Abstract
The seed maturation programme occurs only during the late phase of embryo development, and repression of the maturation genes is pivotal for seedling development. However, mechanisms that repress the expression of this programme in vegetative tissues are not well understood. A genetic screen was performed for mutants that express maturation genes in leaves. Here, it is shown that mutations affecting SDG8 (SET DOMAIN GROUP 8), a putative histone methyltransferase, cause ectopic expression of a subset of maturation genes in leaves. Further, to investigate the relationship between SDG8 and the Polycomb Group (PcG) proteins, which are known to repress many developmentally important genes including seed maturation genes, double mutants were made and formation of somatic embryos was observed on mutant seedlings with mutations in both SDG8 and EMF2 (EMBRYONIC FLOWER 2). Analysis of histone methylation status at the chromatin sites of a number of maturation loci revealed a synergistic effect of emf2 and sdg8 on the deposition of the active histone mark which is the trimethylation of Lys4 on histone 3 (H3K4me3). This is consistent with high expression of these genes and formation of somatic embryos in the emf2 sdg8 double mutants. Interestingly, a double mutant of sdg8 and vrn2 (vernalization2), a paralogue of EMF2, grew and developed normally to maturity. These observations demonstrate a functional cooperative interplay between SDG8 and an EMF2-containing PcG complex in maintaining vegetative cell identity by repressing seed genes to promote seedling development. The work also indicates the functional specificities of PcG complexes in Arabidopsis.
Keywords: Arabidopsis, embryonic programme, EMF2, histone methylation, PcG proteins, SDG8, seed maturation genes, somatic embryos, VRN2
Introduction
Seed maturation is a highly coordinated developmental phase when storage reserves, including seed storage proteins (SSPs), are synthesized and accumulated to high levels. The maturation genes need to be repressed, however, in order to allow seedling development to occur. Indeed, these genes are not observed to be expressed in vegetative organs of the plant (Vicente-Carbajosa and Carbonero, 2005). Research in the past decade with the model plant Arabidopsis has led to the identification of repressors of seed maturation genes in vegetative organs (reviewed in Zhang and Ogas, 2009), including chromatin-remodelling ATPases PICKLE and BRAHMA (Henderson et al., 2004; Li et al., 2005; Tang et al., 2008), polycomb group (PcG) proteins (Moon et al., 2003; Chanvivattana et al., 2004; Schubert et al., 2005; Makarevich et al., 2006; Kim et al., 2010), and histone deacetylases HDA6 and HDA19 (Tanaka et al., 2008). This indicates the crucial roles for chromatin-based mechanisms in the repression process. Despite this progress, our knowledge remains fragmented, and thus continued efforts are needed to identify the additional factors involved and to build an integrated genetic network.
In Arabidopsis, ABI3, FUS3, LEC1, and LEC2 are master regulators of seed maturation (Giraudat et al., 1992; Lotan et al., 1998; Luerssen et al., 1998; Stone et al., 2001), and they regulate each other (Kagaya et al., 2005b; To et al., 2006). ABI3, FUS3, and LEC2 are closely related members of a plant-specific B3-domain transcription factor family. LEC1 encodes a novel homologue of the CCAAT-binding factor HAP3 subunit. Loss-of-function mutations in ABI3, FUS3, and LEC1 give rise to pleiotropic seed phenotypes including significant reduction of SSPs. These regulatory genes are predominantly expressed in the seeds. When misexpressed in vegetative tissues, they are able to induce ectopic expression of the SSP genes and even somatic embryos (Parcy et al., 1994; Lotan et al., 1998; Stone et al., 2001; Gazzarrini et al., 2004; Kagaya et al., 2005a; Santos Mendoza et al., 2005; Braybrook et al., 2006).
The nucleosome is the basic unit of chromatin and it is composed of an octamer of four core histones (H3, H4, H2A, and H2B) around which 147 bp of DNA are wrapped. The N-terminal ‘tails’ of the core histones are unstructured and are frequently found modified by various enzymes (Kouzarides, 2007). These modifications have important implications in transcriptional activities of the genes with which they are associated. Some modifications are often found associated with actively transcribed genes [e.g. the trimethylation of histone 3 at Lys4 (H3K4me3) and acetylation], and are thus considered as active marks; whilst some other modifications are frequently found associated with silenced genes (e.g. H3K27me3, H3K9me3, and deacetylation), and thus are considered as repressive marks (Kouzarides, 2007). Histone modifications do not all act independently, but rather can antagonize or promote one another (Fischle et al., 2003; Suganuma and Workman, 2008).
The repressive H3K27me3 mark is deposited by PcG proteins. The PcG genes were first identified genetically in Drosophila through their role in controlling homeotic gene expression, and have long been one of the premier models for deciphering chromatin mechanisms during development (Schwartz and Pirrotta, 2007, 2008; Simon and Kingston, 2009). The PcG proteins form two main classes of complexes, PcG Repressive Complex 1 (PRC1) and PRC2. PRC2 contains the Enhancer of Zeste [E(z)], the methyltransferase, Suppressor of Zeste 12 [Su(z)12], Extra Sex Combs (Esc), and p55. PRC2 is responsible for placing the H3K27me3 mark, whereas PRC1 is commonly viewed as a direct executor of silencing at target genes. PRC2 components are conserved in plants, and three PRC2 complexes have been identified in Arabidopsis. The EMF2 (EMBRYONIC FLOWER 2)-containing PRC2 and the VRN2-containing PRC2 mainly function in vegetative and floral development, and the third one plays important roles in the seed (Calonje and Sung, 2006; Pien and Grossniklaus, 2007; Schatlowski et al., 2008). Little is known about PRC1 in plants, but recent studies have identified putative PRC1 components in Arabidopsis (Calonje et al., 2008; Sanchez-Pulido et al., 2008; Xu and Shen, 2008; Bratzel et al., 2010). Arabidopsis plants with mutations that destroy the activities of either PRC2 or PRC1 complexes lost cell identity control and thus exhibited massive growth of somatic embryo-like structures (Chanvivattana et al., 2004; Schubert et al., 2005; Makarevich et al., 2006; Bratzel et al., 2010).
Here, it is shown that mutations affecting SDG8, a histone methyltransferase, resulted in the ectopic expression of seed maturation genes in leaves. Further, the genetic relationship between the SDG8 and the PcG gene EMF2 in repressing seed traits was investigated, followed by analysis of the histone modification status at seed maturation loci. The observed changes of the histone methylation marks in mutant backgrounds provide an explanation for the synergism of SDG8 and EMF2 in repressing seed gene expression.
Materials and methods
Plant material, growth conditions, and genotype analysis
Seeds of mutants were obtained from the ABRC and INRA, unless otherwise indicated. Seeds were vernalized at 4°C for 3d. Then the seeds were sowed on soil or on agar plates containing 4.3 g l−1 Murashige and Skoog nutrient mix (Sigma-Aldrich), 1.5% sucrose, 0.5 g l−1 MES, pH 5.7 with KOH, and 0.8% agar. Plants were grown under 16 h light (22 °C)/8 h dark (20 °C) cycles. Homozygous T-DNA insertion mutants were identified by PCR.
