of shoot organs (Gomez-Mena and Sablowski, 2008), shoot apical meristem activity (Rutjens et al., 2009), and repression of flowering via activation of FLOWERING LOCUS C (FLC) (Proveniers et al., 2007). Moreover, ectopic expression of ATH1 resulted in plants with irregular internodes (Cole et al., 2006; Gomez-Mena and Sablowski, 2008; Rutjens et al., 2009).
In this study, it is reported that ath1 is a suppressor of the AS2 overexpression lines, which produce shortened, downward-pointing pedicels. It is demonstrated that ATH1 acts in the KNAT2 pathway to regulate pedicel development. Furthermore, it is shown that ATH1 physically interacts with KNAT2, and the ATH1–KNAT2 protein complex is required for normal pedicel morphology.
Materials and methods
Plant materials and growth conditions
Seeds of isoginchaku-2D (iso-2D), as2-5D, ath1-1, bp-9, knat6-1, knat2-5, knat6-1 bp-9, and knat2-5 bp-9 were kindly provided by M. Matsui, R. S. Poethig, R. Sablowski, S. Hake, and V. Pautot, respectively (Mele et al., 2003; Nakazawa et al., 2003; Gomez-Mena and Sablowski, 2008; Ragni et al., 2008; Wu et al., 2008). Seeds of pBP:GUS and p35S:BP were obtained from the Arabidopsis Biological Resource Center (ABRC). Plants were grown on soil as previously described (Chen et al., 2000).
Plant genetics and map-based cloning
To construct double and triple mutants, candidate plants in the F2 progeny of each cross were genotyped by polymerase chain reaction (PCR). To generate iso-2D suppressors, >4000 iso-2D seeds (Columbia-0) were mutagenized with ethyl methanesulphonate (0.2%). The mutagenized seeds (M1) were planted in soil, and plants with impaired iso-2D phenotypes were identified in the M2 generation. Mapping of the suppressor locus was performed by analysis of an F2 population from a cross between one of the suppressor mutants and the polymorphic Landsberg erecta (Ler) plants. The suppressor locus was mapped to the proximal arm of chromosome 4, between two simple sequence length polymorphim (SSLP) markers nga1139 and F1N20 in a 4700 kb region. Because the ATH1 locus is located in this region and the ath1 mutants show similar phenotypes to certain plants in the F2 mapping population, the ATH1 locus was thus sequenced and it was confirmed that ath1 is the suppressor of the iso-2D phenotypes (see Results).
Quantitative reverse transcription-PCR (qRT-PCR)
RNA extraction was performed as described previously (Xu et al., 2003) using inflorescences from plants ∼5 weeks old, and reverse transcription was performed using a kit (Fermentas, Lithuania). Quantitative PCR was performed in the presence of the double-stranded DNA-specific dye SYBR green following the manufacturer’s instructions (TOYOBO, Japan), with the following gene-specific primers: 5′-CCTCCAAACCGTTTCCTTCT-3′ and 5′-TTTA TGCATTGCTTGGCTCATCA-3′ for ATH1; 5′-CTTTGAGGCT CGACAACA-3′ and 5′-TAATGCAACTCCCACCAC-3′ for BP; 5′-GAACTCGCTACCGCTTTGTCCTC-3′ and 5′-ATCGCGGTCAT TGCTTCTTTGT-3′ for KNAT2; 5′-CGAGTCAGACAAGAA CTC-3′ and 5′-GGATCTCTACATGCAAGC-3′ for KNAT4; 5′-CTCCGCCGGTGAAAATCGTGT-3′ and 5′-GGTTCCGTAGC TGCATCTCAATCT-3′ for KNAT6; 5′-ATGAAAGAGAGACAA CGTTGG-3′ and 5′-GGGGCGGTCTAATCTGCAA-3′ for AS1; 5′-ATGGCATCTTCTTCAACAAAC-3′ and 5′-AGACGGATCAA CAGTACGGC-3′ for AS2; and 5′-TGGCATCA(T/C)ACTTTCTACAA-3′ and 5′-CCACCACT(G/A/T)AGCACAATGTT-3′ for ACTIN.