Map-based cloning of essp4
Mutant essp4 was isolated from the same genetic screening as essp1 and essp3 (Tang et al., 2008; Lu et al., 2010). For genetic mapping of the essp4 mutation, mutant plants from the Col background were crossed with wild-type plants of the Ler ecotype. A total of 836 homozygous essp4 mutants were selected from an F2 segregating population. Genomic DNA extracted from these seedlings was used for PCR-based mapping with simple sequence polymorphism markers, and the essp4 locus was mapped to an ∼120 kb genomic interval on bacterial artificial chromosome (BAC) F22K20, T14N5, and F2P24 at the bottom of chromosome 1 (28 965–29 084 kb). Sequencing of the genomic region revealed a mutation in At1g77300.
Histochemical GUS and fat red staining
The modified β-glucuronidase (GUS) staining solution (0.5 mg ml−1 5-bromo-4-chloro-3-indolyl-glucuronide, 20% methanol, 0.01 M TRIS-HCl, pH 7.0) (Tang et al., 2008) was used. Seedlings immersed in GUS staining solution were placed under vacuum for 15 min, and then incubated at 37 °C overnight. The staining solution was removed and samples were cleared by sequential changes of 75% and 95% ethanol. Fat red staining was performed by incubating samples in a saturated solution of Sudan red 7B (Sigma) in 70% ethanol for 1 h at room temperature. Samples were then rinsed with 70% ethanol (Bratzel et al., 2010).
Microarray hybridization and data analysis
Total RNA was isolated in three biological replicates from leaves of 2-week-old wild-type (βCGpro:GUS) and mutant (essp4/sdg8-5 and sdg8-2) seedlings grown on MS (Murashige and Skoog) agar plates (1.5% sucrose), using an RNeasy Plant Mini kit (Qiagen). Labelling, hybridization, and detection were performed at the McGill University and Genome Quebec Innovation Centre (http://genomequebec.mcgill.ca). The Affymetrix Arabidopsis ATH1-whole genome array, containing 22 810 probe sets representing ∼24 000 genes, was used. The raw MAS 5.0 data files obtained from scanned array images are then imported into GeneSpring 7.3.1 (Silicon Genetics). Only genes with Present (P) calls were included in the analysis. Raw signals of each gene were normalized with the median of all measurements on the chip. The average normalized value of the signal intensity for each gene in three replicate hybridization experiments for the wild type (βCGpro:GUS) and two replicate hybridization experiments for sdg8 (sdg8-2, dg8-5) was adopted as the expression value of the gene. Expression data were analysed by a one-way analysis of variance (ANOVA) model to identify differentially regulated transcripts. False discovery rate (FDR) multiple testing corrections were calculated based on the P-value generated from the one-way ANOVA. Using the FDR at 5% that corresponds to a P-value of 0.05, only statistically significant genes that were regarded as differentially regulated only if their fold change was 2.0 for up-regulated and 0.5-fold for down-regulated were selected. The microarray data have been deposited with the NCBI Gene Expression Omnibus data repository (http:/www.ncbi.nlm.nih.gov/geo) under accession number GSE29771.
Gene expression and SDS–PAGE analysis
Plants grown on MS medium were used for gene expression and SDS–PAGE analyses. Reverse transcription-PCR (RT-PCR), real-time PCR, and RNA blot analyses were preformed as described previously (Tang et al., 2008). Extra PCR primers used in this work are listed in Supplementary Table S3 available at JXB online. SDS–PAGE was carried out to profile seed storage proteins as described by Hou et al. (2005).
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as previously described (Tang et al., 2008) using leaves from 13-day-old plants grown on an MS agar plate for the wild type and single mutants, while 13- to 16-day-old seedlings or 30-day-old somatic embryos were used for the sdg8 emf2 double mutant. Chromatin from 0.3 g of leaves or somatic embryos was used for one immunoprecipitation with H3K27me3 (Millipore, 07-449) or H3K4me3 (Millipore, 07-473) antibodies, or no antibody as a mock. Input DNA, immunoprecipitated DNA, or mock DNA was subjected to quantitative PCR for quantifying ChIP enrichment. Ta3 and Actin2/7 were amplified as controls for a repressed and an actively expressed locus, respectively. RT-PCR analysis was used to confirm that Ta3 is not detectable in both wild-type and sdg8 mutant leaves, while Actin2/7 is uniformly expressed (data not shown). The relative amount of ChIP DNA was first deducted by background mock DNA and then calculated as a percentage of input DNA.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1g77300 (SDG8), AT5G51230 (EMF2), AT4G16845 (VRN2), AT3G20740 (FIE), At3g24650 (ABI3), At3g26790 (FUS3), At1g21970 (LEC1), AT1G28300 (LEC2), At4g27140 (At2S1), At4g27150 (At2S2), At4g27160 (At2S3), At4g27170 (At2S4), and At5g54740 (At2S5).
Results
Identification of SDG8 as repressor of a seed gene promoter
A genetic screen has recently been conducted to identify mutants exhibiting ectopic expression of a soybean conglycinin (7S storage protein) gene promoter–GUS transgene (βCGpro:GUS) (Tang et al., 2008; Lu et al., 2010). This article reports the characterization of one of the mutants identified from the screen, initially named essp4. The essp4 mutant plants exhibited strong ectopic GUS activity in leaves, not detectable in other organs (Fig. 1A, B). In addition, the mutant plants had pleiotropic developmental defects, such as early flowering, more branches, shorter siliques, and fewer seeds (Fig. 1C–G).
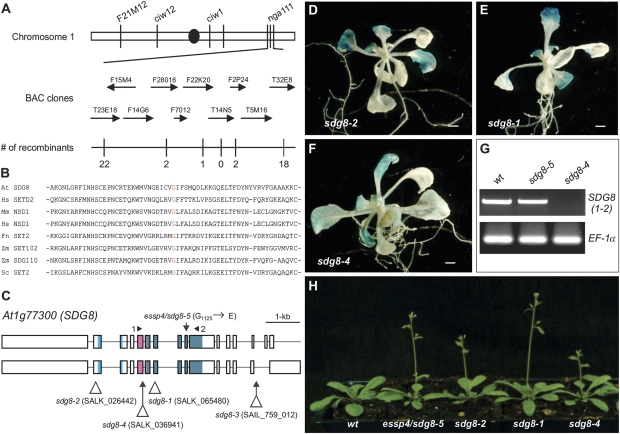
Fig. 1.
Phenotypes of the essp4 mutant. (A, B) GUS phenotypes of the essp4 mutant grown on agar at two different growth phases. (C–E) Comparison of the essp4 mutant with the wild type (βCGpro:GUS) at bolting and mature phases, respectively. (F, G) Comparison of essp4 siliques with that of the wild type (βCGpro:GUS). (This figure is available in colour at JXB online.)
The essp4 mutation is a recessive mutation and mapped to a genomic interval of ∼120 kb on the bottom of chromosome 1 (Fig. 2A). To identify the molecular lesion in essp4, the genomic region was amplified by PCR and sequenced. A single point mutation was identified in SDG8/EFS (At1g77300), potentially leading to a missense mutation at the amino acid level, from Gly1125 to Glu1125. The amino acid residue affected by the essp4 mutation is a highly conserved residue in the SET domain across kingdoms (Fig. 2B).
Fig. 2.
Map-based cloning of essp4. (A) Fine genetic mapping with PCR-based markers located the essp4 locus to the bottom of chromosome 1, on BAC clone T14N5. The numbers of recombination events out of the total numbers of chromosomes examined (1536) are indicated. (B) Alignment of amino acid sequences of SET domains from Arabidopsis (At), human (Hs), mouse (Mm), fungus (Fn), maize (Zm), and yeast (Sc). (C) Structure of the SDG8/ESSP4 gene and the location of mutation/T-DNA insertion sites of sdg8 alleles. Boxes and lines represent exons and introns, respectively. The shaded boxes represent the conserved protein domains (from left to right): CW (cysteine and tryptophan conserved), AWS (associated with SET), and SET. (D–F) GUS phenotypes of three T-DNA insertion alleles. Shown here is a representative F2 progeny from each of the crosses of the corresponding T-DNA allele with the βCGpro:GUS line. (G) RT-PCR analysis of the expression of SDG8 in the wild type and sdg8 mutants. The primers used are indicated in (C) and elongation factor 1α was used as an internal control. (H) Comparison of sdg8 mutant plants with the wild type at bolting. (This figure is available in colour at JXB online.)
SDG8 has recently been reported by several groups to be a regulator of diverse growth and developmental processes, including flowering timing and shoot branching (Zhao et al., 2005; Dong et al., 2008; Xu et al., 2008; Cazzonelli et al., 2009; Grini et al., 2009; Ko et al., 2010). The reported sdg8 mutant phenotypes are similar to those of the essp4 mutant. To confirm that essp4 is allelic to SDG8, T-DNA insertion lines, sdg8-1, sdg8-2, and sdg8-4, were obtained, and plants homozygous for the T-DNA insertions were crossed with βCGpro:GUS. In the F2 generation, about a quarter of the plants showed the ectopic GUS phenotype concomitant with other morphological phenotypes (Fig. 2C–H). These data strongly suggest that ESSP4 is SDG8.
Expression of 2S albumin genes and other embryogenesis-related genes in sdg8 mutant leaves
To obtain an overview of the effects of the sdg8 mutations on endogenous seed storage protein genes and other seed genes, a transcript profiling analysis was performed to compare gene expression at the whole genome level in mutant (sdg8-5/essp4 and sdg8-2) and wild-type (βCGpro:GUS) leaves. Total RNA was isolated from leaves of mutant and wild-type plants grown on MS agar for 2 weeks, and labelled RNAs were hybridized to the Affymetrix Arabidopsis ATH1 gene chip whole genome array. As listed in Supplementary Tables S1 and S2 at JXB online, 1299/1132 and 352/382 genes were significantly up- and down-regulated in sdg8-5 (essp4)/sdg8-2 (≥2.0-fold; FDR ≤0.05), respectively. Importantly, among the up-regulated genes are a subset of seed storage protein genes, At2S2, At2S3, At2S5, and At7S1 (Table 1). Also among the up-regulated genes are a number of other nutrient reserve-related genes, such as those encoding lipid transfer proteins (LTPs) and late embryogenesis abundant (LEA) proteins (Table 1). Moreover, a group of genes that have been previously shown to be required for normal embryo development (EMB; Tzafrir et al., 2003, 2004; www.seedgenes.org) are also among the genes whose mRNAs were significantly elevated in mutant leaves (Table 1). The EMB genes are a group of genes encoding proteins with diverse functions in embryogenesis. Lastly, it is worth mentioning that the transcript of the gibberellin 2-oxidase gene (AtGA2ox2, At1g30040) is highly elevated in mutant leaves (Supplementary Table S1, S2). AtGAox2 is one of the five C19-GA 2-oxidases which constitute a major GA inactivation pathway in Arabidopsis (Yamauchi et al., 2007; Rieu et al., 2008). In contrast, fewer genes were reported to be affected in two recent studies using 6- and 10-day-old seedlings, and no ectopic expression of seed storage protein genes was detected (Xu et al., 2008; Cazzonelli et al., 2009), suggesting a development stage-dependent regulation of these genes.
Table 1.
Selected seed-related genes up-regulated in essp4 leaves as revealed by microarray analysis
| Gene identification | Locus | Fold elevated |
| Seed storage proteins | ||
| 2S seed storage protein 2 (At2S2) | At4g27150 | 1391.98 |
| 2S seed storage protein 3 (At2S3) | At4g27160 | 12.39 |
| 2S seed storage protein 5 (At2S5) | At5g54740 | 89.18 |
| Cupin family protein (At7S1) | At4g36700 | 452.18 |
| Other storage proteins | ||
| Lipid transfer protein 6 (LTP6) | At3g08770 | 46.65 |
| Lipid transfer protein 3 (LTP3) | At5g59320 | 11.24 |
| Non-specific lipid transfer protein 2 (LTP2) | At2g38530 | 10.84 |
| Lipid transfer protein 4 (LTP4) | At5g59310 | 9.04 |
| Lipid transfer protein family protein (LTP) | At4g12490 | 7.97 |
| Lipid transfer protein family protein (LTP) | At3g18280 | 6.93 |
| Lipid transfer protein family protein (LTP) | At4g22490 | 6.81 |
| Lipid transfer protein family protein (LTP) | At4g22470 | 4.47 |
| Lipid transfer protein family protein (LTP) | At4g12500 | 4.09 |
| Lipid transfer protein family protein (LTP) | At5g64080 | 3.30 |
| Lipid transfer protein family protein (LTP-a) | At1g62500 | 3.02 |
| Lipid transfer protein family protein (LTP) | At4g12480 | 2.98 |
| Lipid transfer protein family protein (LTP) | At1g48750 | 2.31 |
| Lipid transfer protein family protein (LTP) | At1g55260 | 2.17 |
| Lipoxygenase (LOX2) | At3g45140 | 10.07 |
| Late embryogenesis abundant domain-containing protein (LEA) | At3g17520 | 48.82 |
| Late embryogenesis abundant 3 family protein (LEA3) | At1g02820 | 9.71 |
| Embryo-specific protein-related | At5g62210 | 8.11 |
| Embryo-abundant protein-related | At2g41380 | 3.75 |
| EMB genes | ||
| Proline-rich extensin-like family protein (RSH) | At1g21310 | 37.85 |
| Oligopeptide transporter OPT family protein (AtOPT3) | At4g16370 | 19.24 |
| DNA-directed DNA polymerase epsilon catalytic subunit putative (POL2B/TIL2) | At2g27120 | 17.65 |