Yeast two-hybrid analysis
Full-length cDNA fragments of KNAT2 and KNAT6 and the N-terminal portion of ATH1 were PCR-amplified using the following primers: 5′-agatctATGGATAGAATGTGTGGTTTCC-3′ and 5′-gtcgacCTCGGTAAAGAATGTTTCATT-3′ for KNAT2; 5′-agatctATGGATGGAATGTACAATTTCC-3′ and 5′-gtcgacTTCCTC GGTAAAGAATGATCCA-3′ for KNAT6; and 5′-gagctcggatccATGGACAACAACAACAACAAC-3′ and 5′-gtcgacTTAAGGT CTCCAAATCTGATGGTTC 3′ for the ATH1 N-terminal portion. In each of the above primer sequences, the lower case letters represent additional nucleotides to introduce restriction sites. All constructs were verified by sequencing. Construct combinations pGADT7-KNAT2/pGBKT7-ATH1-N and pGADT7-KNAT6/pGBKT7-ATH1-N were co-transformed into the yeast strain PJ69-4A, and the interaction between the proteins was determined according to the manufacturer’s recommendation (Clontech, USA).
Transient protein expression and bimolecular fluorescence complementation (BiFC) assay
Full-length cDNA fragments of ATH1, KNAT2, and KNAT6 were subcloned into the plant transformation vector pC131 under the control of a 35S promoter and with a 3′ in-frame fusion to sequences encoding yellow fluorescent protein (YFP), using the following primers: 5′-gagctcggatccATGGACAACAACAACAACAAC-3′ and 5′-tctagagtcgacTTTATGCATTGCTTGGCTCATC-3′ for ATH1; 5′-agatctATGGATAGAATGTGTGGTTTCC-3′ and 5′-gtcgacCTCG GTAAAGAATGTTTCATT-3′ for KNAT2; and 5′-agatctATGGAT GGAATGTACAATTTCC-3′ and 5′-gtcgacTTCCTCGGTAAAGAATGATCCA-3′ for KNAT6. Fragments of YFP were truncated at residue 155 (designated YN and YC) as previously described (Kerppola, 2006), using the following primers: 5′-gtcgacGGAGGAGGCTCAGCGGACTACAAAGATGACGATGACAAAATGGTGAGCAAGGGCGAGGA-3′ and 5′-gagctcTTAGGCCATGATAT AGACGTTGT-3′ for YN; and 5′-gtcgacGGAGGAGGCTCAGCGGACTACAAAGATGACGATGACAAAGACAAGCAGAAGAAC GGCAT-3′ and 5′-gagctcTTACTTGTACAGCTCGTCCATGC-3′ for YC. In each of the above primer sequences, the lower case letters represent additional nucleotides to introduce restriction sites; and the italic letters represent a FLAG tag. Sequences between restriction sites and the FLAG tag encode the GGGS linker peptide to facilitate the association between target proteins and YN/YC as previously described (Kerppola, 2006). All constructs were verified by sequencing. To construct BiFC plasmids, the YN fragment was inserted into the C-terminus of KNAT2 and KNAT6, and the YC into the C terminus of ATH1 of the above constructs to replace the YFP fragment. Leaves of 4- to 8-week-old Nicotiana benthamiana plants were co-infiltrated with strains containing P19, a viral silencing suppressor gene (Voinnet et al., 2003), and localization of the BiFC fluorescence was observed 2–7 d after infiltration using a confocal laser scanning microscope (LSM 510 META, ZEISS, Germany).
Results
Genetic screening for suppressors of the AS2 overexpression line iso-2D
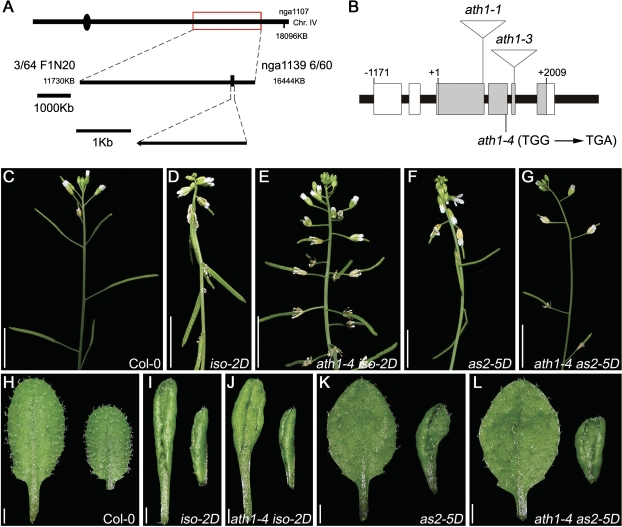
Previous data showed that overexpression of the Arabidopsis AS2 gene results in two types of abnormal phenotypes: (i) all leaves become adaxialized and are curled upwards; and (ii) inflorescences produce shortened and downward-pointing pedicels, which are similar to those of the bp mutants (Douglas et al., 2002; Venglat et al., 2002; Lin et al., 2003; Xu et al., 2003). To identify the regulatory network relating to the abnormal pedicel phenotypes when AS2 is overexpressed, a genetic screening for suppressors of the AS2 overexpression phenotypes was conducted, using a stable AS2 