| Zinc finger protein-related (EMB2454) | At3g18290 | 10.33 |
| Homeobox protein SHOOT MERISTEMLESS (STM) | At1g62360 | 6.89 |
| DNA-directed DNA polymerase epsilon catalytic subunit putative (EMB2284) | At1g08260 | 5.54 |
| RNA polymerase sigma subunit SigE (sigE)/sigma-like factor (SIG5) | At5g24120 | 4.62 |
| Heat shock protein putative (EMB1956) | At2g04030 | 2.68 |
| Syntaxin-related protein KNOLLE (KN)/syntaxin 111 (SYP111) | At1g08560 | 2.59 |
| Pre-mRNA splicing factor putative (EMB2444) | At2g18510 | 2.56 |
| Transducin family protein/WD-40 repeat family protein (TOZ) | At5g16750 | 2.53 |
| Hypothetical protein (EMB1692) | At5g62990 | 2.52 |
| NLI-interacting factor (NIF) family protein (EMB1860) | At1g55900 | 2.25 |
| Ubiquitin-specific protease 14 putative (UBP14/TTN6) | At3g20630 | 2.22 |
| Acetyl-CoA carboxylase 1 (ACC1) | At1g36160 | 2.20 |
| Expressed protein (EMB1974) | At3g07060 | 2.20 |
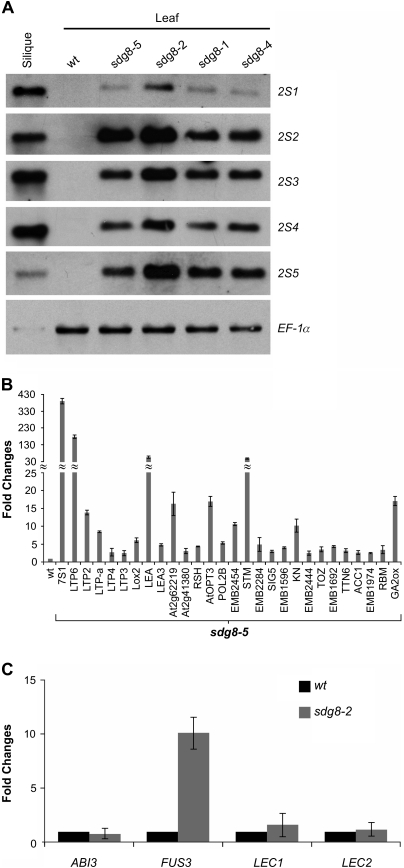
The DNA microarray results listed in Table 1 were validated and are shown in Fig. 3. Since the 2S genes do not contain introns, RNA-blot analysis was used to examine their expression. Although the 2S1 and 2S4 RNAs were not detected in the microarray experiments, they were detectable by northern analysis (Fig. 3A). In addition, the other three T-DNA insertion mutants, sdg8-1, sdg8-2, and sdg8-4, also exhibited strong expression of 2S genes (Fig. 3A), providing further evidence that ESSP4 is SDG8. For the other genes listed in Table 1, data from real-time quantitative RT-PCR (qRT-PCR) experiments validated the microarray results (Fig. 3B). RNAs of the master regulators of seed maturation, ABI3, FUS3, LEC1, and LEC2, were also examined by qRT-PCR, although they were not detected in the microarray experiment. As shown in Fig. 3C, with the exception of FUS3, none of these RNAs is detected in sdg8-2 leaves.
Fig. 3.
Expression analysis of seed maturation genes in essp4 mutant leaves. (A) RNA blot analysis of the expression of the five 2S genes in leaves of four sdg8 mutants grown for 14 d on MS agar. Wild-type (Col) leaves and siliques were used as negative and positive controls, respectively. The same amount of RNA was used for each blot. Elongation factor 1α was used as loading control. (B) Real-time quantitative RT-PCR (qRT-PCR) validation of the expression in sdg8-5 leaves of seed-related genes revealed in the DNA microarray analysis. RNAs from leaves of 14-day-old plants grown on MS agar were used for PCR. Only those validated by qRT-PCR are shown here. Wild-type (βCGpro:GUS) RNA levels are designed as 1-fold. The expression of Actin-8 was used as an internal control. The mean and standard error (SE) were determined from three biological replicates. Bars represent SEs. (C) qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 genes in seedlings (aerial portion) of sdg8-2 mutants grown for 14 d on MS agar. Wild-type (Col) RNA levels are designed as 1-fold. The expression of Actin-8 was used as an internal control. The mean and SE were determined from three biological replicates, each of which was conducted in triplicate.
Formation of somatic embryos on sdg8 emf2 double mutant seedlings
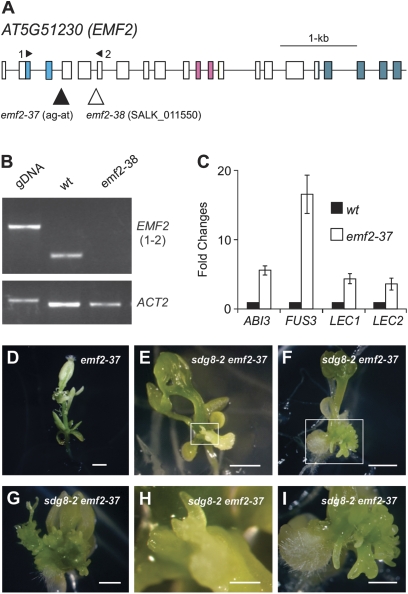
The identification of SDG8, a histone methyltransferase, as a moderate repressor of seed genes provided an opportunity to study its functional interplay with the PcG proteins on seed maturation genes. Evidence for a role for PRC2 in repressing seed genes is strong, including double mutant studies that demonstrated the formation of somatic embryos in double mutants deficient for both of the redundant PRC2 subunits, CURLY LEAF (CLF)/SWINGER (SWN) or EMF2/VRN2 (Chanvivattana et al., 2004; Schubert et al., 2005; Makarevich et al., 2006). However, previous reports on the ectopic expression of seed genes in the emf2 single mutant were not conclusive (Moon et al., 2003; Kim et al., 2010). To clarify this, two new alleles of emf2, designated as emf2-37 and emf2-38 (SALK_011550) (Fig. 4A), were obtained. The emf2-37 allele is a single nucleotide mutation which is predicted to disrupt mRNA splicing. emf2-38 is a T-DNA insertion knock-out allele (Fig. 4A, B). Both the two new emf2 mutant alleles displayed similar morphological phenotypes to those described previously (Yoshida et al., 2001; Moon et al., 2003). Transcript levels of the four master regulators were examined for 15-day-old emf2-37 seedlings as shown in Fig. 4C. Clearly, FUS3 was expressed and the other three transcripts were also detected.
Fig. 4.
Phenotypes of the sdg8-2 emf2-37 double mutants. (A) Structure of the EMF2 gene and the location of mutation/T-DNA insertion sites of emf2 alleles. Boxes and lines represent exons and introns, respectively. The shaded boxes represent the conserved protein domains (from left to right): conserved N-terminal basic domain, C2H2-type zinc finger domain, and C-terminal acidic-W/M domain. The mutation in emf2-37 is ‘G’ to ‘T’ at base pair 20 8247 27 on chromosome 5. (B) RT-PCR analysis of the expression of EMF2 in the wild type and emf2-38 mutants. The primers used are indicated in (A). Genomic DNA (gDNA) was included as a size control for RT-PCR products, and Actin2 was used as an internal control. (C) qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 genes in seedlings (aerial portion) of emf2-37 mutants grown for 15 d on MS agar. Wild-type (Col) RNA levels are designed as 1-fold. The expression of Actin-8 was used as an internal control. The mean and standard error were determined from three biological replicates, each of which was conducted in triplicate. (D–I) Morphological phenotypes of emf2-37 single (D) and sdg8-2 emf2-37 double mutants at different growth phases on MS agar (E, 16 d; F, 25 d; G, 32 d). (H) and (I) are close-up images of the boxed areas in (E) and (F), respectively. Bar=1 mm. (This figure is available in colour at JXB online.)
To investigate the genetic relationship between the two moderate repressor genes, EMF2 and SDG8, emf2 sdg8 double mutants were generated and their phenotypes were examined. Two null alleles of sdg8, sdg8-1 and sdg8-2 (Fig. 2), were crossed with emf2-37 and emf2-38. Since emf2-37/38 are sterile, heterozygous (EMF2 emf2-37) plants were used to cross with sdg8 plants. In the F2 generation, EMF2 emf2-37/sdg8-2 sdg8-2 progeny plants were identified by genotyping, and F3 seeds harvested. The F3 seeds were plated on MS agar, mutant segregation data were generated and the phenotypes were observed. Approximately a quarter of the F3 seedlings were tiny and were emf2-37 emf2-37/sdg8-2 sdg8-2 plants as confirmed by emf2-37 genotyping; and ∼50% (113/220) of these started forming somatic embryo-like structures in just over 2 weeks after germination (Fig. 4D–I). In most of the cases, the somatic embryos were found at the bottom of the aerial portion of the plant near the cotyledons (Fig. 4F). Other allele combinations of sdg8-1 emf2-38 exhibited a similar phenotype (data not shown). This observation demonstrates the synergistic genetic interaction of SDG8 and EMF2 in repressing embryonic traits.
High level expression of seed maturation genes in sdg8 emf2 seedlings
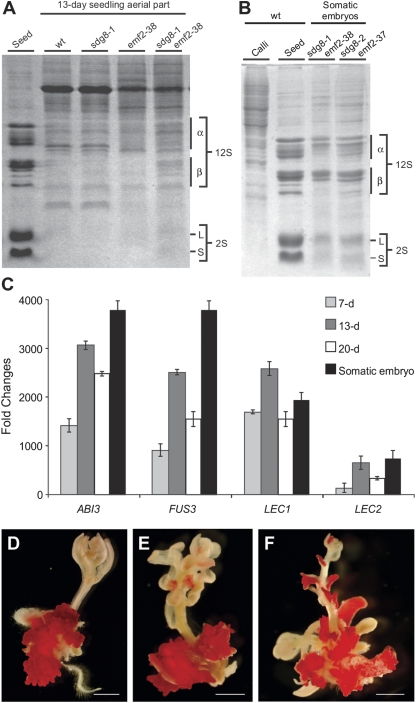
Next, expression of seed maturation genes in the emf2-37/38 sdg8-1/2 double mutants was examined. First, the expression and accumulation of seed storage proteins in 13-day-old double mutants (aerial portions) were profiled by SDS–PAGE analysis. As shown in Fig. 5A, both the 12S cruciferins and the 2S napins are clearly expressed and accumulated in the double mutants, but not detectable in either the sdg8-1/2 or the emf2-37/38 single mutants. The somatic embryos formed on the double mutants, as expected, exhibited essentially the same profiles of seed storage proteins as those of seeds (Fig. 5B). As a control, calli induced from the wild-type background were also analysed and displayed very different protein profiles, supporting the identity of the somatic embryos formed on the double mutants. Consistent with the seed storage protein profiling results, the maturation master regulators were also highly expressed in the double mutants. The transcript levels of the four master regulators were analysed by qRT-PCR for somatic embryos and seedlings (aerial parts) collected at three developmental stages: 7, 13, and 20 d. All the samples exhibited very high expression of the master regulators. Among the three time points, 13-day-old seedlings exhibited the highest expression. The somatic embryos had an even higher level of expression for all the master regulator genes with the exception of LEC1 which was slightly lower than that of the 13-day-old seedlings (Fig. 5C). In contrast, the transcripts of the master regulators in the sdg8-2 and emf2-37 single mutant seedlings were a few orders of magnitude lower than those in the double mutants (Figs 3C, 4C). In addition, the sdg8-2 emf2-37 double mutant was also stained with the neutral lipid dye fat red and, as shown in Fig. 5D–F, the somatic embryos were all stained, but not the other organs, indicating the high level accumulation of seed storage-specific triacylglycerols in somatic embryos. These results further support the identity of somatic embryos formed on the double mutant and strongly suggest a synergistic, rather than a simple additive, genetic interaction between emf2 and sdg8 on seed maturation genes.
Fig. 5.
Expression of seed maturation genes in sdg8 emf2 double mutants. (A, B) SDS–PAGE analysis of seed storage proteins in seedlings (aerial portion) (A) and somatic embryos (B) from sdg8-1/2 emf2-37/38 double mutants. Wild-type (Col) seeds were used as positive controls, and leaves and calli induced from wild-type plants were used as negative controls. (C) qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 genes in somatic embryos and aerial portions of seedlings of sdg8-2 emf2-37 double mutants at various time points on MS agar (7, 13, and 20 d). Wild-type (Col) RNA levels are designed as 1-fold. The expression of Actin-8 was used as an internal control. The mean and standard error were determined from three biological replicates, each of which was conducted in triplicate. (D–F) Fat red staining of 25-day-old sdg8-2 emf2-37 mutants grown on MS agar. Scale bar=1 mm. (This figure is available in colour at JXB online.)
No synergistic genetic interaction between SDG8 and VRN2 in repressing embryonic traits
Since EMF2 and VRN2 are redundant in seed gene repression as reported previously (Chanvivattana et al., 2004; Schubert et al., 2005), it was also investigated whether there is a synergistic genetic relationship between SDG8 and VRN2 in repressing seed genes. For that, a new mutant allele of VRN2 was obtained, designated vrn2-2 (FLAG_376E07), which contains a T-DNA insertion in the 10th intron and results in the disruption of the transcript (Fig. 6A, B). Homozygous vrn2-2 plants were crossed with emf2-37 EMF2 heterozygous plants, emf2-37 EMF2/vrn2-2 vrn2-2 progeny were identified in the F2 generation, and selfed F3 seeds were collected. The F3 seeds were plated on MS agar, the mutant genotype assessed, and the phenotypes observed. Approximately a quarter of the F3 seedlings were tiny and were emf2-37 emf2-37/vrn2-2 vrn2-2 plants as confirmed by emf2-37 genotyping. The majority of these homozygous double mutant plants (75/96, ∼80%) started forming somatic embryo-like structures in just over 2 weeks after germination and later developed into massive somatic embryos (Fig. 6D–F). This observation is consistent with published observations (Chanvivattana et al., 2004; Schubert et al., 2005) and demonstrates that vrn2-2 is a true loss-of-function allele. The sdg8 vrn2 double mutants were made and their phenotype examined. Approximately 1000 F2 seedlings (sdg8-1 sdg8-1/vrn2-2 vrn2-2) were examined and none displayed any phenotype resembling those of the sdg8-2 emf2-37 double mutants (Fig. 6G). Another allele combination (sdg8-2 sdg8-2/vrn2-2 vrn2-2) showed similar results. These results suggest that VRN2 plays a different role from that of EMF2 in repressing seed genes during seedling development.