overexpression line, iso-2D (Nakazawa et al., 2003). One mutant showing compromise of the downward-pointing pedicel was identified, and was further crossed with the wild-type Col-0 to obtain the suppressor single mutant. The isolated single mutant showed similar phenotypes to those of a previously reported mutant arabidopsis thaliana homeobox gene1 (ath1) (Quaedvlieg et al., 1995; Gomez-Mena and Sablowski, 2008). In addition, the mutation locus was mapped to a region between genetic markers F1N20 and nga1139 on chromosome 4, where the ATH1 locus is positioned (Fig. 1A). The ATH1 locus in the suppressor mutant was thus sequenced and a single nucleotide substitution from G to A was found in the second exon, resulting in an earlier stop codon in the ATH1 gene (Fig. 1B). In addition, an allelism test was performed by crossing the suppressor mutant to the previously characterized ath1-1 mutant (Proveniers et al., 2007; Gomez-Mena and Sablowski, 2008), and all F1 plants showed the ath1 phenotypes (Supplementary Fig. S1 available at JXB online). Hence, it is concluded that the suppressor is a new ath1 allele, which was renamed ath1-4.
Fig. 1.
Abnormal pedicel phenotypes caused by AS2 overexpression were impaired in the ath1 mutant background. (A) Map-based cloning to localize an AS2 suppressor gene on chromosome 4, between markers F1N20 and nga1139. (B) Structure of the ATH1 gene. Grey and white boxes indicate protein-coding regions and untranslated regions (UTRs), respectively. (C–G) Inflorescence structures of wild type (C), iso-2D (D), ath1-4 iso-2D (E), as2-5D (F), and ath1-4 as2-5D (G). (H–L) The fifth (left) and sixth (right) rosette leaves of 19-day-old wild-type (H), iso-2D (I), ath1-4 iso-2D (J), as2-5D (K), and ath1-4 as2-5D (L) plants. Note that only phenotypes of the downward-pointing pedicels but not the up-curled rosette leaves in iso-2D and as2-5D were rescued in ath1-4 iso-2D and ath1-4 as2-5D. Bars=1 cm in C–G, and 0.1 cm in H–L.
ATH1 plays an important role in pedicel development
Wild-type Arabidopsis plants form inflorescences that bear flowers and fruits with upward-pointing pedicels (Fig. 1C), whereas pedicels from the iso-2D inflorescence are drastically shortened with a downward-pointing orientation (Fig. 1D). Compared with the wild-type and iso-2D inflorescences, the pedicel orientation of the ath1-4 iso-2D inflorescence was partially rescued, showing a horizontal or only slightly downward-pointing orientation (Fig. 1E). In addition, the shortened pedicels in iso-2D were also largely rescued in the ath1-4 iso-2D double mutant (Fig. 1E). To confirm further that removal of the ATH1 gene can rescue the pedicel phenotypes of AS2 overexpression, ath1-4 was crossed to another AS2 overexpression allele, as2-5D (Fig. 1F) (Wu et al., 2008), and a previously generated p35S:AS2 transgenic line, which has the typical bp-like pedicel phenotypes (Xu et al., 2003). Both ath1-4 as2-5D (Fig. 1G) and p35S:AS2/ath1-4 (data not shown) plants displayed the rescued pedicels, similar to ath1-4 iso-2D. These results indicate that ATH1 plays a role in formation of normal pedicel morphology. Although the abnormal pedicel phenotype was rescued in ath1-4 iso-2D and ath1-4 as2-5D, the up-curled rosette leaves caused by the iso-2D and as2-5D mutations remained hyponastic (Fig. 1H–L). These results indicate that the ATH1 function is required only for the pathway controlling pedicel morphology, but not for the pathway regulating leaf polarity establishment.