Fig. 6.
Characterization of a new vrn2 allele and phenotype of the sdg8 vrn2 double mutants. (A) Structure of the VRN2 gene and the location of the T-DNA insertion site of the vrn2-2 allele. Boxes and lines represent exons and introns, respectively. The shaded boxes represent the conserved protein domains (from left to right): the conserved N-terminal basic domain, the C2H2-type zinc finger domain, and the C-terminal acidic-W/M domain. (B) RT-PCR analysis of the expression of VRN2 in the wild type and the vrn2-2 mutant. The primers used are indicated in (A). Genomic DNA (gDNA) was included as a size control for RT-PCR products, and Actin2 was used as an internal control. (C) Phenotype comparison of the vrn2-2 mutant at 25 d with the wild type (Ws ecotype). (D–F) Morphological phenotypes of the emf2-37 vrn2-2 double mutants grown on MS agar (D and E, 30 d; F, 20 d). Bar=1 mm. (G) Phenotype comparison of the sdg8-2 vrn2-2 double mutant with the sdg8-2 and vrn2-2 single mutants at 30 d. (This figure is available in colour at JXB online.)
Histone methylation status at seed genes in sdg8 single and sdg8 emf2 double mutants
To understand the molecular mechanisms underlying the sdg8-2 and the sdg8-2 emf2-37 mutant phenotypes, ChIP experiments were performed to examine the histone methylation status changes at several seed maturation genes in the mutant backgrounds. Recent data suggest that SDG8 may mediate the deposition of H3K36me3/me2 at a few genomic loci while it may also be responsible for placing H3K9me3 at some other loci (Zhao et al., 2005; Dong et al., 2008; Xu et al., 2008). Based on these published observations, first the status of H3K36me2/me3 was examined, and no changes in these two modifications were observed between mutants and wild-type plants. This result is consistent with a recent global mapping of H3K36me2 in wild-type Arabidopsis which did not detect any significant enrichment of this mark at seed genes (Oh et al., 2008). Next, the status of the H3K9me3 mark at several seed genes in sdg8 mutants was examined, and again no obvious changes were observed.
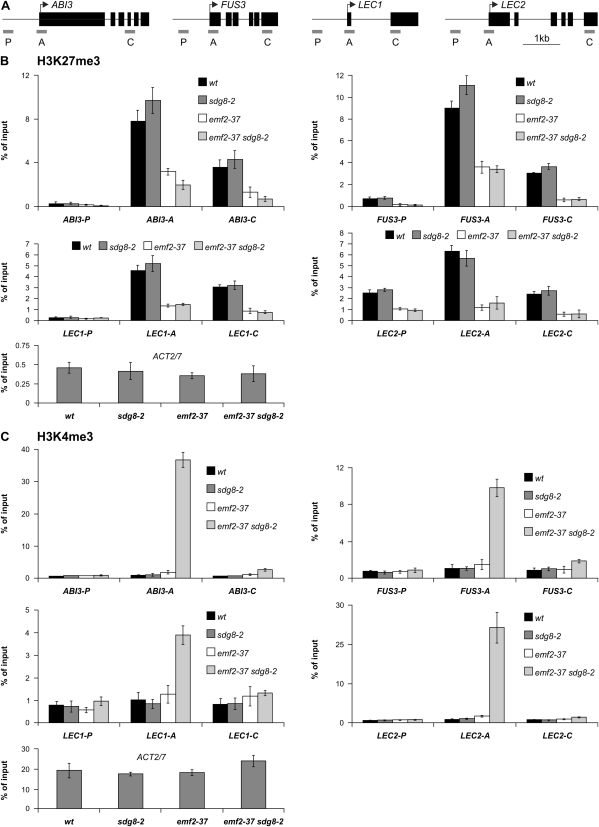
Further, the changes of histone marks in emf2-37 sdg8-2 double mutants were examined to search for clues to the synergistic interaction between emf2-37 and sdg8-2. It was reasoned that, to allow for the seed programme to develop in the double mutant, there must be cross-talk between H3K27me3 and the one placed by SDG8, assuming that SDG8 acts directly at seed genes. The cross-talk would result in (i) mutual promotion of the removal of the two repressive marks, thus clearing the way for the active machinery; and/or (ii) promotion of the deposition of active histone marks to recruit transcriptional activators. To test the first possibility, the levels of H3K27me3 in all the genetic backgrounds were examined. As shown in Fig. 7B, there was no change of this mark in sdg8-2 relative to the wild type and no further decrease in emf2-37 sdg8-2 double mutants relative to emf2-37 single mutants, suggesting that SDG8 does not affect PRC2 activity. Then the status of the most common active mark H3K4me3 was examined and a dramatic elevation of the active mark in emf2-37 sdg8-2 double mutants was observed at the transcription start site of the master regulator genes, particularly those of ABI3 and LEC2 (Fig. 7C). No changes were detected in the sdg8-2 single mutant and only a slight enrichment in the emf2-37 single mutant at the transcription start site of the master regulator genes relative to the wild type. Thus, the ChIP results are consistent with the observed synergistic genetic interaction between emf2-37 and sdg8-2, and suggest that only when both genes are disrupted could the active mark H3K4me3 be deposited to a high level and consequently lead to the full ectopic expression of the seed maturation programme.
Fig. 7.
ChIP analyses of H3K27me3 and H3K4me3 levels at seed maturation loci in sdg8-2, emf2-37, and sdg8-2 emf2-37 mutants. (A) Structures of the four master regulator genes and locations of primers used for quantitative ChIP-PCR analyses. Boxes and lines represent exons and introns, respectively. (B, C) Relative levels of H3K27me3 and H3K4me3 at four maturation loci. After ChIP, three different regions of each locus (as indicated in A) were analysed by qPCR. The results show the recovery of immunoprecipitated material with anti-H3K27me3 or anti-H3K4me3 antibodies (IP) as a percentage of input after deduction of background DNA (no antidoby mock control). For the wild type, and emf2-37 and sdg8-2 single mutants, the aerial parts from 13-day-old plants grown on MS agar plates were used. For the sdg8-2 emf2-37 double mutant, both 13- to 16-day-old seedlings (one biological replicate) and 30-day-old somatic embryos (two biological replicates) were used in the H3K4me3 assay and only somatic embryos were used in the H3K27me3 assay. ACT2/7 is shown as a control locus. Error bars represent the standard deviation from the mean of three biological replications.
Discussion
How does SDG8 act to repress seed genes?
The genetic and molecular evidence presented here clearly indicates a role for SDG8 in the repression of seed maturation genes in seedlings (Figs. 1, 2, Table 1). SDG8 is a predicted histone methyltransferase based on its SET domain, and indeed it has been demonstrated to have H3 methyltransferase activity in vitro (Dong et al., 2008). However, recombinant SDG8 could not methylate recombinant H3 or synthetic H3 peptides, thus preventing the determination of specific lysine residues in H3 methylated by SDG8 in vitro (Dong et al., 2008; Xu et al., 2008; Ko et al., 2010). Nevertheless, in vivo data, including immunobloting and ChIP analyses, show that SDG8 may mediate the placement of H3K36me2/me3, H3K9me3, and H3K4me3. This is consistent with structural and phylogenetic analyses that grouped SDG8 and four other SDGs in a clade together with the H3K36-specific histone methyltransferases found in fungi and mammals (Xu et al., 2008). SDG8 also has homology with Drosophila Ash1, which can methylate Lys4 and Lys9 in H3 (Beisel et al., 2002; Dong et al., 2008). In the ChIP experiment, no reduction in the abundance of H3K36me3 or H3K9me3 was detected at seed genes in the mutant relative to the wild type. It is tempting to speculate that, even with the lack of the in vitro determination of its specific activity, there might be an as yet unidentified histone methylation activity of SDG8 that plays a role in repressing seed genes. Meanwhile, it is also possible that SDG8 acts indirectly to repress seed gene expression, for example by repressing a positive regulator. Although interesting, this hypothesis is at the present time hard to test since so many genes are affected in the sdg8 mutant and no well-characterized activator of seed maturation genes is available for such a test. In addition, the up-regulation of AtGA2ox2 might also contribute to the derepression of embryonic genes by lowering the level of GA in seedlings. GA is, however, also known to promote flowering, and thus a possible decrease in the GA level in sdg8 is expected to cause delayed flowering. That is in contrast to the observed early flowering phenotype of sdg8 plants. Future investigation is needed to understand this apparent conflict, but the sdg8 flowering phenotype is probably an outcome of multiple factors, and GA is only one of them.
Roles of PcG proteins in repressing seed genes
PRC2 components are conserved in plants and animals. In Arabidopsis, some PRC2 components are encoded by multigene families; for example, MEDEA (MEA), CLF, and SWN are E(z) homologues (Goodrich et al., 1997; Grossniklaus et al., 1998; Chanvivattana et al., 2004; Hennig et al., 2003), and EMF2, FERTILIZATION INDEPENDENT SEED2 (FIS2), and VRN2 are Su(z)12 homologues (Chaudhury et al., 1997; Gendall et al., 2001; Yoshida et al., 2001). In contrast, there is only one Arabidopsis homologue of ESC, which is the Fertilization Independent Endosperm (FIE) gene (Ohad et al., 1999; Kinoshita et al., 2001). The MEA–FIS complex is believed to function mainly in the seed, whereas the other two have roles in other aspects of development. Previous genetic evidence has demonstrated the essential roles of Arabidopsis PRC2 components in repressing seed genes, exemplified by the formation of somatic embryos on clf swn and emf2 vrn2 double mutants (Chanvivattana et al., 2004; Schubert et al., 2005; Makarevich et al., 2006) and a FIE-rescued-fie mutant seedling (Kinoshita et al., 2001). This genetic evidence demonstrates that a functional PRC2 is required for repression of the seed programme in seedlings. Recent genome-wide mapping of H3K27me3 in Arabidopsis identified a large number of genes (∼4400, ∼15% of all genes) that are marked by H3K27me3 (Zhang et al., 2007; Oh et al., 2008). Most of these genes are expressed at a low level throughout development or are expressed in a tissue-specific manner, including the seed-specific genes. These data are consistent with the pleiotropic phenotypes observed for PcG mutants and further indicate a central role for PcG proteins in repressing seed genes.
The differential roles of the two Su(z)12 homologues, EMF2 and VRN2, in repressing seed genes remain to be understood. The phenotype of the emf2-37 vrn2-2 double mutant, namely formation of somatic embryos on seedlings, suggests a redundant role for the two PcG proteins in repressing seed programmes; whereas the fact that the sdg8-2 vrn2-2 double mutant did not exhibit such a phenotype suggests a more important role for EMF2 than for VRN2 at the seed maturation loci. The outcomes of a genetic screen for sdg8-2 enhancers also appear to support a special role for EMF2: four new alleles of emf2, but none of the other PcG genes, have been recovered in screens for mutants forming somatic embryos. In addition, the sdg8 clf double mutant was also generated but no somatic embryo formation was observed, further suggesting a special role for EMF2 among PRC2 components in repressing seed genes.
Future work is needed to gain detailed understanding of how PcG functions at the seed maturation loci. Questions to be answered include how PRC2 is recruited to specific maturation loci and what is the biochemical composition of the EMF2-containing PRC2. In Drosophila, specific regulatory elements called the Polycomb Response Elements (PREs) are the sites of recruitment. The Drosophila PREs are also binding sites of the Trithorax protein (TRX), a H3K4 methyltranferase that acts to antagonize PcG repression. PcG complex binding is a dynamic process, sensitive to the antagonistic action of TrxG complexes as well as to positive or negative input from other transcription factors. The functional state of the PcG target is probably determined by the equilibrium between all these activities (Schwartz and Pirrotta, 2008). Future efforts are required to identify plant PREs and the DNA-binding PcG recruiters, or other alternative recruiting mechanisms such as those mediated by non-coding RNAs (Guenther and Young, 2010; Margueron and Reinberg, 2010).
Synergy of SDG8 and EMF2 at seed genes
The formation of somatic embryos on the emf2-37 sdg8-2 seedlings indicates a synergistic genetic interaction between EMF2 and SDG8 in repressing seed genes during vegetative development. The ChIP data show that the active histone mark H3K4me3 is enriched only in the double mutant, which is consistent with the observed synergistic genetic interaction. One possible explanation is the potential cross-talk between H3K27me3 and the putative unknown histone mark placed by SDG8, assuming that SDG8 acts directly at seed maturation loci. Chromatin modifications may act alone or in concert in a context-dependent manner to facilitate or repress chromatin-mediated processes (Fischle et al., 2003; Suganuma and Workman, 2008; Lee et al., 2010). The relationship between H3K27me3 and the one placed by SDG8 at seed gene chromatin loci still remains to be investigated. However, it is tempting to speculate that a reduction of both marks provides the correct chromatin context to allow the placement of H3K4me3 at seed genes. Alternatively, the double mutant phenotype could be an outcome of synergistic interaction between loss of H3K27me3 in emf2-37 and misexpression of a putative positive regulator(s) in sdg8-2.
The next question is how the active H3K4me3 mark is deposited following the loss of the repressive histone marks. This includes what enzymes are responsible and under what conditions. In Drosophila, Trx functions as an antagonist of PcG-mediated gene silencing and its main activity is correlated with H3K4 methylation, particularly H3K4me3. In Arabidopsis, there are five Trx homologues that have been identified (Avramova, 2009), of which ARABIDOPSIS HOMOLOG OF TRITHORAX 1 (ATX1) has been shown to have specific methylation activity for H3K4me3 and is required for placing the mark at several genes (Saleh et al., 2007, 2008; Pien et al., 2008). However, it still has not been determined whether ATX1 is responsible for the H3K4me3 at seed genes and, if not, which of the other ATXs is responsible.