Rescue of pedicel phenotypes in ath1 iso-2D is not because of recovery of BP expression
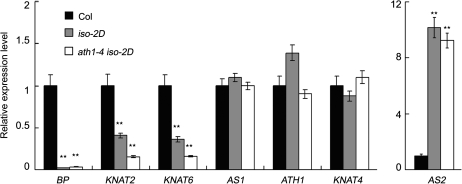
Because AS2 overexpression is known to repress BP in the inflorescence (Lin et al., 2003; Xu et al., 2003) and the bp mutant itself bears downward-pointing pedicels, it was hypothesized that the BP expression level might be recovered in the ath1-4 iso-2D inflorescence. Expression of BP and several other related genes was thus analysed in iso-2D single and ath1-4 iso-2D double mutants by qRT-PCR. Surprisingly, although the downward-pointing pedicel phenotypes of iso-2D were suppressed in the ath1-4 iso-2D plants, the BP expression remained at a very low level in ath1-4 iso-2D, which was markedly below the wild-type level (Fig. 2). On the other hand, levels of KNAT2 and KNAT6 transcripts were also reduced in both iso-2D and ath1-4 iso-2D compared with those in the wild type. As controls, while the AS2 transcript level was elevated in both iso-2D and ath1-4 iso-2D, the AS1, ATH1, and KNAT4 levels in iso-2D and ath1-4 iso-2D showed no significant changes compared with those in the wild type (Fig. 2). To confirm further that ath1-4 iso-2D does not affect BP expression, a pBP:GUS transgenic line was crossed to iso-2D and ath1-4 iso-2D, respectively, to generate the isogenic pBP:GUS/iso-2D and pBP:GUS/ath1-4 iso-2D sibling plants for further analyses. In wild-type plants, β-glucuronidase (GUS) staining accumulated in the pedicel, especially at the junction between the pedicel and flower (Supplementary Fig. S2A, D at JXB online). In contrast, GUS signals were barely detected in inflorescences and flowers of pBP:GUS/iso-2D and pBP:GUS/ath1-4 iso-2D plants (Supplementary Fig. S2B, C, E, F), consistent with the qRT-PCR results.
Fig. 2.
qRT-PCR analysis of transcript levels of BP, KNAT2, KNAT6, AS1, ATH1, KNAT4, and AS2 in wild-type Col-0, iso-2D, and ath1-4 iso-2D inflorescences. Quantification was normalized to that of ACTIN and then to the value of wild-type Col-0, whose value was arbitrarily fixed at 1.0. Each cDNA sample was made in triplicate, and the consistent results from two separately prepared RNA samples were used. Bars show the standard deviation, and double asterisks show significant statistical differences by t-test (P < 0.01).
It was previously known that BP acts to repress KNAT2 and KNAT6 to ensure normal inflorescence architecture (Ragni et al., 2008). Therefore, either loss of BP function or loss of repression for KNAT2 and/or KNAT6 could result in the downward-pointing pedicel phenotype. Whether overexpression of BP could rescue the downward-pointing pedicel in AS2 overexpression lines was investigated by crossing a p35S:BP transgenic plant to iso-2D and as2-5D mutants, respectively, and the F1 isogenic populations were analysed. Compared with those of iso-2D/+ and as2-5D/+ (Fig. 3A, C), the downward-pointing pedicels of both p35S:BP/+ iso-2D/+ (Fig. 3B) and p35S:BP/+ as2-5D/+ (Fig. 3D) were ameliorated. These results indicate that the pedicel phenotypes of iso-2D and as2-5D are, indeed, the result of lack of BP, and the insufficient down-regulation of KNAT2 and KNAT6 might be the major reason for the abnormal inflorescence architecture in the mutants.
Fig. 3.
The overexpression of BP rescues the pedicel phenotypes caused by gain-of-function mutations of the AS2 gene. (A–D) Inflorescence structures of iso-2D/+ (A), iso-2D/+ p35S:BP/+ (B), as2-5D/+ (C), and as2-5D/+ p35S:BP/+ (D). Note that the abnormal pedicel phenotypes caused by overexpression of AS2 were rescued in iso-2D/+ p35S:BP/+ and as2-5D/+ p35S:BP/+ plants. Note that iso-2D/+, p35S:BP/+, and as2-5D/+ indicates heterozygous for iso-2D, p35S:BP, and as2-5D, respectively. Bars=1 cm. (This figure is available in colour at JXB online.)
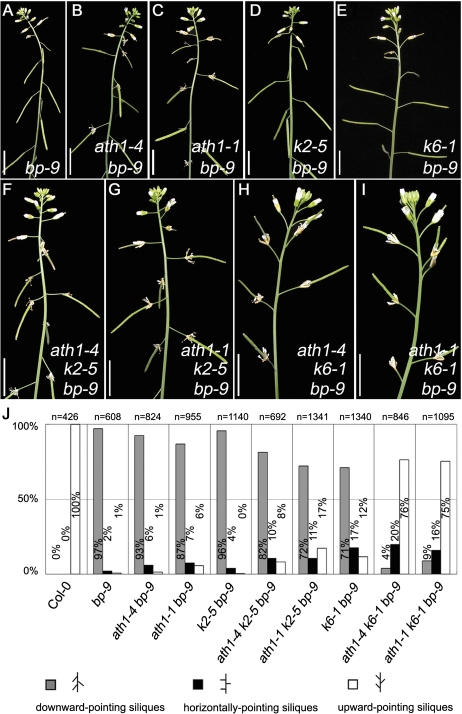
Removal of both ATH1 and KNAT6 rescues the bp inflorescence phenotype
To investigate further genetic interaction between BP and ATH1 in morphological control, ath1-4 and ath1-1 were introduced into the bp-9 mutant, which produces downward-pointing pedicels. The abnormal bp-9 inflorescence (Fig. 4A, J) was almost unaffected in ath1-4 bp-9 and ath1-1 bp-9 double mutants (Fig. 4B, C, J). It was previously reported that knat6, but not knat2, can partially rescue the bp downward-pointing pedicel phenotype, and the knat2 knat6 bp triple mutant produces completely normal upward-pointing pedicels (Ragni et al., 2008) (Fig. 4D, E, J). The present genetic data showed that the pedicel phenotype of both ath1-4 knat2-5 bp-9 and ath1-1 knat2-5 bp-9 triple mutants (Fig. 4F, G, J) was similar to those in the knat2-5 bp-9 double mutant (Fig. 4D, J), in which most pedicels were downward pointing. Remarkably, both ath1-4 knat6-1 bp-9 and ath1-1 knat6-1 bp-9 (Fig. 4H, I, J) substantially rescued the bp-9 pedicel phenotype, with only a small proportion of pedicels showing the horizontal or downward-pointing phenotype. These results strongly suggest that ATH1 and KNAT2 might function in the same pathway, which is separate from the KNAT6 pathway, to regulate pedicel phenotype.
Fig. 4.
bp pedicel phenotypes were rescued in the ath1 and knat6 double mutation backgrounds. (A–I) Inflorescence structures of bp-9 (A), ath1-4 bp-9 (B), ath1-1 bp-9 (C), knat2-5 bp-9 (D), knat6-1 bp-9 (E), ath1-4 knat2-5 bp-9 (F), ath1-1 knat2-5 bp-9 (G), ath1-4 knat6-1 bp-9 (H), and ath1-1 knat6-1 bp-9 (I). (J) Statistical analysis of pedicel orientation. ‘n’ indicates the number of pedicels scored. k2-5, knat2-5; k6-1, knat6-1. Bars=1 cm in A–I. (This figure is available in colour at JXB online.)
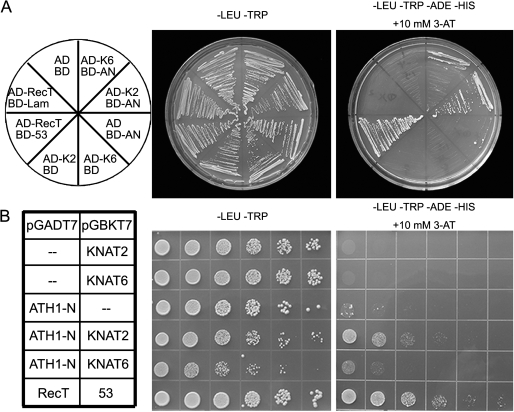
ATH1 physically interacts with KNAT2
Several recent studies revealed that, through their N-terminal domain, a number of BELL family proteins are able to form complexes with KNOX family proteins (Bellaoui et al., 2001; Muller et al., 2001; Smith et al., 2002). To determine whether ATH1 physically interacts with KNAT2, a yeast two-hybrid assay was performed. The data showed that co-expression of the N-terminal domain of ATH1 and full-length KNAT2 promoted expression of the reporter genes, resulting in cells able to grow on media lacking tryptophan, leucine, adenine, and histidine (Fig. 5A, B). However, in the present experimental conditions, the protein–protein interaction by co-expression of the N-terminal domain of ATH1 and full-length KNAT6 appeared very weak, as compared with the RecT-Lam and RecT-53 negative and positive controls, respectively (Fig. 5A, B).
Fig. 5.