The findings presented here demonstrate that partial loss of the H3K27me3 mark, when combined with the sdg8 mutation, has similar consequence to the complete abolishment of the repressive mark, namely high level deposition of H3K4me3 and full derepression of embryonic traits. This is in contrast to the observation that loss-of-function emf2 mutation causes a dramatic embryonic flower phenotype but only a weak derepression of seed genes. Together, these observations point to an important role for the interplay between PcG and other histone methylation activities in determining the PcG targeting specificity and ultimate transcriptional status of PcG target genes in plants.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Genes up- and down-regulated in sdg8-5/essp4 mutant leaves.
Table S2. Genes up- and down-regulated in sdg8-2 mutant leaves.
Table S3. PCR primers used in this work.
Acknowledgments
We thank the ABRC and INRA for seeds of T-DNA insertion lines; Alex Molnar for help with preparing the figures; Ida van Grinsven for sequencing service; Gang Tian for Fig. 5D–F; and Agriculture and Agri-Food Canada A-base and Canadian Crop Genomics Initiative (to YC), Genome Prairie/Genome Canada (to YC, EWTT and WAK), and NSERC (to SJR) for funding.
References
- Avramova Z. Evolution and pleiotropy of TRITHORAX function in Arabidopsis. International Journal of Developmental Biology. 2009;53:371–381. doi: 10.1387/ijdb.082664za. [DOI] [PubMed] [Google Scholar]
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature. 2002;419:857–862. doi: 10.1038/nature01126. [DOI] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Current Biology. 2010;20:1853–1859. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proceedings of the National Academy of Sciences, USA. 2006;103:3468–3473. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje M, Sanchez R, Chen L, Sung ZR. EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. The Plant Cell. 2008;20:277–291. doi: 10.1105/tpc.106.049957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje M, Sung ZR. Complexity beneath the silence. Current Opinion in Plant Biology. 2006;9:530–537. doi: 10.1016/j.pbi.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. The Plant Cell. 2009;21:39–53. doi: 10.1105/tpc.108.063131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ. Fertilization-independent seed development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1997;94:4223–4228. doi: 10.1073/pnas.94.8.4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Ma DP, Li J. The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochemical and Biophysical Research Communications. 2008;373:659–664. doi: 10.1016/j.bbrc.2008.06.096. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Current Opinion in Cell Biology. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Developmental Cell. 2004;7:373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell. 2001;107:525–535. doi: 10.1016/s0092-8674(01)00573-6. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature. 1997;386:44–51. doi: 10.1038/386044a0. [DOI] [PubMed] [Google Scholar]
- Grini PE, Thorstensen T, Alm V, Vizcay-Barrena G, Windju SS, Jørstad TS, Wilson ZA, Aalen RB. The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One. 2009;4:e7817. doi: 10.1371/journal.pone.0007817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada JP, Hoeppner MA, Gagliano WB. Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Young RA. Repressive transcription. Science. 2010;329:150–151. doi: 10.1126/science.1193995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiology. 2004;134:995–1005. doi: 10.1104/pp.103.030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schönrock N, Gruissem W. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development. 2003;130:2555–2565. doi: 10.1242/dev.00470. [DOI] [PubMed] [Google Scholar]
- Hou A, Liu K, Catawatcharakul N, Tang X, Nguyen V, Keller WA, Tsang EW, Cui Y. Two naturally occurring deletion mutants of 12S seed storage proteins in Arabidopsis thaliana. Planta. 2005;222:512–520. doi: 10.1007/s00425-005-1555-z. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant and Cell Physiology. 2005a;46:300–311. doi: 10.1093/pcp/pci031. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant and Cell Physiology. 2005b;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- Kim SY, Zhu T, Sung ZR. Epigenetic regulation of gene programs by EMF1 and EMF2 in Arabidopsis. Plant Physiology. 2010;152:516–528. doi: 10.1104/pp.109.143495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL. Polycomb repression of flowering during early plant development. Proceedings of the National Academy of Sciences, USA. 2001;98:14156–14161. doi: 10.1073/pnas.241507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, Noh B, Noh YS. Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO Journal. 2010;29:3208–3215. doi: 10.1038/emboj.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HC, Chuang K, Henderson JT, Rider SD, Jr, Bai Y, Zhang H, Fountain M, Gerber J, Ogas J. PICKLE acts during germination to repress expression of embryonic traits. The Plant Journal. 2005;44:1010–1022. doi: 10.1111/j.1365-313X.2005.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. The Plant Journal. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S. FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. The Plant Journal. 1998;15:755–764. doi: 10.1046/j.1365-313x.1998.00259.x. [DOI] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Reports. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nature Reviews Genetics. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YH, Chen L, Pan RL, Chang HS, Zhu T, Maffeo DM, Sung ZR. EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. The Plant Cell. 2003;15:681–693. doi: 10.1105/tpc.007831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. The Plant Cell. 1999;11:407–416. doi: 10.1105/tpc.11.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. The Plant Cell. 1994;6:1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inzé D, Avramova Z, Dean C, Grossniklaus U. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. The Plant Cell. 2008;20:580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochimica et Biophysica Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Rieu I, Eriksson S, Powers SJ, et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. The Plant Cell. 2008;20:2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Research. 2007;35:6290–6296. doi: 10.1093/nar/gkm464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Yilmaz M, et al. The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. The Plant Cell. 2008;20:568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M. RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics. 2008;9:308. doi: 10.1186/1471-2164-9-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Letters. 2005;579:4666–4670. doi: 10.1016/j.febslet.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Schatlowski N, Creasey K, Goodrich J, Schubert D. Keeping plants in shape: polycomb-group genes and histone methylation. Seminars in Cell and Developmental Biology. 2008;19:547–553. doi: 10.1016/j.semcdb.2008.07.019. [DOI] [PubMed] [Google Scholar]
- Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Current Opinion in Plant Biology. 2005;8:553–561. doi: 10.1016/j.pbi.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature Reviews Genetics. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Current Opinion in Cell Biology. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature Reviews Molecular Cell Biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proceedings of the National Academy of Sciences, USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiology. 2008;146:149–161. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Hou A, Babu M, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiology. 2008;147:1143–1157. doi: 10.1104/pp.108.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. The Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. The Arabidopsis SeedGenes Project. Nucleic Acids Research. 2003;31:90–93. doi: 10.1093/nar/gkg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, et al. Identification of genes required for embryo development in Arabidopsis. Plant Physiology. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. International Journal of Developmental Biology. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen WH. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Current Biology. 2008;18:1966–1971. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Molecular and Cell Biology. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Takeda-Kamiya N, Hanada A, Ogawa M, Kuwahara A, Seo M, Kamiya Y, Yamaguchi S. Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant and Cell Physiology. 2007;48:555–561. doi: 10.1093/pcp/pcm023. [DOI] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S. EMBRYONIC FLOWER2, a novel polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. The Plant Cell. 2001;13:2471–2481. doi: 10.1105/tpc.010227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ogas J. An epigenetic perspective on developmental regulation of seed genes. Molecular Plant. 2009;2:610–627. doi: 10.1093/mp/ssp027. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nature Cell Biology. 2005;7:1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.