ATH1 physically interacts with KNAT2 and KNAT6 in yeast cells. (A, B) Yeast two-hybrid assay shows that the N-terminal domain of ATH1 is able to interact with KNAT2, while the interaction between the N-terminal domain of ATH1 and KNAT6 appeared very weak. The RecT-Lam and RecT-53 pair served as the negative and positive controls, respectively. Yeast cultures with a 1:10 dilution series were plated on the media from left to right (B). K2, KNAT2; K6, KNAT6; and AN, N-terminal domain of ATH1.
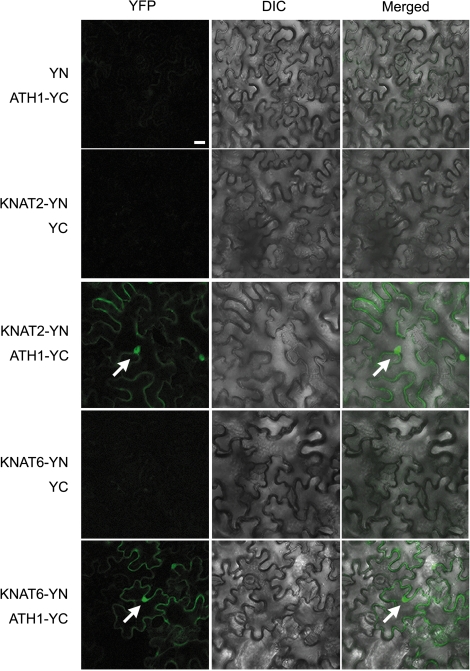
A BiFC assay was performed to investigate further the interaction between ATH1 and KNAT2 or KNAT6 in planta. The KNAT2–YN and ATH1–YC pair or the KNAT6–YN and ATH1–YC pair was co-expressed in Nicotiana benthamiana leaves, with YN and ATH1–YC, KNAT2–YN and YC, and KNAT6–YN and YC pairs serving as negative controls (Fig. 6). The data showed that the presence of KNAT2–YN and ATH1–YC, or KNAT6–YN and ATH1–YC, in tobacco cells produced YFP signals in both the cytoplasm and nuclei (Fig. 6), whereas all the negative controls displayed no fluorescence signal. These results indicate that ATH1 potentially has the ability to bind KNAT2 and KNAT6 in plant cells.
Fig. 6.
Bimolecular fluorescence complementation (BiFC) assay shows that ATH1 is able to associate with both KNAT2 and KNAT6. White arrows indicate nuclei. All images are of the same magnification. DIC, differential interference contrast; YN, N-terminal domain of YFP; YC, C-terminal domain of YFP. Bars=20 μm.
Discussion
Formation of inflorescence architecture appears to be a complex developmental process, requiring a number of regulatory components, including those in the class-I KNOX genes. In this work, ATH1 functions in modulating pedicel morphology are reported, adding a new factor to the present regulatory network of inflorescence architecture. In addition, the data reveal the protein–protein interaction between ATH1 and KNAT2, and that the protein complex may act to fulfil the task of regulating pedicel development.
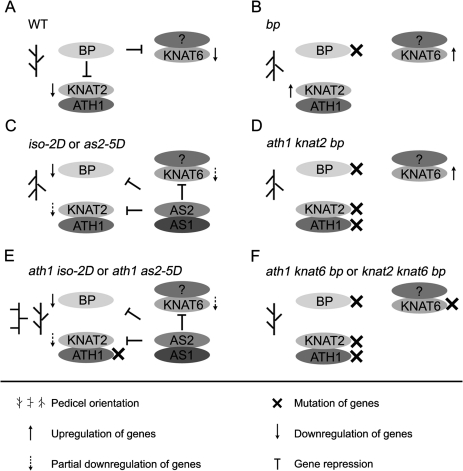
Based on previous data and the results obtained in this study, genetic action models for genes that regulate pedicel phenotypes are proposed (Fig. 7). According to the previous model, for a normal pedicel shape the BP gene must down-regulate two redundant genes, KNAT2 and KNAT6 (Fig. 7A) (Ragni et al., 2008). In contrast, the bp mutation causes derepression of both KNAT2 and KNAT6, and the increased expression of these two genes resulted in the downward-pointing pedicels (Ragni et al., 2008) (Fig. 7B). In the iso-2D and as2-5D mutants, the increased AS1–AS2 function represses BP, KNAT2, and KNAT6. However, because down-regulation of BP in turn derepresses KNAT2 and KNAT6, the transcript levels of these two genes are only reduced moderately (Fig. 7C). The data from gene expression analyses also reveal that the downward-pointing pedicel phenotype relies not only on the increased KNAT2 and KNAT6 transcripts, but, more importantly, the functional balance between BP and KNAT2/6. Both KNAT2 and KNAT6 transcript levels were actually reduced in iso-2D; however, because the BP level in iso-2D was even more severely reduced, the downward-pointing pedicel phenotype of the iso-2D mutant is evident.
Fig. 7.
Action models for ATH1, BP, KNAT2, KNAT6, and AS1-AS2 in regulation of pedicel orientation.
In the ath1 knat2 bp triple mutants, removal of KNAT2 is equivalent to removal of ATH1, and the KNAT6 transcripts must increase due to bp mutation. Therefore, the pedicel orientation is either downward pointing or only slightly recovered (Fig. 7D). In ath1 iso-2D and ath1 as2-5D double mutants, BP is repressed and the KNAT2 pathway is blocked completely because of the ath1 mutation (Fig. 7E). In this case, the reduced KNAT6 function only weakly affects pedicel orientation, and the pedicels display a horizontal or normal orientation. Finally, the ath1 knat6 bp triple mutation (Fig. 7F) is equivalent to the knat2 knat6 bp triple mutation, in which pedicel defects caused by the bp mutation could be largely or completely rescued. It would be interesting to validate the models by changing KNAT2 and KNAT6 expression levels in the bp, iso-2D, or as2-5D backgrounds in the future.
Heterodimers between several BELL and class-I KNOX proteins have demonstrated important roles in regulating plant inflorescence architecture or other developmental processes. The identified heterodimers include PNY–BP, PNF–BP, PNY–STM, and PNF–STM (Byrne et al., 2003; Smith and Hake, 2003; Bhatt et al., 2004; Kanrar et al., 2006). In this study, the results of genetic analyses and yeast two-hybrid and BiFC assays also support the assumption that ATH1 and KNAT2 form a heterodimer to regulate pedicel development. It was found that ATH1 and KNAT6 can also form a protein complex, although the protein–protein interaction in the yeast two-hybrid assay was fairly weak. It is proposed that, because of their similar protein structure, heterodimer formation between the BELL and the KNOX family proteins is common in artificial experimental conditions. However, endogenous protein dimerization may follow different rules. To determine the endogenous protein complex for the two families, genetic evidence is important. Based on the genetic analysis, pedicel phenotypes of ath1 knat6 bp are almost normal as compared with that of the bp mutant, whereas the abnormal pedicels in ath1 knat2 bp were only very weakly rescued. These results indicate that, although the BiFC assay showed ATH1 and KNAT6 interaction, this heterodimer may not exist in planta or is not the major player in regulation of pedicel morphology.
Interaction between KNOX and BELL proteins in plants was proposed to guide correct protein subcellular localization. For example, the nuclear localization of STM relies on its interacting with the BELL proteins ATH1, PNY, and BEL1-like homeodomain 3 (BLH3) (Cole et al., 2006; Rutjens et al., 2009). Likewise, previous studies showed that cellular localization of PNY also relies on protein interaction with its KNOX homeodomain partner (Bhatt et al., 2004). It is possible that the established ATH1–KNAT2 dimer may bring the protein complex to its correct subcellular position. The heterodimer may also help to recognize the promoter sequence of specific downstream genes during inflorescence development. A recent study showed that molecular regulation of the haploid–diploid transition in the unicellular green soil alga Chlamydomonas reinhardti requires functioning of the Gsp1–Gsm1 protein heterodimer. Gsp1 and Gsm1 correspond to the Arabidopsis BELL and KNOX proteins, respectively. These two proteins are contributed by gametes of plus and minus mating types, respectively, physically interact, and translocate from the cytosol to the nucleus upon gametic fusion (Lee et al., 2008). Differing from the monomer Gsp1 and Gsm1 in the gametes, this heterodimer in a diploid background initiates gamete development, probably through recognizing and regulating distinct targets. Although ATH1 appears to influence multiple aspects of plant development, its roles may be defined by the presence and function of specific interacting partners (e.g. ATH1/KNAT2 in pedicel development).
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The iso-2D suppressor corresponds to the ATH1 gene.
Figure S2. pBP:GUS staining in wild-type Col-0 (A, D), iso-2D (B, E), and ath1-4 iso-2D (C, F).
Acknowledgments
We thank M. Matsui, R. S. Poethig, R. Sablowski, S. Hake, V. Pautot, and the ABRC for providing Arabidopsis seeds, J. Huang for marker primers of genetic mapping, Y. Fang for YFP vector, J. Zhang for helpful discussion about the BiFC experiment, and X. Gao for confocal observations. This work was supported by grants from the Chief Scientist Program of Shanghai Institutes for Biological Sciences, the Chinese National Scientific Foundation (31071064), and Shanghai Institutes for Biological Sciences (2009KIP205).
References
- Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrandiz C, Martinez-Laborda A. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development. 2007;134:2663–2671. doi: 10.1242/dev.02864. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. The Plant Cell. 2001;13:2455–2470. doi: 10.1105/tpc.010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. VAAMANA—a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene. 2004;328:103–111. doi: 10.1016/j.gene.2003.12.033. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Groover AT, Fontana JR, Martienssen RA. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development. 2003;130:3941–3950. doi: 10.1242/dev.00620. [DOI] [PubMed] [Google Scholar]
- Chen C, Wang S, Huang H. LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis. 2000;26:42–54. doi: 10.1002/(sici)1526-968x(200001)26:1<42::aid-gene7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Cole M, Nolte C, Werr W. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Research. 2006;34:1281–1292. doi: 10.1093/nar/gkl016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Halliday KJ, Harberd NP, Whitelam GC. The rosette habit of Arabidopsis thaliana is dependent upon phytochrome action: novel phytochromes control internode elongation and flowering time. The Plant Journal. 1996;10:1127–1134. doi: 10.1046/j.1365-313x.1996.10061127.x. [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. The Plant Cell. 2002;14:547–558. doi: 10.1105/tpc.010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mena C, Sablowski R. ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. The Plant Cell. 2008;20:2059–2072. doi: 10.1105/tpc.108.059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. The Plant Cell. 2008;20:48–58. doi: 10.1105/tpc.107.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. The role of KNOX genes in plant development. Annual Review of Cell and Developmental Biology. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- Hamant O, Pautot V. Plant development: a TALE story. Comptes Rendus Biologie. 2010;333:371–381. doi: 10.1016/j.crvi.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Kanrar S, Onguka O, Smith HM. Arabidopsis inflorescence architecture requires the activities of KNOX–BELL homeodomain heterodimers. Planta. 2006;224:1163–1173. doi: 10.1007/s00425-006-0298-9. [DOI] [PubMed] [Google Scholar]
- Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nature Protocols. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lin H, Joo S, Goodenough U. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell. 2008;133:829–840. doi: 10.1016/j.cell.2008.04.028. [DOI] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. The Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. The Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzella MA, Bertero D, Casal JJ. Temperature-dependent internode elongation in vegetative plants of Arabidopsis thaliana lacking phytochrome B and cryptochrome 1. Planta. 2000;210:497–501. doi: 10.1007/PL00008157. [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake S. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes and Development. 2003;17:2088–2093. doi: 10.1101/gad.1120003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. In vitro interactions between barley TALE homeodomain proteins suggest a role for protein–protein associations in the regulation of Knox gene function. The Plant Journal. 2001;27:13–23. doi: 10.1046/j.1365-313x.2001.01064.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, Suzuki K, Muto S, Matsui M. Activation tagging, a novel tool to dissect the functions of a gene family. The Plant Journal. 2003;34:741–750. doi: 10.1046/j.1365-313x.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- Proveniers M, Rutjens B, Brand M, Smeekens S. The Arabidopsis TALE homeobox gene ATH1 controls floral competency through positive regulation of FLC. The Plant Journal. 2007;52:899–913. doi: 10.1111/j.1365-313X.2007.03285.x. [DOI] [PubMed] [Google Scholar]
- Quaedvlieg N, Dockx J, Rook F, Weisbeek P, Smeekens S. The homeobox gene ATH1 of Arabidopsis is derepressed in the photomorphogenic mutants cop1 and det1. The Plant Cell. 1995;7:117–129. doi: 10.1105/tpc.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Gunl M, Pautot V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. The Plant Cell. 2008;20:888–900. doi: 10.1105/tpc.108.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. The Plant Journal. 2009;58:641–654. doi: 10.1111/j.1365-313X.2009.03809.x. [DOI] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proceedings of the National Academy of Sciences, USA. 2002;99:9579–9584. doi: 10.1073/pnas.092271599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Campbell BC, Hake S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Current Biology. 2004;14:812–817. doi: 10.1016/j.cub.2004.04.032. [DOI] [PubMed] [Google Scholar]
- Smith HM, Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. The Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2002;99:4730–4735. doi: 10.1073/pnas.072626099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proceedings of the National Academy of Sciences, USA. 2008;105:16392–16397. doi: 10.1073/pnas.0803997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.